Modulating T Cell Responses by Targeting CD3
Abstract
Simple Summary
Abstract
1. Mobilizing the Immune Response in Context of Different Immunotherapeutic Strategies
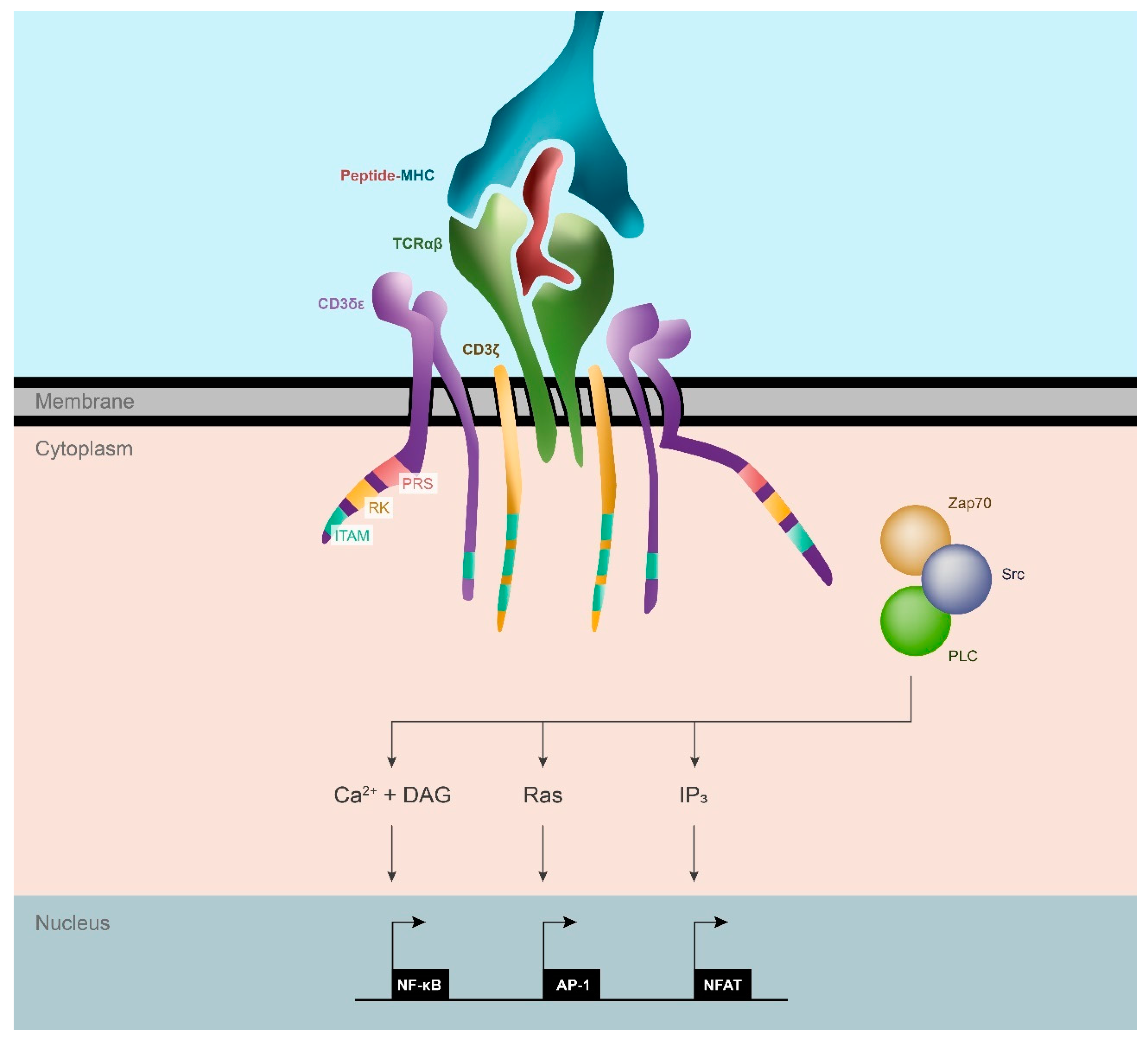
2. The T Cell Receptor: Intercepting Signals for T Cell Activation
2.1. Structure of the TCR/CD3 Complex
2.1.1. TCR Chains
2.1.2. CD3 Subunits
2.2. Signaling Motifs in the CD3 Chains Protein Complex
2.3. TCR Triggering
3. Strategies to Modulate T Cell Responses Targeting CD3
3.1. CD3 Agonistic Therapies to Rescue Function of T Cells
3.2. Anti-CD3 mAbs
3.3. The Importance of Providing CD3-Mediated Signaling In Situ: Bi-Specific T-Cell Engagers (BiTEs)
3.4. Aptamers as a Novel Class of CD3 Modulators
3.5. Targeting CD3 Complex with Small Molecules to Modulate T Cell Activation
3.6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| % | Percentage |
| °C | Degrees Celsius |
| < | Less than |
| > | Greater than |
| 145-2C11 | Mouse ati-CD3 antibody |
| 2-DG | 2-Deoxy-D-glucose; glucose analog |
| 3D | three dimensional |
| 4-1BB | Costimulatory Receptor |
| α | alpha |
| β | beta |
| δ | delta |
| ε | epsilon |
| γ | gamma |
| ζ | zeta |
| αβ | alpha-beta TCR chains |
| γδ | gamma-delta TCR chains |
| δε | delta-epsilon CD3 subunit |
| γε | gamma-epsilon CD3 subunit |
| ζζ | zeta-zeta CD3 subunit |
| ADCC | Antibody-dependent cellular cytotoxicity |
| AP-1 | Activator protein; key transcription factor involved in T cell activation |
| APC | Antigen Presenting Cell |
| aPD-1 | Anti- PD-1 antibody; immune checkpoint blockade |
| aPDL-1 | Anti- PD-L1 antibody; immune checkpoint blockade |
| AX-024 | T cell inhibitor |
| BCMA | B-cell maturation antigen; Tumor associated antigen in hematological |
| BCR | B Cell Receptor, expressed on B cells |
| BiTE | Bispecific T Cell Engager; CD3Ab-TAA bispecific antibody |
| BRS | Basic-Rich Stretch; signaling motif in CD3/TCR subunits |
| Ca2+ | Calcium ions; released upon T cell activation |
| CAR T cells | Chimeric Antigen Receptor T cells; Modified T Cell based cancer immunotherapy |
| CD123 | Tumor associated antigen in hematological cancers |
| CD19 | Tumor associated antigen in hematological cancers |
| CD20 | Tumor associated antigen in hematological cancers |
| CD27 | Tumor associated antigen in hematological cancers |
| CD28 | Costimulatory Receptor |
| CD3 | Cd3 signalling subunit of the CD3-TCR complex on T cells |
| CD3+ | CD3 expressing |
| CD33 | Tumor associated antigen in hematological cancers |
| CD38 | Tumor associated antigen in hematological cancers |
| CD39 | Enzyme that converts ATP to ADP; ATP/adenosine pathway |
| CD3ɛ−/− | CD3 knock out |
| CD3-TCR | CD3-T Cell Receptor complex expressed on T Cells, composed of 1:1:1:1 ratio TCRαβ:CD3γε:CD3δε:CD3ζζ subunits |
| CD3δ | CD3 delta |
| CD3ε | CD3 epsilon |
| CD3γ | CD3 gamma |
| CD3ζ | CD3 zeta |
| CD3γε | Gamma-Epsilon heterodimeric subunit of the TCR/CD3 receptor complex |
| CD3δε | Delta-Epsilon heterodimeric subunit of the TCR/CD3 receptor complex |
| CD3ζζ | Zeta-Zeta homodimeric subunit of the TCR/CD3 receptor complex |
| CD4+ | CD4+ Helper T cell |
| CD40 | Costimulatory Receptor |
| CD43 | Protein tyrosine phosphatase that favors dephosphrylation of ITAMs |
| CD45 | Protein tyrosine phosphatase that favors dephosphrylation of ITAMs |
| CD47 | “Don’t eat me” signal expressed on tumor cells to prevent phagocytosis by macrophages |
| CD69 | Early T cell activation marker |
| CD73 | Enzyme that converts AMP to Adenosine; ATP/adenosine pathway |
| CD79αβ | Signalling subunit of the B Cell Receptor |
| CD8+ | CD8+ Cytotoxic T cell |
| CDC | complement-dependent cytotoxicity |
| CDRs | Complementarity-determining regions on T cells; involved in pMHC binding |
| CEA | Carcinoembryonic antigen; a type of tumor associated antigen |
| CNS | Central Nervous System |
| CRISPR/Cas9 | clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9; genomic engineering technique |
| CTLA-4 | Negative regulator of the immune response; Exhaustion marker on T cells |
| DAG | Diacylglycerol kinase involved in T cell activation signaling |
| DGKα | Diacylglycerol Kinase Alpha |
| DLL3 | Delta-like ligand 3; tumor associated antigen in neuroendocrine tumors |
| DNA | Deoxyribonucleic acid; Double stranded nucleic acid composed of the bases A, T, G and C |
| EAE | Experimental autoimmune encephalomyelitis |
| EGFRviii | Epidermal growth factor receptor variant III; tumor associated antigen in glioblastoma |
| EM | Electron Microscopy |
| EpCAM | Epithelial Cell Adhesion Molecule; tumor associated antigen |
| Fab | Antigen-binding fragment on antibody |
| Fc | Fragment crystallizable; Antigen non-binding fragment on antibody |
| FDA | U.S. Food and Drug Administration |
| FYN | src superfamily protein tyrosine kinases involved in ITAM phosphorylation |
| G4.18 | Anti-CD3 antibody |
| GMP | Good manufacturing practice; production quality regulations for clinical products and therapies |
| gp100 | Cognate peptide for PMEL T cells |
| HCV | Hepatitis C Virus |
| HER-2 | Human epidermal growth factor receptor 2; tumor associated antigen in breast cancers |
| HIF1-α | Hypoxia-inducible factor 1-alpha; key transcription factor that drives hypoxia |
| HLA | Human leukocyte antigens |
| IBD | inflammatory bowel disease |
| ICB | Immune Checkpoint Blocakde |
| ICOS | Inducible T-cell COStimulator; Costimulatory Receptor |
| IFN-γ | Interferon Gamma; T cell activation cytokine |
| IL-10 | Interleukin-10; anti-inflammatory cytokine |
| ImmTAC | Immune mobilizing monoclonal T-cell receptors against cancer |
| IP3 | Inositol trisphosphate |
| ITAM | Immunoreceptor tyrosine-based activation motif; involved in activation of T cells |
| lL-2 | Iinterleukin-2; T cell activation cytokine |
| LCK | src superfamily protein tyrosine kinases involved in ITAM phosphorylation |
| LIGS | Ligand-Guided Selection; a type of modified SELEX |
| mAb | Monoclonal antibody |
| MHC | Major Histocompatibility Complex; proteins involved in self-discrimination |
| miRNAs | microRNA; non-coding ssRNA molecule |
| MS | Multiple Sclerosis |
| MUC16 | Mucin 16; tumor associated antigen |
| NASH | nonalcoholic steatohepatitis |
| NCK | Non-catalytic region of tyrosine kinase; second messenger in T cell signalling |
| NFAT | Nuclear factor of activated T-cells; key transcription factor involved in T cell activation |
| NF-κB | Nuclear Factor kappa-light-chain-enhancer of activated B cells; key transcription factor involved in T cell activation |
| nM | Nanomolar |
| NOD | Nucleotide oligomerization domain |
| NSCLC | Non-small-cell lung carcinoma |
| OKT3 | Human Anti-CD3 antibody |
| OX-40 | Costimulatory Receptor |
| PD-1 | Programmed cell death protein 1; negative regulator of the immune response; Exhaustion marker on T cells |
| PDL-1 | Programmed death-ligand 1; negative regulator of the immune response |
| PLC | Phospholipase C; second messenger in T cell activation signaling |
| PLP139–151 EAE | PLP139–151 peptide-induced Experimental autoimmune encephalomyelitis |
| pMHC | Peptide-Major Histocompatibility Complex; cognate ligand for the T cell Receptor |
| PRS | Proline-Rich Stretch; signaling motif in CD3/TCR subunits |
| PSMA | Prostate-Specific Membrane Antigen; tumor associated antigen in prostate cancer |
| pSMAC | Peripheral supramolecular activation complex; macromolecular structure of TCRs in activated T cells |
| PTPN2 | Protein tyrosine phosphatase non-receptor type 2 |
| PTPN22 | Protein tyrosine phosphatase non-receptor type 22 |
| RAG1/2 | Recombination-activating gene 1/2 |
| Ras | Rat sarcoma virus protein; small GTPase |
| RNA | Riboxynucleic Acid; single stranded oligonucleotide containing the bases Adenine, Guanine, Uracil and Cytosine. |
| scFvs | Single Chain Variable Fragment; fusion protein of heavy and variable chains of the antigen binding arm of an antibody |
| SCID | Severe combined immunodeficiency |
| SELEX | Systematic Evolution of Ligands by Exponential Evolution; the process through which aptamers are identified |
| SH.3 | Src Homology 3 (SH3) domains; signaling motif present in protein tyrosine kinases |
| SHP1 | Src homology region 2 domain-containing phosphatase-1 |
| siRNAs | Small interfering RNA; non-coding RNA used to silence genes |
| SJL/J | Swiss Jim Lambert EAE mice |
| Src | src superfamily protein tyrosine kinases involved in ITAM phosphorylation |
| SSTR2 | Somatostatin receptor 2; Tumor associated antigen in pancreatic cancer |
| TCR | T Cell Receptor; expressed on T cells |
| TCR-CD3 | CD3-T Cell Receptor complex expressed on T Cells, composed of 1:1:1:1 ratio TCRαβ:CD3γε:CD3δε:CD3ζζ subunits |
| TCR α-β | alpha beta T Cell |
| TCR-scFv | fusion protein containing a TCR and a single chain variable fragment |
| TCRα | alpha chain of the TCR |
| TCRβ | beta chain of the TCR |
References
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, C.; Saz, A.; Fornaguera, C.; Borrós, S. Cancer Immunotherapies Revisited: State of the Art of Conventional Treatments and next-Generation Nanomedicines. Cancer Gene Ther. 2021, 28, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Shema, E.; Loi, S.; Galluzzi, L. Intratumoral Heterogeneity in Cancer Progression and Response to Immunotherapy. Nat. Med. 2021, 27, 212–224. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the Tumor Microenvironment in PD-L1/PD-1-Mediated Tumor Immune Escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef]
- Meraviglia-Crivelli, D.; Zheleva, A.; Barainka, M.; Moreno, B.; Villanueva, H.; Pastor, F. Therapeutic Strategies to Enhance Tumor Antigenicity: Making the Tumor Detectable by the Immune System. Biomedicines 2022, 10, 1842. [Google Scholar] [CrossRef]
- Meraviglia-Crivelli, D.; Villanueva, H.; Zheleva, A.; Villalba-Esparza, M.; Moreno, B.; Menon, A.P.; Calvo, A.; Cebollero, J.; Barainka, M.; de los Mozos, I.R.; et al. IL-6/STAT3 Signaling in Tumor Cells Restricts the Expression of Frameshift-Derived Neoantigens by SMG1 Induction. Mol. Cancer 2022, 21, 211. [Google Scholar] [CrossRef]
- Dong, D.; Zheng, L.; Lin, J.; Zhang, B.; Zhu, Y.; Li, N.; Xie, S.; Wang, Y.; Gao, N.; Huang, Z. Structural Basis of Assembly of the Human T Cell Receptor–CD3 Complex. Nature 2019, 573, 546–552. [Google Scholar] [CrossRef]
- Call, M.E.; Pyrdol, J.; Wiedmann, M.; Wucherpfennig, K.W. The Organizing Principle in the Formation of the T Cell Receptor-CD3 Complex. Cell 2002, 111, 967–979. [Google Scholar] [CrossRef]
- Xu, X.; Li, H.; Xu, C. Structural Understanding of T Cell Receptor Triggering. Cell Mol. Immunol. 2020, 17, 193–202. [Google Scholar] [CrossRef]
- Sušac, L.; Vuong, M.T.; Thomas, C.; von Bülow, S.; O’Brien-Ball, C.; Santos, A.M.; Fernandes, R.A.; Hummer, G.; Tampé, R.; Davis, S.J. Structure of a Fully Assembled Tumor-Specific T Cell Receptor Ligated by PMHC. Cell 2022, 185, 3201–3213.e19. [Google Scholar] [CrossRef]
- Reth, M. Antigen Receptor Tail Clue. Nature 1989, 338, 383–384. [Google Scholar] [CrossRef]
- Müller, B.; Cooper, L.; Terhorst, C. Interplay between the Human TCR/CD3ϵ and the B-Cell Antigen Receptor Associated Ig-β (B29). Immunol. Lett. 1995, 44, 97–103. [Google Scholar] [CrossRef]
- Mariuzza, R.A.; Agnihotri, P.; Orban, J. The Structural Basis of T-Cell Receptor (TCR) Activation: An Enduring Enigma. J. Biol. Chem. 2020, 295, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Pettmann, J.; Abu-Shah, E.; Kutuzov, M.; Wilson, D.B.; Dustin, M.L.; Davis, S.J.; van der Merwe, P.A.; Dushek, O. T Cells Exhibit Unexpectedly Low Discriminatory Power and Can Respond to Ultra-Low Affinity Peptide-MHC Ligands. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gálvez, J.; Gálvez, J.J.; García-Peñarrubia, P. Is TCR/PMHC Affinity a Good Estimate of the T-Cell Response? An Answer Based on Predictions from 12 Phenotypic Models. Front. Immunol. 2019, 10, 349. [Google Scholar] [CrossRef]
- Rickert, R.C. New Insights into Pre-BCR and BCR Signalling with Relevance to B Cell Malignancies. Nat. Rev. Immunol. 2013, 13, 578–591. [Google Scholar] [CrossRef] [PubMed]
- Ngoenkam, J.; Schamel, W.W.; Pongcharoen, S. Selected Signalling Proteins Recruited to the T-Cell Receptor-CD3 Complex. Immunology 2018, 153, 42–50. [Google Scholar] [CrossRef]
- Karjalainen, K. High Sensitivity, Low Affinity-Paradox of T-Cell Receptor Recognition. Curr. Opin. Immunol. 1994, 6, 9–12. [Google Scholar] [CrossRef]
- Alarcon, B.; Ley, S.C.; Sanchez-Madrid, F.; Blumberg, R.S.; Ju, S.T.; Fresno, M.; Terhorst, C. The CD3-γ and CD3-δ Subunits of the T Cell Antigen Receptor Can Be Expressed within Distinct Functional TCR/CD3 Complexes. EMBO J. 1991, 10, 903–912. [Google Scholar] [CrossRef]
- DeJarnette, J.B.; Sommers, C.L.; Huang, K.; Woodside, K.J.; Emmons, R.; Katz, K.; Shores, E.W.; Love, P.E. Specific Requirement for CD3ε in T Cell Development. Proc. Natl. Acad. Sci. USA 1998, 95, 14909–14914. [Google Scholar] [CrossRef] [PubMed]
- Letourneur, F.; Klausner, R.D. Activation of T Cells by a Tyrosine Kinase Activation Domain in the Cytoplasmic Tail of CD3 ε. Science 1992, 255, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Kuhns, M.S.; Davis, M.M.; Garcia, K.C. Deconstructing the Form and Function of the TCR/CD3 Complex. Immunity 2006, 24, 133–139. [Google Scholar] [CrossRef]
- Bettini, M.L.; Chou, P.-C.; Guy, C.S.; Lee, T.; Vignali, K.M.; Vignali, D.A.A. Cutting Edge: CD3 ITAM Diversity Is Required for Optimal TCR Signaling and Thymocyte Development. J. Immunol. 2017, 199, 1555–1560. [Google Scholar] [CrossRef] [PubMed]
- Tjon, J.M.L.; Verbeek, W.H.M.; Kooy-Winkelaar, Y.M.C.; Nguyen, B.H.; van der Slik, A.R.; Thompson, A.; Heemskerk, M.H.M.; Schreurs, M.W.J.; Dekking, L.H.A.; Mulder, C.J.; et al. Defective Synthesis or Association of T-Cell Receptor Chains Underlies Loss of Surface T-Cell Receptor CD3 Expression in Enteropathy-Associated T-Cell Lymphoma. Blood 2008, 112, 5103–5110. [Google Scholar] [CrossRef]
- Hall, C.; Berkhout, B.; Alarcon, B.; Sancho, J.; Wileman, T.; Terhorst, C. Requirements for Cell Surface Expression of the Human TCR/CD3 Complex in Non-T Cells. Int. Immunol. 1991, 3, 359–368. [Google Scholar] [CrossRef]
- Dave, V.P. Hierarchical Role of CD3 Chains in Thymocyte Development. Immunol. Rev. 2009, 232, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Dave, V.P. Role of CD3ε-Mediated Signaling in T-Cell Development and Function. Crit. Rev. Immunol. 2011, 31, 73–84. [Google Scholar] [CrossRef]
- Gil, D.; Schamel, W.W.A.; Montoya, M.; Sánchez-Madrid, F.; Alarcón, B. Recruitment of Nck by CD3ϵ Reveals a Ligand-Induced Conformational Change Essential for T Cell Receptor Signaling and Synapse Formation. Cell 2002, 109, 901–912. [Google Scholar] [CrossRef]
- Minguet, S.; Schamel, W.W.A.; Alarcon, B.; Höfer, T. The Allostery Model of TCR Regulation. J. Immunol. Ref. 2021, 198, 47–52. [Google Scholar] [CrossRef]
- Alarcón, B.; Gil, D.; Delgado, P.; Schamel, W.W.A. Initiation of TCR Signaling: Regulation within CD3 Dimers. Immunol. Rev. 2003, 191, 38–46. [Google Scholar] [CrossRef]
- Huynh, H.T.; Nelson, A.D.; Hoffmann, M.; Abergel, M.; Hu, S.; Alarcon, B.; Schrum, A.G.; Gil, D. Molecular Mechanisms Underlying T Cell Co-Potentiation by Anti-CD3 Fab Fragments. J. Immunol. 2020, 204, 246. [Google Scholar] [CrossRef]
- Tailor, P.; Tsai, S.; Shameli, A.; Serra, P.; Wang, J.; Robbins, S.; Nagata, M.; Szymczak-Workman, A.L.; Vignali, D.A.A.; Santamaria, P. The Proline-Rich Sequence of CD3ε as an Amplifier of Low-Avidity TCR Signaling. J. Immunol. 2008, 181, 243–255. [Google Scholar] [CrossRef]
- Mingueneau, M.; Sansoni, A.; Grégoire, C.; Roncagalli, R.; Aguado, E.; Weiss, A.; Malissen, M.; Malissen, B. The Proline-Rich Sequence of CD3ε Controls T Cell Antigen Receptor Expression on and Signaling Potency in Preselection CD4+CD8+ Thymocytes. Nat. Immunol. 2008, 9, 522–532. [Google Scholar] [CrossRef]
- Hartl, F.A.; Beck-Garcìa, E.; Woessner, N.M.; Flachsmann, L.J.; Cárdenas, R.M.-H.V.; Brandl, S.M.; Taromi, S.; Fiala, G.J.; Morath, A.; Mishra, P.; et al. Noncanonical Binding of Lck to CD3ε Promotes TCR Signaling and CAR Function. Nat. Immunol. 2020, 21, 902–913. [Google Scholar] [CrossRef]
- Hartl, F.A.; Ngoenkam, J.; Beck-Garcia, E.; Cerqueira, L.; Wipa, P.; Paensuwan, P.; Suriyaphol, P.; Mishra, P.; Schraven, B.; Günther, S.; et al. Cooperative Interaction of Nck and Lck Orchestrates Optimal TCR Signaling. Cells 2021, 10, 834. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Alarcon, B.; Wileman, T.; Terhorst, C. The T Cell Receptor/CD3 Complex: A Dynamic Protein Ensemble. Annu. Rev. Immunol. 1988, 6, 629–662. [Google Scholar] [CrossRef]
- de la Hera, A.; Muller, U.; Olsson, C.; Isaaz, S.; Tunnacliffe, A. Structure of the T Cell Antigen Receptor (TCR): Two CD3ε Subunits in a Functional TCR/CD3 Complex. J. Exp. Med. 1991, 173, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Huppa, J.B.; Ploegh, H.L. The α Chain of the T Cell Antigen Receptor Is Degraded in the Cytosol. Immunity 1997, 7, 113–122. [Google Scholar] [CrossRef]
- Huppa, J.B.; Ploegh, H.L. In Vitro Translation and Assembly of a Complete T Cell Receptor-CD3 Complex. J. Exp. Med. 1997, 186, 393–403. [Google Scholar] [CrossRef]
- Delgado, P.; Alarcón, B. An Orderly Inactivation of Intracellular Retention Signals Controls Surface Expression of the T Cell Antigen Receptor. J. Exp. Med. 2005, 201, 555–566. [Google Scholar] [CrossRef]
- Recio, M.J.; Moreno-Pelayo, M.A.; Kiliç, S.S.; Guardo, A.C.; Sanal, O.; Allende, L.M.; Pérez-Flores, V.; Mencía, A.; Modamio-Høybjør, S.; Seoane, E.; et al. Differential Biological Role of CD3 Chains Revealed by Human Immunodeficiencies. J. Immunol. 2007, 178, 2556–2564. [Google Scholar] [CrossRef]
- Weissman, A.M.; Frank, S.J.; Orloff, D.G.; Mercep, M.; Ashwell, J.D.; Klausner, R.D. Role of the Zeta Chain in the Expression of the T Cell Antigen Receptor: Genetic Reconstitution Studies. EMBO J. 1989, 8, 3651–3656. [Google Scholar] [CrossRef]
- Hayes, S.M.; Love, P.E. Stoichiometry of the Murine Γδ T Cell Receptor. J. Exp. Med. 2006, 203, 47–52. [Google Scholar] [CrossRef]
- Muñoz-Ruiz, M.; Pérez-Flores, V.; Garcillán, B.; Guardo, A.C.; Mazariegos, M.S.; Takada, H.; Allende, L.M.; Kilic, S.S.; Sanal, O.; Roifman, C.M.; et al. Human CD3γ, but Not CD3δ, Haploinsufficiency Differentially Impairs Γδ versus Aβ Surface TCR Expression. BMC Immunol. 2013, 14, 3. [Google Scholar] [CrossRef]
- Fischer, A.; de Saint Basile, G.; le Deist, F. CD3 Deficiencies. Curr. Opin. Allergy Clin. Immunol. 2005, 5, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Erman, B.; Fırtına, S.; Fışgın, T.; Bozkurt, C.; Çipe, F.E. Biallelic Form of a Known CD3E Mutation in a Patient with Severe Combined Immunodeficiency. J. Clin. Immunol. 2020, 40, 539–542. [Google Scholar] [CrossRef]
- de saint Basile, G.; Geissmann, F.; Flori, E.; Uring-Lambert, B.; Soudais, C.; Cavazzana-Calvo, M.; Durandy, A.; Jabado, N.; Fischer, A.; Deist, F. le Severe Combined Immunodeficiency Caused by Deficiency in Either the δ or the ε Subunit of CD3. J. Clin. Investig. 2004, 114, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.L.; Lauritsen, J.P.H.; Cooney, M.; Parrott, R.E.; Sajaroff, E.O.; Win, C.M.; Keller, M.D.; Carpenter, J.H.; Carabana, J.; Krangel, M.S.; et al. T-B+NK+ Severe Combined Immunodeficiency Caused by Complete Deficiency of the CD3ζ Subunit of the T-Cell Antigen Receptor Complex. Blood 2007, 109, 3198–3206. [Google Scholar] [CrossRef]
- Pitcher, L.A.; van Oers, N.S.C. T-Cell Receptor Signal Transmission: Who Gives an ITAM? Trends Immunol. 2003, 24, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H. T Cell Receptors, Mechanosensors, Catch Bonds and Immunotherapy. Prog. Biophys. Mol. Biol 2020, 153, 23–27. [Google Scholar] [CrossRef]
- Stone, J.D.; Chervin, A.S.; Kranz, D.M. T-Cell Receptor Binding Affinities and Kinetics: Impact on T-Cell Activity and Specificity. Immunology 2009, 126, 165–176. [Google Scholar] [CrossRef]
- Zhu, C.; Chen, W.; Lou, J.; Rittase, W.; Li, K. Mechanosensing through Immunoreceptors. Nat. Immunol. 2019, 20, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; van der Merwe, P.A. The Kinetic-Segregation Model: TCR Triggering and Beyond. Nat. Immunol. 2006, 7, 803–809. [Google Scholar] [CrossRef]
- Swamy, M.; Beck-Garcia, K.; Beck-Garcia, E.; Hartl, F.A.; Morath, A.; Yousefi, O.S.; Dopfer, E.P.; Molnár, E.; Schulze, A.K.; Blanco, R.; et al. A Cholesterol-Based Allostery Model of T Cell Receptor Phosphorylation. Immunity 2016, 44, 1091–1101. [Google Scholar] [CrossRef]
- Minguet, S.; Swamy, M.; Alarcón, B.; Luescher, I.F.; Schamel, W.W.A. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity 2007, 26, 43–54. [Google Scholar] [CrossRef]
- Trebak, M.; Kinet, J.P. Calcium Signalling in T Cells. Nat. Rev. Immunol. 2019, 19, 154–169. [Google Scholar] [CrossRef]
- Kaye, J.; Janeway, C.A. The Fab Fragment of a Directly Activating Monoclonal Antibody That Precipitates a Disulfide-Linked Heterodimer from a Helper T Cell Clone Blocks Activation by Either Allogeneic Ia or Antigen and Self-Ia. J. Exp. Med. 1984, 159, 1397–1412. [Google Scholar] [CrossRef]
- Boniface, J.J.; Rabinowitz, J.D.; Wülfing, C.; Hampl, J.; Reich, Z.; Altman, J.D.; Kantor, R.M.; Beeson, C.; McConnell, H.M.; Davis, M.M. Initiation of Signal Transduction through the T Cell Receptor Requires the Multivalent Engagement of Peptide/MHC Ligands. Immunity 1998, 9, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Luengo, A.; Gui, D.Y.; vander Heiden, M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef]
- Kim, H.; Peng, G.; Hicks, J.M.; Weiss, H.L.; van Meir, E.G.; Brenner, M.K.; Yotnda, P. Engineering Human Tumor-Specific Cytotoxic T Cells to Function in a Hypoxic Environment. Mol. Ther. 2008, 16, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Vigano, S.; Alatzoglou, D.; Irving, M.; Ménétrier-Caux, C.; Caux, C.; Romero, P.; Coukos, G. Targeting Adenosine in Cancer Immunotherapy to Enhance T-Cell Function. Front. Immunol. 2019, 10, 925. [Google Scholar] [CrossRef] [PubMed]
- Hay, N. Reprogramming Glucose Metabolism in Cancer: Can It Be Exploited for Cancer Therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Lu, F.; Fei, Q.; Yu, X.; Xiong, P.; Yu, X.; Dang, Y.; Hou, Z.; Lin, W.; Lin, X.; et al. Single-Cell RNA Sequencing Reveals Compartmental Remodeling of Tumor-Infiltrating Immune Cells Induced by Anti-CD47 Targeting in Pancreatic Cancer. J. Hematol. Oncol. 2019, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Waiser, J.; Duerr, M.; Budde, K.; Rudolph, B.; Wu, K.; Bachmann, F.; Halleck, F.; Schönemann, C.; Lachmann, N. Treatment of Acute Antibody-Mediated Renal Allograft Rejection with Cyclophosphamide. Transplantation 2017, 101, 2545–2552. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, C.; Weiner, H.L. Therapeutic Anti-CD3 Monoclonal Antibodies: From Bench to Bedside. Immunotherapy 2016, 8, 889–906. [Google Scholar] [CrossRef]
- Xu, D.; Alegre, M.L.; Varga, S.S.; Rothermel, A.L.; Collins, A.M.; Pulito, V.L.; Hanna, L.S.; Dolan, K.P.; Parren, P.W.H.I.; Bluestone, J.A.; et al. In Vitro Characterization of Five Humanized OKT3 Effector Function Variant Antibodies. Cell Immunol. 2000, 200, 16–26. [Google Scholar] [CrossRef]
- Hickey, J.W.; Dong, Y.; Chung, J.W.; Salathe, S.F.; Pruitt, H.C.; Li, X.; Chang, C.; Fraser, A.K.; Bessell, C.A.; Ewald, A.J.; et al. Engineering an Artificial T-Cell Stimulating Matrix for Immunotherapy. Adv. Mater. 2019, 31, 1807359. [Google Scholar] [CrossRef]
- Cheung, A.S.; Zhang, D.K.Y.; Koshy, S.T.; Mooney, D.J. Scaffolds That Mimic Antigen-Presenting Cells Enable Ex Vivo Expansion of Primary T Cells. Nat. Biotechnol. 2018, 36, 160–169. [Google Scholar] [CrossRef]
- Leo, O.; Foo, M.; Sachs, D.H.; Samelson, L.E.; Bluestone, J.A. Identification of a Monoclonal Antibody Specific for a Murine T3 Polypeptide. Proc. Natl. Acad. Sci. USA 1987, 84, 1374–1378. [Google Scholar] [CrossRef]
- Tran, G.T.; Carter, N.; He, X.Y.; Spicer, T.S.; Plain, K.M.; Nicolls, M.; Hall, B.M.; Hodgkinson, S.J. Reversal of Experimental Allergic Encephalomyelitis with Non-Mitogenic, Non-Depleting Anti-CD3 MAb Therapy with a Preferential Effect on Th 1 Cells That Is Augmented by IL-4. Int. Immunol. 2001, 13, 1109–1120. [Google Scholar] [CrossRef]
- Kohm, A.P.; Williams, J.S.; Bickford, A.L.; McMahon, J.S.; Chatenoud, L.; Bach, J.-F.; Bluestone, J.A.; Miller, S.D. Treatment with Nonmitogenic Anti-CD3 Monoclonal Antibody Induces CD4+ T Cell Unresponsiveness and Functional Reversal of Established Experimental Autoimmune Encephalomyelitis. J. Immunol. 2005, 174, 4525–4534. [Google Scholar] [CrossRef] [PubMed]
- Waldron-Lynch, F.; Henegariu, O.; Deng, S.; Preston-Hurlburt, P.; Tooley, J.; Flavell, R.; Herold, K.C. Teplizumab Induces Human Gut-Tropic Regulatory Cells in Humanized Mice and Patients. Sci. Transl. Med. 2012, 4, 118ra12. [Google Scholar] [CrossRef] [PubMed]
- Herold, K.C.; Hagopian, W.; Auger, J.A.; Poumian-Ruiz, E.; Taylor, L.; Donaldson, D.; Gitelman, S.E.; Harlan, D.M.; Xu, D.; Zivin, R.A.; et al. Anti-CD3 Monoclonal Antibody in New-Onset Type 1 Diabetes Mellitus. N. Engl. J. Med. 2002, 346, 1692–1698. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.; Eriksson, L.; Nowak, C.; Teixeira, P.F.; Widman, M.; Lindqvist, A.; Casas, R.; Lind, M.; Hannelius, U. Phase III, Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial to Evaluate the Efficacy and Safety of RhGAD65 to Preserve Endogenous Beta Cell Function in Adolescents and Adults with Recently Diagnosed Type 1 Diabetes, Carrying the Genetic HLA DR3-DQ2 Haplotype: The DIAGNODE-3 Study Protocol. BMJ Open 2022, 12, e061776. [Google Scholar] [CrossRef]
- Bisikirska, B.; Colgan, J.; Luban, J.; Bluestone, J.A.; Herold, K.C. TCR Stimulation with Modified Anti-CD3 MAb Expands CD8+ T Cell Population and Induces CD8+CD25+ Tregs. J. Clin. Investig. 2005, 115, 2904–2913. [Google Scholar] [CrossRef]
- Belghith, M.; Bluestone, J.A.; Barriot, S.; Mégret, J.; Bach, J.-F.; Chatenoud, L. TGF-β-Dependent Mechanisms Mediate Restoration of Self-Tolerance Induced by Antibodies to CD3 in Overt Autoimmune Diabetes. Nat. Med. 2003, 9, 1202–1208. [Google Scholar] [CrossRef]
- Keymeulen, B.; Vandemeulebroucke, E.; Ziegler, A.G.; Mathieu, C.; Kaufman, L.; Hale, G.; Gorus, F.; Goldman, M.; Walter, M.; Candon, S.; et al. Insulin Needs after CD3-Antibody Therapy in New-Onset Type 1 Diabetes. N. Engl. J. Med. 2005, 352, 2598–2608. [Google Scholar] [CrossRef]
- Plevy, S.; Salzberg, B.; van Assche, G.; Regueiro, M.; Hommes, D.; Sandborn, W.; Hanauer, S.; Targan, S.; Mayer, L.; Mahadevan, U.; et al. A Phase I Study of Visilizumab, a Humanized Anti-CD3 Monoclonal Antibody, in Severe Steroid-Refractory Ulcerative Colitis. Gastroenterology 2007, 133, 1414–1422. [Google Scholar] [CrossRef]
- Newman, M.J.; Benani, D.J. A Review of Blinatumomab, a Novel Immunotherapy. J. Oncol. Pharm. Pract. 2016, 22, 639–645. [Google Scholar] [CrossRef]
- Locatelli, F.; Zugmaier, G.; Rizzari, C.; Morris, J.D.; Gruhn, B.; Klingebiel, T.; Parasole, R.; Linderkamp, C.; Flotho, C.; Petit, A.; et al. Effect of Blinatumomab vs Chemotherapy on Event-Free Survival among Children with High-Risk First-Relapse B-Cell Acute Lymphoblastic Leukemia: A Randomized Clinical Trial. JAMA 2021, 325, 843–854. [Google Scholar] [CrossRef]
- Fu, Y.; Xiao, W.; Mao, Y. Recent Advances and Challenges in Uveal Melanoma Immunotherapy. Cancers 2022, 14, 3094. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Tebentafusp: First Approval. Drugs 2022, 82, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Zumrut, H.E.; Batool, S.; Argyropoulos, K.V.; Williams, N.; Azad, R.; Mallikaratchy, P.R. Integrating Ligand-Receptor Interactions and In Vitro Evolution for Streamlined Discovery of Artificial Nucleic Acid Ligands. Mol. Ther. Nucleic. Acids. 2019, 17, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Miodek, A.; Mourlane, F.; Bauche, C.; Vaillant, R. Anti-CD3 aptamers for use in cell targeting and labeling. U.S. Patent Application No. 17/629,943, 11 August 2022. [Google Scholar]
- Poltorak, M.; Arndt, B.; Kowtharapu, B.S.; Reddycherla, A.V.; Witte, V.; Lindquist, J.A.; Schraven, B.; Simeoni, L. TCR Activation Kinetics and Feedback Regulation in Primary Human T Cells. Cell Commun. Signal. 2013, 11, 4. [Google Scholar] [CrossRef]
- Arndt, B.; Poltorak, M.; Kowtharapu, B.S.; Reichardt, P.; Philipsen, L.; Lindquist, J.A.; Schraven, B.; Simeoni, L. Analysis of TCR Activation Kinetics in Primary Human T Cells upon Focal or Soluble Stimulation. J. Immunol. Methods 2013, 387, 276–283. [Google Scholar] [CrossRef]
- Ellenhorn, J.D.I.; Hirsch, R.; Schreiber, H.; Bluestone, J.A. In Vivo Administration of Anti-CD3 Prevents Malignant Progressor Tumor Growth. Science 1988, 242, 569–571. [Google Scholar] [CrossRef]
- Kung, P.C.; Goldstein, G.; Reinherz, E.L.; Schlossman, S.F. Monoclonal Antibodies Defining Distinctive Human T Cell Surface Antigens. Science 1979, 206, 347–349. [Google Scholar] [CrossRef]
- Weiner, L.M.; Murray, J.C.; Shuptrine, C.W. Antibody-Based Immunotherapy of Cancer. Cell 2012, 148, 1081–1084. [Google Scholar] [CrossRef]
- LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases; 2012 Muromonab-CD3. Available online: https://www.ncbi.nlm.nih.gov/books/NBK548590/ (accessed on 18 December 2022).
- Chatenoud, L.; Thervet, E.; Primo, J.; Bach, J.F. Anti-CD3 Antibody Induces Long-Term Remission of Overt Autoimmunity in Nonobese Diabetic Mice. Proc. Natl. Acad. Sci. USA 1994, 91, 123–127. [Google Scholar] [CrossRef]
- Lúdvíksson, B.R.; Ehrhardt, R.O.; Strober, W. TGF-Beta Production Regulates the Development of the 2,4,6-Trinitrophenol-Conjugated Keyhole Limpet Hemocyanin-Induced Colonic Inflammation in IL-2-Deficient Mice. J. Immunol. 1997, 159, 3622–3628. [Google Scholar] [CrossRef]
- Hughes, C.; Wolos, J.A.; Giannini, E.H.; Hirsch, R. Induction of T Helper Cell Hyporesponsiveness in an Experimental Model of Autoimmunity by Using Nonmitogenic Anti-CD3 Monoclonal Antibody. J. Immunol. 1994, 153, 3319–3325. [Google Scholar] [CrossRef] [PubMed]
- Goto, R.; You, S.; Zaitsu, M.; Chatenoud, L.; Wood, K.J. Delayed Anti-CD3 Therapy Results in Depletion of Alloreactive T Cells and the Dominance of Foxp3+ CD4+ Graft Infiltrating Cells. Am. J. Transpl. 2013, 13, 1655–1664. [Google Scholar] [CrossRef]
- You, S.; Zuber, J.; Kuhn, C.; Baas, M.; Valette, F.; Sauvaget, V.; Sarnacki, S.; Sawitzki, B.; Bach, J.-F.; Volk, H.-D.; et al. Induction of Allograft Tolerance by Monoclonal CD3 Antibodies: A Matter of Timing. Am. J. Transpl. 2012, 12, 2909–2919. [Google Scholar] [CrossRef] [PubMed]
- Nicolls, M.R.; Aversa, G.G.; Pearce, N.W.; Spinelli, A.; Berger, M.F.; Gurley, K.E.; Hall, B.M. Induction of Long-Term Specific Tolerance to Allografts in Rats by Therapy with an Anti-CD3-like Monoclonal Antibody. Transplantation 1993, 55, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Ilan, Y.; Zigmond, E.; Lalazar, G.; Dembinsky, A.; ben Ya’acov, A.; Hemed, N.; Kasis, I.; Axelrod, E.; Zolotarov, L.; Klein, A.; et al. Oral Administration of OKT3 Monoclonal Antibody to Human Subjects Induces a Dose-Dependent Immunologic Effect in T Cells and Dendritic Cells. J. Clin. Immunol. 2010, 30, 167–177. [Google Scholar] [CrossRef]
- Halota, W.; Ferenci, P.; Kozielewicz, D.; Dybowska, D.; Lisovoder, N.; Samira, S.; Shalit, I.; Ellis, R.; Ilan, Y. Oral Anti-CD3 Immunotherapy for HCV-Nonresponders Is Safe, Promotes Regulatory T Cells and Decreases Viral Load and Liver Enzyme Levels: Results of a Phase-2a Placebo-Controlled Trial. J. Viral. Hepat. 2015, 22, 651–657. [Google Scholar] [CrossRef]
- Lalazar, G.; Mizrahi, M.; Turgeman, I.; Adar, T.; ben Ya’acov, A.; Shabat, Y.; Nimer, A.; Hemed, N.; Zolotarovya, L.; Lichtenstein, Y.; et al. Oral Administration of OKT3 MAb to Patients with NASH, Promotes Regulatory T-Cell Induction, and Alleviates Insulin Resistance: Results of a Phase IIa Blinded Placebo-Controlled Trial. J. Clin. Immunol. 2015, 35, 399–407. [Google Scholar] [CrossRef]
- Labrijn, A.F.; Janmaat, M.L.; Reichert, J.M.; Parren, P.W.H.I. Bispecific Antibodies: A Mechanistic Review of the Pipeline. Nat. Rev. Drug Discov. 2019, 18, 585–608. [Google Scholar] [CrossRef]
- Benonisson, H.; Altıntaş, I.; Sluijter, M.; Verploegen, S.; Labrijn, A.F.; Schuurhuis, D.H.; Houtkamp, M.A.; Sjef Verbeek, J.; Schuurman, J.; van Hall, T. CD3-Bispecific Antibody Therapy Turns Solid Tumors into Inflammatory Sites but Does Not Install Protective Memory. Mol. Cancer 2019, 18, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Liu, M.; Zhang, Y.; Wang, X. Bispecific T Cell Engagers: An Emerging Therapy for Management of Hematologic Malignancies. J. Hematol. Oncol. 2021, 14, 75. [Google Scholar] [CrossRef]
- Middelburg, J.; Kemper, K.; Engelberts, P.; Labrijn, A.F.; Schuurman, J.; van Hall, T. Overcoming Challenges for CD3-Bispecific Antibody Therapy in Solid Tumors. Cancers 2021, 13, 287. [Google Scholar] [CrossRef] [PubMed]
- Bortoletto, N.; Scotet, E.; Myamoto, Y.; D’Oro, U.; Lanzavecchia, A. Optimizing Anti-CD3 Affinity for Effective T Cell Targeting against Tumor Cells. Eur. J. Immunol. 2002, 32, 3102–3107. [Google Scholar] [CrossRef]
- Singh, A.; Dees, S.; Grewal, I.S. Overcoming the Challenges Associated with CD3+ T-Cell Redirection in Cancer. Br. J. Cancer 2021, 124, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Goebeler, M.; Bargou, R.C. T Cell-Engaging Therapies—BiTEs and Beyond. Nat. Rev. Clin. Oncol. 2020, 17, 418–434. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.M.; Molina-Mendiola, C.; Nelson, A.D.; Parks, C.A.; Reyes, E.E.; Hansen, M.J.; Rajagopalan, G.; Pease, L.R.; Schrum, A.G.; Gil, D. Co-Potentiation of Antigen Recognition: A Mechanism to Boost Weak T Cell Responses and Provide Immunotherapy In Vivo. Sci. Adv. 2015, 1, e1500415. [Google Scholar] [CrossRef] [PubMed]
- Shigdar, S.; Schrand, B.; Giangrande, P.H.; de Franciscis, V. Aptamers: Cutting Edge of Cancer Therapies. Mol. Ther. 2021, 29, 2396–2411. [Google Scholar] [CrossRef] [PubMed]
- Sola, M.; Menon, A.P.; Moreno, B.; Meraviglia-Crivelli, D.; Soldevilla, M.M.; Cartón-García, F.; Pastor, F. Aptamers Against Live Targets: Is In Vivo SELEX Finally Coming to the Edge? Mol. Nucleic. Acids. 2020, 21, 192–204. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX Optimization of Aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef]
- Soldevilla, M.M.; Villanueva, H.; Pastor, F. Aptamers: A Feasible Technology in Cancer Immunotherapy. J. Immunol. Res. 2016, 2016, 1083738. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Soldevilla, M.M.; Hervas, S.; Villanueva, H.; Lozano, T.; Rabal, O.; Oyarzabal, J.; Lasarte, J.J.; Bendandi, M.; Inoges, S.; de Cerio, A.L.D.; et al. Identification of LAG3 High Affinity Aptamers by HT-SELEX and Conserved Motif Accumulation (CMA). PLoS ONE 2017, 12, e0185169. [Google Scholar] [CrossRef] [PubMed]
- Gelinas, A.D.; Davies, D.R.; Janjic, N. Embracing Proteins: Structural Themes in Aptamer-Protein Complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Gold, L. SELEX: How It Happened and Where It Will Go. J. Mol. Evol. 2015, 81, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Hamm, J.; Libri, D.; Tavitian, B.; de Franciscis, V. Nucleic Acid Aptamers in Cancer Medicine. FEBS Lett. 2002, 528, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Freage, L.; Jamal, D.; Williams, N.B.; Mallikaratchy, P.R. A Homodimeric Aptamer Variant Generated from Ligand-Guided Selection Activates the T Cell Receptor Cluster of Differentiation 3 Complex. Mol. Nucleic. Acids. 2020, 22, 167–178. [Google Scholar] [CrossRef]
- Pandey, P.R.; Young, K.H.; Kumar, D.; Jain, N. RNA-Mediated Immunotherapy Regulating Tumor Immune Microenvironment: Next Wave of Cancer Therapeutics. Mol. Cancer 2022, 21, 58. [Google Scholar] [CrossRef]
- Borroto, A.; Reyes-Garau, D.; Jiménez, M.A.; Carrasco, E.; Moreno, B.; Martínez-Pasamar, S.; Cortés, J.R.; Perona, A.; Abia, D.; Blanco, S.; et al. First-in-Class Inhibitor of the T Cell Receptor for the Treatment of Autoimmune Diseases. Sci. Transl. Med. 2016, 8, 370ra184. [Google Scholar] [CrossRef] [PubMed]
- Borroto, A.; Arellano, I.; Blanco, R.; Fuentes, M.; Orfao, A.; Dopfer, E.P.; Prouza, M.; Suchànek, M.; Schamel, W.W.; Alarcón, B. Relevance of Nck-CD3 Epsilon Interaction for T Cell Activation In Vivo. J. Immunol. 2014, 192, 2042–2053. [Google Scholar] [CrossRef]
- Castro-Sanchez, P.; Teagle, A.R.; Prade, S.; Zamoyska, R. Modulation of TCR Signaling by Tyrosine Phosphatases: From Autoimmunity to Immunotherapy. Front. Cell Dev. Biol. 2020, 8, 608747. [Google Scholar] [CrossRef]
- Arranz-Nicolás, J.; Ogando, J.; Soutar, D.; Arcos-Pérez, R.; Meraviglia-Crivelli, D.; Mañes, S.; Mérida, I.; Ávila-Flores, A. Diacylglycerol Kinase α Inactivation Is an Integral Component of the Costimulatory Pathway That Amplifies TCR Signals. Cancer Immunol. Immunother. 2018, 67, 965–980. [Google Scholar] [CrossRef] [PubMed]
| Name | Type of T Cell Modulator | Target | In Vitro Effect | In Vivo Effect |
|---|---|---|---|---|
| OKT3 | Anti-CD3 antibody | Human | Induces activation, proliferation, and cytolytic activity of T cells in vitro [66] | Used to prevent acute rejection in transplants, for the treatment of autoimmune disorders [64,65], and for depleting CD3+ lymphoblastic leukemia populations in vivo [66]. Furthermore, variants of OKT3 are also used to expand T cell adoptive therapy populations ex vivo [67,68]. |
| 145-2C11 | Anti-CD3 antibody | Mouse | Agonistic activity in vitro [69] | Induces immune tolerance in vivo and tolerance towards syngeneic pancreatic islet grafts in preclinical models of diabetes [69] |
| G4.18 | Anti-CD3 antibody | Mouse | Nonmitogenic, nonactivating in vitro [70] | Induces immunotolerance in vivo in preclinical animal models of Multiple Sclerosis (MS) [70,71] |
| Teplizumab | Anti-CD3 antibody | Human | NA | Delays onset, reduces activity of autoreactive T cells, and induces T regulatory cells [72,73,74,75,76] |
| Otelixizumab | Anti-CD3 antibody | Human | NA | Used in the treatment of type 1 diabetes—improves preservation of the β cells mass in the pancreas [77] |
| Visilizumab | Anti-CD3 antibody | Human | NA | Used in the treatment of severe corticosteroid-refractory ulcerative colitis [78] |
| Blinatumomab | CD19-directed CD3 T-cell engager | Human | NA | Used in the treatment of acute lymphocytic leukemia [79,80] |
| Tebentafusp | gp100 peptide-HLA-A*02:01 directed T cell receptor (TCR) CD3 T cell engager (immune mobilizing monoclonal T-cell receptors against cancer (ImmTAC)) | Human | NA | Used in the treatment of uveal melanoma and malignant melanoma [81,82] |
| CD3-specific DNA Aptamer generated via LIGS | human CD3ε complex on Jurkat cells | Human | Robust binding to human T cells and induction of CD69, a T cell activation marker [83] | NA |
| CD3-specific RNA aptamer | Recombinant human CD3ε/γ and CD3ε/δ subunits | Human | Binding to T cells. They do not show the ability to activate the T cells [84] | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menon, A.P.; Moreno, B.; Meraviglia-Crivelli, D.; Nonatelli, F.; Villanueva, H.; Barainka, M.; Zheleva, A.; van Santen, H.M.; Pastor, F. Modulating T Cell Responses by Targeting CD3. Cancers 2023, 15, 1189. https://doi.org/10.3390/cancers15041189
Menon AP, Moreno B, Meraviglia-Crivelli D, Nonatelli F, Villanueva H, Barainka M, Zheleva A, van Santen HM, Pastor F. Modulating T Cell Responses by Targeting CD3. Cancers. 2023; 15(4):1189. https://doi.org/10.3390/cancers15041189
Chicago/Turabian StyleMenon, Ashwathi Puravankara, Beatriz Moreno, Daniel Meraviglia-Crivelli, Francesca Nonatelli, Helena Villanueva, Martin Barainka, Angelina Zheleva, Hisse M. van Santen, and Fernando Pastor. 2023. "Modulating T Cell Responses by Targeting CD3" Cancers 15, no. 4: 1189. https://doi.org/10.3390/cancers15041189
APA StyleMenon, A. P., Moreno, B., Meraviglia-Crivelli, D., Nonatelli, F., Villanueva, H., Barainka, M., Zheleva, A., van Santen, H. M., & Pastor, F. (2023). Modulating T Cell Responses by Targeting CD3. Cancers, 15(4), 1189. https://doi.org/10.3390/cancers15041189









