AI-Powered Diagnosis of Skin Cancer: A Contemporary Review, Open Challenges and Future Research Directions
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Contribution of this Survey
- This survey comprehensively discusses the application of various machine learning and deep learning methods in the implementation of skin cancer diagnosis.
- There is a discussion of new techniques in skin lesion detection such as deep belief networks and extreme learning machines, along with the traditional Computational Intelligence techniques such as random forests, recurrent neural networks, and k-nearest neighbors, etc.
- There is a designated tabular summary of works on the deep learning and machine learning techniques used for skin cancer diagnosis and detection. The tabulated summary also includes key contributions and limitations for the same.
- There is a classification of various types of skin cancer based on tumor characteristics that have been elucidated for a deeper understanding of the problem statement.
- The study also describes various open challenges present and future research directions for further improvements in the field of skin cancer diagnosis.
1.2. Survey Methodology
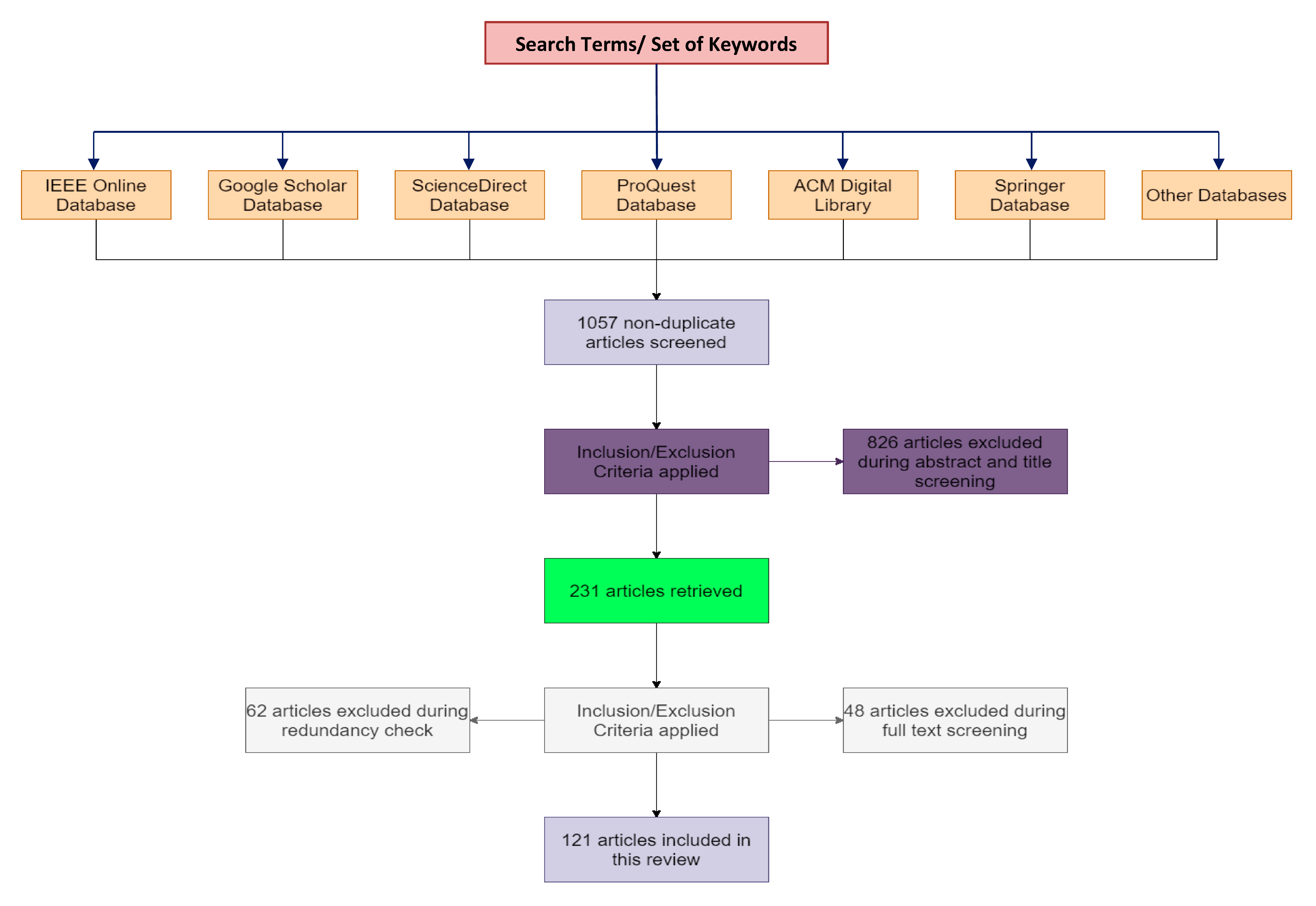
1.2.1. Search Strategy and Literature Sources
1.2.2. Inclusion Criteria
1.2.3. Elimination Criteria
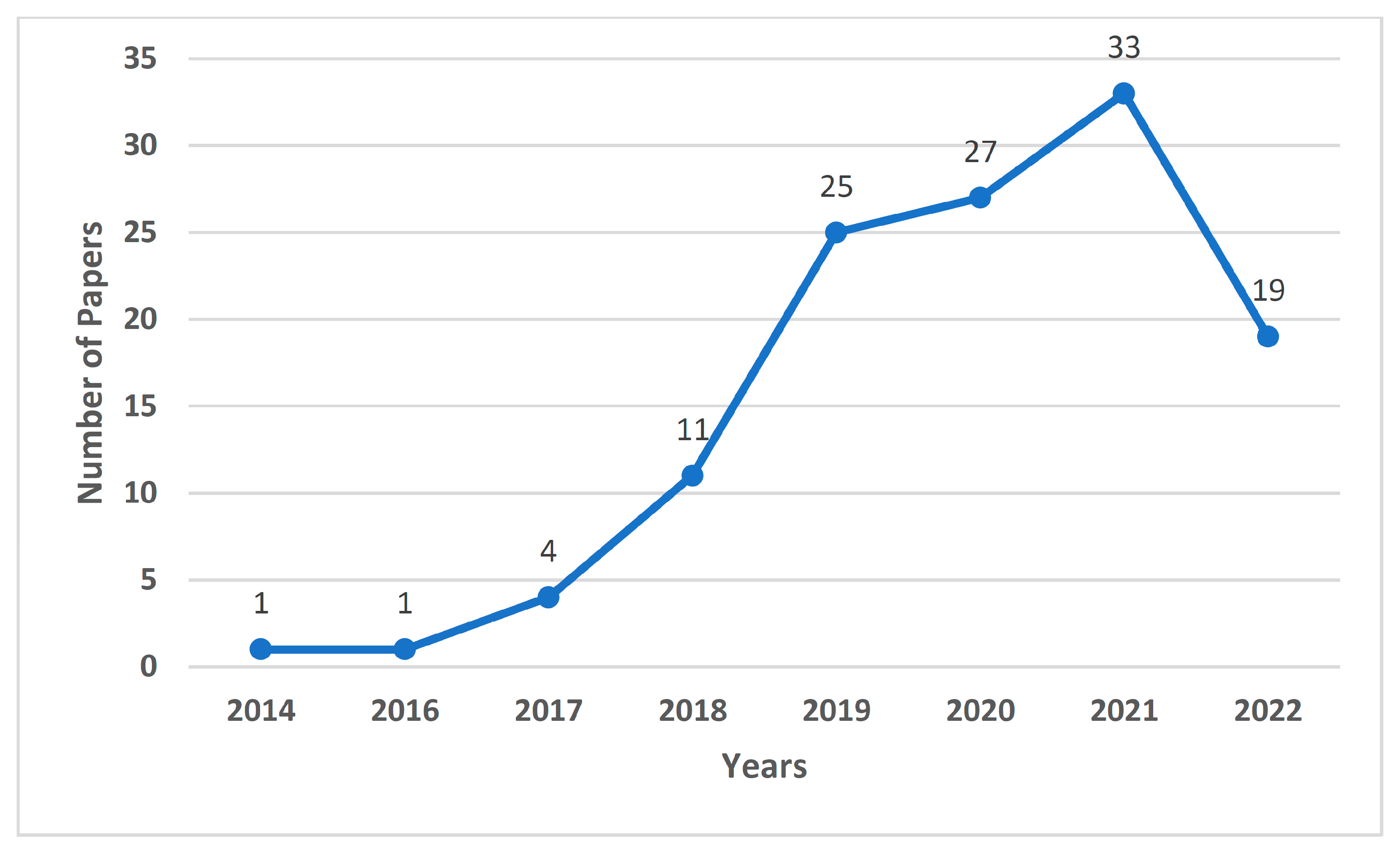
1.2.4. Results
1.3. Structure of this Review
2. Skin Cancer
- 1.
- Basal cell carcinoma: this type of cancer affects and originates from the basal cells. Basal cell carcinoma comes from keratinocytes, which are found in the epidermis. These may invade the entire epidermal thickness.
- 2.
- Squamous cell carcinoma: this subdivision deals with the uncontrollable growth of the abnormal squamous cells present in the root. Squamous cells are flat cells that are found in the tissue that constitutes the surface of the skin, and the lining of vital organs such as the respiratory organs, digestive tracts, and hollow organs of the body.
- 3.
- Melanoma: this form of cancer develops when melanocytes start to grow abnormally. Melanocytes are the cells that can become melanoma. Melanoma can develop anywhere in the skin, while it can also form in other parts of the body such as the eyes, mouth, and genitals, etc.
2.1. Skin Cancer Classification
2.1.1. Benign Tumor
2.1.2. Malignant Tumor
2.1.3. Other Tumors
2.2. Skin Cancer Datasets
3. Machine Learning and Deep Learning Models for Skin Cancer Diagnosis
3.1. Need for Machine Learning and Deep Learning Models for Skin Cancer Diagnosis
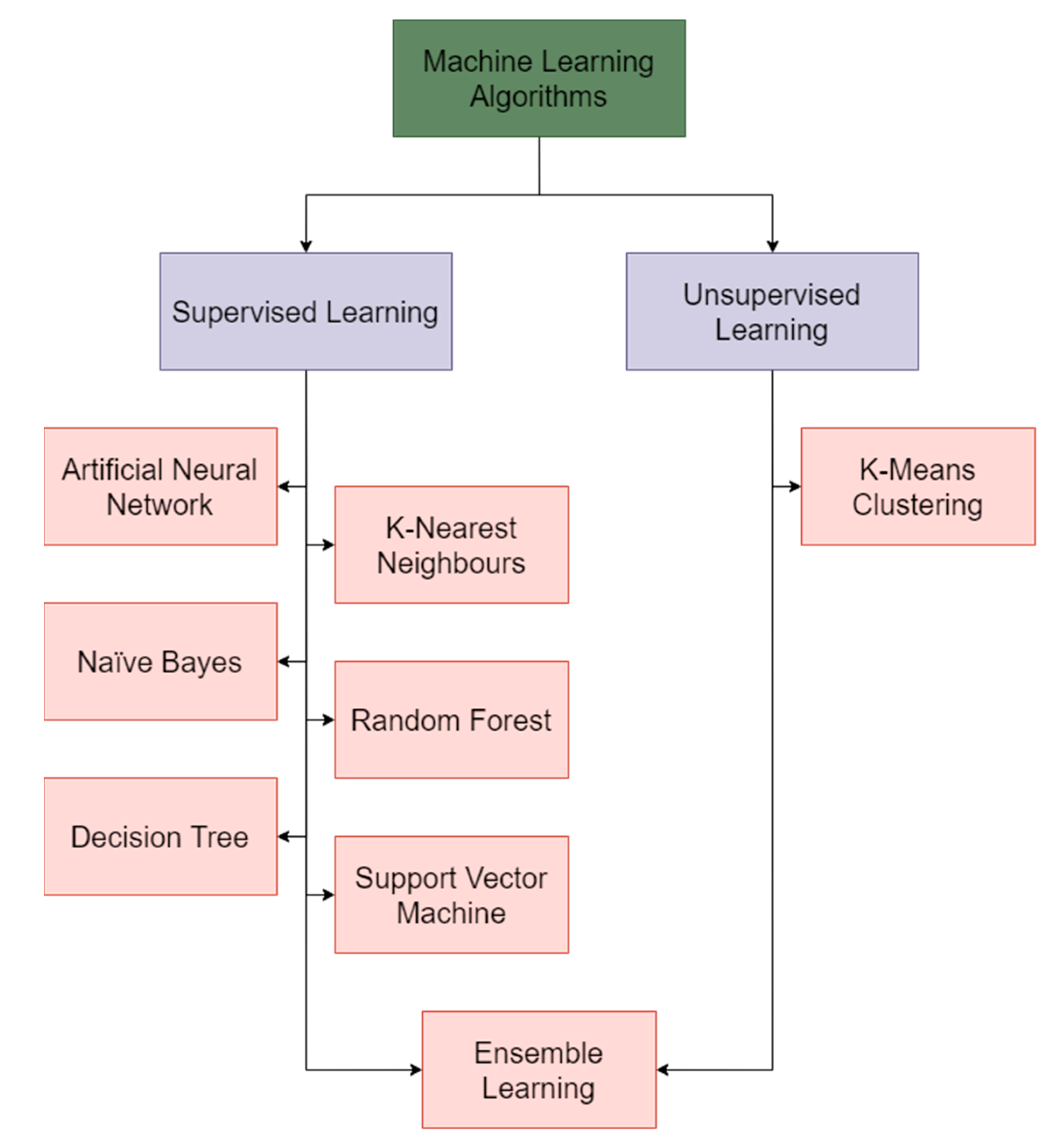
3.2. Machine Learning Techniques
3.2.1. Artificial Neural Networks
3.2.2. Naïve Bayes
3.2.3. Decision Tree
3.2.4. K-Nearest Neighbors
3.2.5. K-Means Clustering
3.2.6. Random Forest
3.2.7. Support Vector Machine
3.2.8. Ensemble Learning
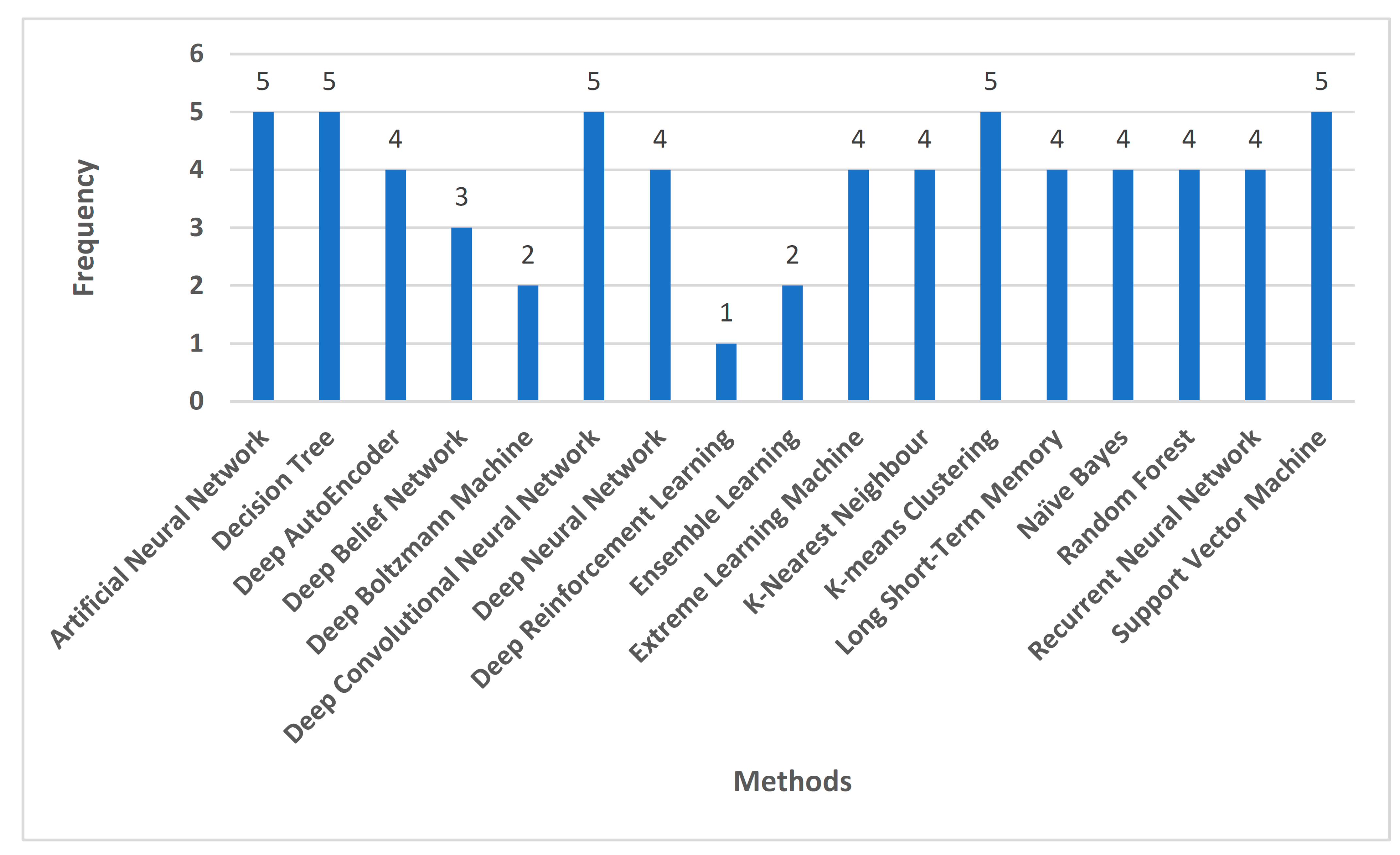
3.2.9. Summary of Machine Learning Techniques
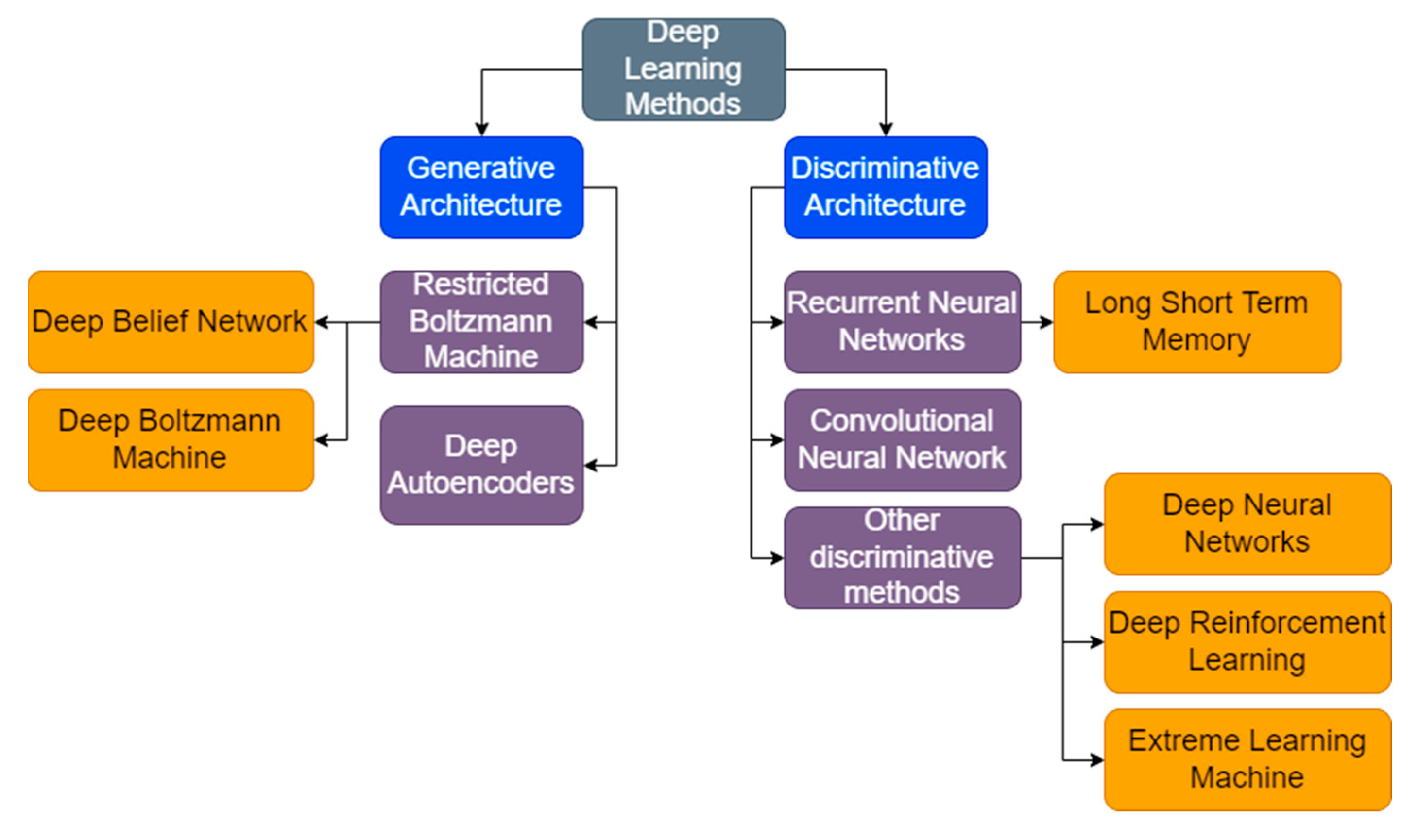
3.3. Deep Learning Techniques
3.3.1. Recurrent Neural Network
3.3.2. Deep Autoencoder
3.3.3. Long Short-Term Memory
3.3.4. Deep Neural Network
3.3.5. Deep Belief Network
3.3.6. Deep Convolutional Neural Network
3.3.7. Deep Boltzmann Machine
3.3.8. Deep Reinforcement Learning
3.3.9. Extreme Learning Machine
3.3.10. Summary of Deep Learning Models
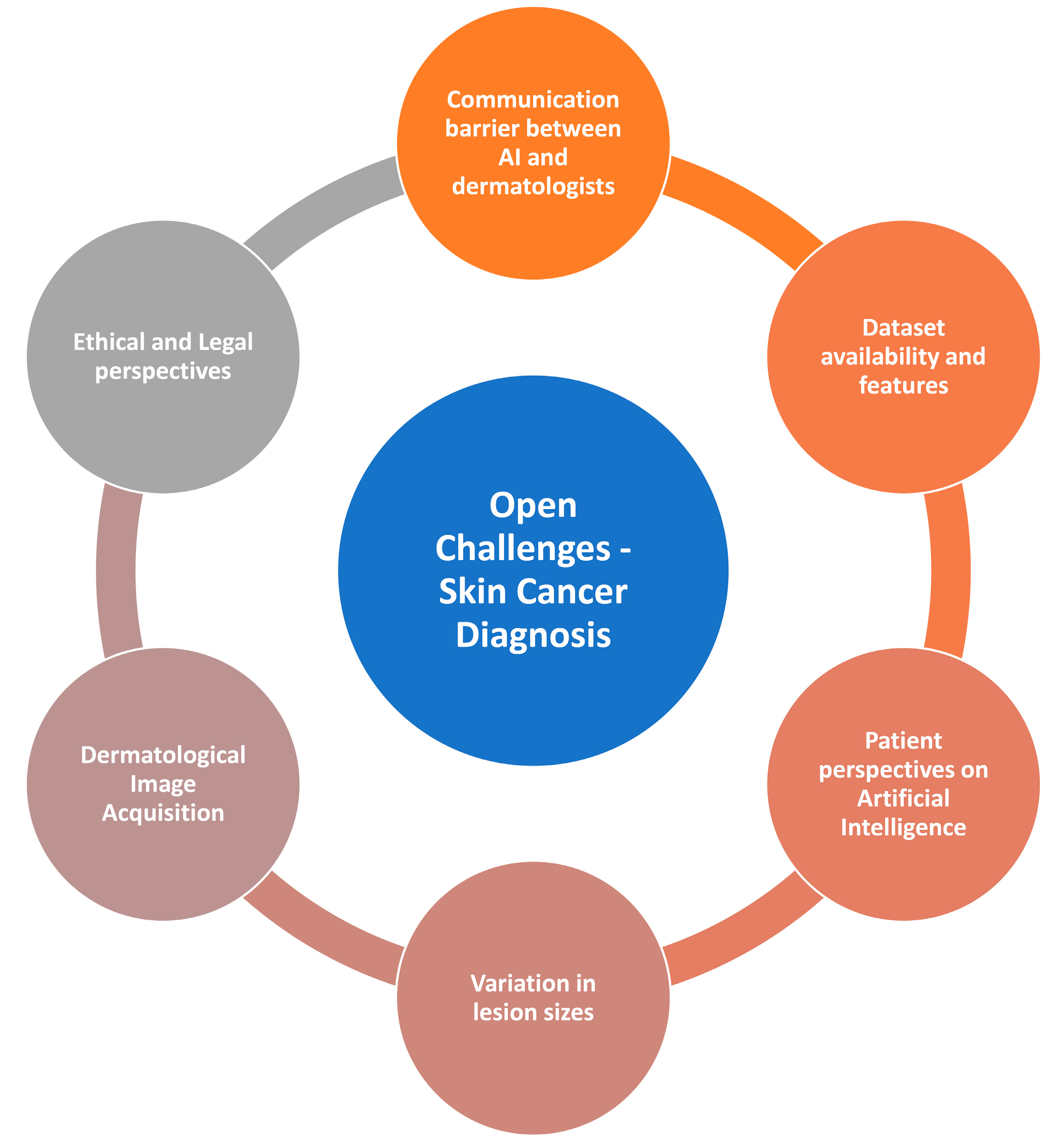
4. Open Challenges in Skin Cancer Diagnosis
4.1. Communication Barrier between AI and Dermatologists
4.2. Dataset Availability and Features
4.3. Patient Perspectives on Artificial Intelligence
4.4. Variation in Lesion Images
4.5. Dermatological Image Acquisition
4.6. Ethical and Legal Perspectives
5. Future Research Directions
5.1. Combining AI with Next-Generation Sequencing for Refining Skin Cancer Diagnosis
5.2. AI-powered Automated Decision Support Systems for Skin Cancer Diagnosis
5.3. Smart Robotics for Skin Cancer Diagnosis
5.4. Wearable Computing for Skin Cancer Diagnosis
5.5. Teledermatology
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Acronym | Definition |
|---|---|
| AI | Artificial Intelligence |
| ANN | Artificial neural network |
| KNN | K-nearest neighbors |
| ABCD | Asymmetry, border, color, diameter |
| SVM | Support vector machine |
| ROC | Receiver Operating Characteristic |
| AUC | Area under curve |
| RNN | Recurrent neural network |
| DHOA | Deer hunting optimization algorithm |
| LSTM | Long short-term memory |
| DBN | Deep belief network |
| CNN | Convolutional neural network |
| DBM | Deep Boltzmann machine |
| RL | Reinforcement learning |
| ELM | Extreme learning machine |
| NGS | Next generation sequencing |
| DNA | Deoxyribonucleic acid |
| RNA | Ribonucleic acid |
| SCC | Squamous cell carcinoma |
References
- Murugan, A.; Nair, S.A.H.; Preethi, A.A.P.; Kumar, K.P.S. Diagnosis of skin cancer using machine learning techniques. Microprocess. Microsyst. 2020, 81, 103727. [Google Scholar] [CrossRef]
- Vijayalakshmi, M.M. Melanoma skin cancer detection using image processing and machine learning. Int. J. Trend Sci. Res. Dev. 2019, 3, 780–784. [Google Scholar]
- Ozkan, I.A.; Koklu, M. Skin lesion classification using machine learning algorithms. Int. J.-Telligent Syst. Appl. Eng. 2017, 5, 285–289. [Google Scholar] [CrossRef]
- Monika, M.K.; Vignesh, N.A.; Kumari, C.U.; Kumar, M.; Lydia, E.L. Skin cancer detection and classification using machine learning. Mater. Today Proc. 2020, 33, 4266–4270. [Google Scholar] [CrossRef]
- Nahata, H.; Singh, S.P. Deep learning solutions for skin cancer detection and diagnosis. In Machine Learning with Health Care Perspective; Springer: Cham, Switzerland, 2020; pp. 159–182. [Google Scholar]
- Das, K.; Cockerell, C.J.; Patil, A.; Pietkiewicz, P.; Giulini, M.; Grabbe, S.; Goldust, M. Machine Learning and Its Application in Skin Cancer. Int. J. Environ. Res. Public Health 2021, 18, 13409. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.-K.; Kaabar, M.K.A.; Martínez, F.; Junejo, A.R.; Ullah, I.; Khan, R. Deep learning in cancer diagnosis and prognosis prediction: A minireview on challenges, recent trends, and future directions. Comput. Math. Methods Med. 2021, 2021, 9025470. [Google Scholar] [CrossRef]
- Munir, K.; Elahi, H.; Ayub, A.; Frezza, F.; Rizzi, A. Cancer Diagnosis Using Deep Learning: A Bibliographic Review. Cancers 2019, 11, 1235. [Google Scholar] [CrossRef]
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial intelligence-based image classification methods for diagnosis of skin cancer: Challenges and opportunities. Comput. Biol. Med. 2020, 127, 104065. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pan, Y.; Zhao, J.; Zhang, L. Skin disease diagnosis with deep learning: A review. Neurocomputing 2021, 464, 364–393. [Google Scholar] [CrossRef]
- Shastry, K.A.; Sanjay, H.A. Cancer diagnosis using artificial intelligence: A review. Artif. Intell. Rev. 2021, 55, 2641–2673. [Google Scholar] [CrossRef]
- Painuli, D.; Bhardwaj, S.; Köse, U. Recent advancement in cancer diagnosis using machine learning and deep learning techniques: A comprehensive review. Comput. Biol. Med. 2022, 146, 105580. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Farooq, M.S.; Khelifi, A.; Abid, A. Malignant Melanoma Classification Using Deep Learning: Datasets, Performance Measurements, Challenges and Opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar] [CrossRef]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Adegun, A.; Viriri, S. Deep learning techniques for skin lesion analysis and melanoma cancer detection: A survey of state-of-the-art. Artif. Intell. Rev. 2020, 54, 811–841. [Google Scholar] [CrossRef]
- Saba, T. Recent advancement in cancer detection using machine learning: Systematic survey of decades, comparisons and challenges. J. Infect. Public Health 2020, 13, 1274–1289. [Google Scholar] [CrossRef]
- Usama, M.; Naeem, M.A.; Mirza, F. Multi-Class Skin Lesions Classification Using Deep Features. Sensors 2022, 22, 8311. [Google Scholar] [CrossRef]
- Bratchenko, I.A.; Bratchenko, L.A.; Khristoforova, Y.A.; Moryatov, A.A.; Kozlov, S.V.; Zakharov, V.P. Classification of skin cancer using convolutional neural networks analysis of Raman spectra. Comput. Methods Programs Biomed. 2022, 219, 106755. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Utikal, J.S.; Grabe, N.; Schadendorf, D.; Klode, J.; Berking, C.; Steeb, T.; Enk, A.H.; von Kalle, C. Skin Cancer Classification Using Convolutional Neural Networks: Systematic Review. J. Med. Internet Res. 2018, 20, e11936. [Google Scholar] [CrossRef]
- Bakos, R.M.; Blumetti, T.P.; Roldán-Marín, R.; Salerni, G. Noninvasive Imaging Tools in the Diagnosis and Treatment of Skin Cancers. Am. J. Clin. Dermatol. 2018, 19, 3–14. [Google Scholar] [CrossRef]
- Wakelin, S.H. Benign skin lesions. Medicine 2021, 49, 443–446. [Google Scholar] [CrossRef]
- Fujisawa, Y.; Otomo, Y.; Ogata, Y.; Nakamura, Y.; Fujita, R.; Ishitsuka, Y.; Fujimoto, M. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumor diagnosis. Br. J. Dermatol. 2019, 180, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, V.; Apalla, Z.; Sotiriou, E.; Papageorgiou, C.; Lazaridou, E.; Vakirlis, S.; Ioannides, D.; Lallas, A. The limitations of dermoscopy: False-positive and false-negative tumors. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Catalano, O.; Roldán, F.A.; Varelli, C.; Bard, R.; Corvino, A.; Wortsman, X. Skin cancer: Findings and role of high-resolution ultrasound. J. Ultrasound 2019, 22, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Jinnai, S.; Yamazaki, N.; Hirano, Y.; Sugawara, Y.; Ohe, Y.; Hamamoto, R. The development of a skin cancer classi-fication system for pigmented skin lesions using deep learning. Biomolecules 2020, 10, 1123. [Google Scholar] [CrossRef] [PubMed]
- Ghazal, T.M.; Hussain, S.; Khan, M.F.; Said, R.A.T.; Ahmad, M. Detection of Benign and Malignant Tumors in Skin Empowered with Transfer Learning. Comput. Intell. Neurosci. 2022, 2022, 4826892. [Google Scholar] [CrossRef]
- Giavina-Bianchi, M.; Cordioli, E.; Dos Santos, A.P. Accuracy of Deep Neural Network in Triaging Common Skin Diseases of Primary Care Attention. Front Med. 2021, 8, 670300. [Google Scholar] [CrossRef]
- Korhonen, N.; Ylitalo, L.; Luukkaala, T.; Itkonen, J.; Häihälä, H.; Jernman, J.; Snellman, E.; Palve, J. Premalignant lesions, basal cell carcinoma and melanoma in patients with cutaneous squamous cell carcinoma. Arch. Dermatol. Res. 2020, 313, 879–884. [Google Scholar] [CrossRef]
- Nauta, M.; Walsh, R.; Dubowski, A.; Seifert, C. Uncovering and Correcting Shortcut Learning in Machine Learning Models for Skin Cancer Diagnosis. Diagnostics 2021, 12, 40. [Google Scholar] [CrossRef]
- Chan, S.; Reddy, V.; Myers, B.; Thibodeaux, Q.; Brownstone, N.; Liao, W. Machine Learning in Dermatology: Current Applications, Opportunities, and Limitations. Dermatol. Ther. 2020, 10, 365–386. [Google Scholar] [CrossRef]
- Zhang, N.; Cai, Y.-X.; Wang, Y.-Y.; Tian, Y.-T.; Wang, X.-L.; Badami, B. Skin cancer diagnosis based on optimized convolutional neural network. Artif. Intell. Med. 2020, 102, 101756. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef]
- Wen, D.; Khan, S.M.; Xu, A.J.; Ibrahim, H.; Smith, L.; Caballero, J.; Zepeda, L.; Perez, C.D.B.; Denniston, A.K.; Liu, X.; et al. Characteristics of publicly available skin cancer image datasets: A systematic review. Lancet Digit. Health 2021, 4, e64–e74. [Google Scholar] [CrossRef] [PubMed]
- Veta, M.; Heng, Y.J.; Stathonikos, N.; Bejnordi, B.E.; Beca, F.; Wollmann, T.; Rohr, K.; Shah, M.A.; Wang, D.; Rousson, M.; et al. Predicting breast tumor proliferation from whole-slide images: The TUPAC16 challenge. Med. Image Anal. 2019, 54, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.N.; Shwe, S.; Mesinkovska, N.A. Current state of machine learning for non-melanoma skin cancer. Arch. Dermatol. Res. 2022, 314, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Murphree, D.H.; Puri, P.; Shamim, H.; Bezalel, S.A.; Drage, L.A.; Wang, M.; Comfere, N. Deep learning for dermatologists: Part I. Fundamental concepts. J. Am. Acad. Dermatol. 2020, 87, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Roffman, D.; Hart, G.; Girardi, M.; Ko, C.J.; Deng, J. Predicting non-melanoma skin cancer via a multi-parameterized artificial neural network. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Sugiarti, S.; Yuhandri, Y.; Na’am, J.; Indra, D.; Santony, J. An artificial neural network approach for detecting skin cancer. Telecommun. Comput. Electron. Control. 2019, 17, 788–793. [Google Scholar] [CrossRef]
- Lopez-Leyva, J.A.; Guerra-Rosas, E.; Alvarez-Borrego, J. Multi-Class Diagnosis of Skin Lesions Using the Fourier Spectral Information of Images on Additive Color Model by Artificial Neural Network. IEEE Access 2021, 9, 35207–35216. [Google Scholar] [CrossRef]
- Xuyi, W.; Seow, H.; Sutradhar, R. Artificial neural networks for simultaneously predicting the risk of multiple co-occurring symptoms among patients with cancer. Cancer Med. 2020, 10, 989–998. [Google Scholar] [CrossRef]
- Sutradhar, R.; Barbera, L. Comparing an Artificial Neural Network to Logistic Regression for Predicting ED Visit Risk Among Patients with Cancer: A Population-Based Cohort Study. J. Pain Symptom Manag. 2020, 60, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alwan, O.F. Skin cancer images classification using naïve bayes. Emergent J. Educ. Discov. Lifelong Learn. 2022, 3, 19–29. [Google Scholar]
- Balaji, V.R.; Suganthi, S.T.; Rajadevi, R.; Kumar, V.K.; Balaji, B.S.; Pandiyan, S. Skin disease detection and seg-mentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement 2020, 163, 107922. [Google Scholar] [CrossRef]
- Mobiny, A.; Singh, A.; Van Nguyen, H. Risk-Aware Machine Learning Classifier for Skin Lesion Diagnosis. J. Clin. Med. 2019, 8, 1241. [Google Scholar] [CrossRef]
- Browning, A.P.; Haridas, P.; Simpson, M.J. A Bayesian Sequential Learning Framework to Parameterise Continuum Models of Melanoma Invasion into Human Skin. Bull. Math. Biol. 2018, 81, 676–698. [Google Scholar] [CrossRef]
- Tanaka, T.; Voigt, M.D. Decision tree analysis to stratify risk of de novo non-melanoma skin cancer following liver transplantation. J. Cancer Res. Clin. Oncol. 2018, 144, 607–615. [Google Scholar] [CrossRef]
- Sun, J.; Huang, Y. Computer aided intelligent medical system and nursing of breast surgery infection. Microprocess. Microsyst. 2020, 81, 103769. [Google Scholar] [CrossRef]
- Quinn, P.L.; Oliver, J.B.; Mahmoud, O.M.; Chokshi, R.J. Cost-Effectiveness of Sentinel Lymph Node Biopsy for Head and Neck Cutaneous Squamous Cell Carcinoma. J. Surg. Res. 2019, 241, 15–23. [Google Scholar] [CrossRef]
- Saba, T.; Khan, M.A.; Rehman, A.; Marie-Sainte, S.L. Region Extraction and Classification of Skin Cancer: A Het-erogeneous framework of Deep CNN Features Fusion and Reduction. J. Med. Syst. 2019, 43, 289. [Google Scholar] [CrossRef]
- Ghiasi, M.M.; Zendehboudi, S. Application of decision tree-based ensemble learning in the classification of breast cancer. Comput. Biol. Med. 2021, 128, 104089. [Google Scholar] [CrossRef]
- Alkhushayni, S.; Al-Zaleq, D.; Andradi, L.; Flynn, P. The Application of Differing Machine Learning Algorithms and Their Related Performance in Detecting Skin Cancers and Melanomas. J. Ski. Cancer 2022, 2022, 2839162. [Google Scholar] [CrossRef]
- Ak, M.F. A Comparative Analysis of Breast Cancer Detection and Diagnosis Using Data Visualization and Machine Learning Applications. Healthcare 2020, 8, 111. [Google Scholar] [CrossRef]
- Sivaraj, S.; Malmathanraj, R.; Palanisamy, P. Detecting anomalous growth of skin lesion using threshold-based segmentation algorithm and Fuzzy K-Nearest Neighbor classifier. J. Cancer Res. Ther. 2020, 16, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Oukil, S.; Kasmi, R.; Mokrani, K.; García-Zapirain, B. Automatic segmentation and melanoma detection based on color and texture features in dermoscopic images. Ski. Res. Technol. 2021, 28, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Mehmood, Z.; Nazir, T.; Naqvi, R.A.; Rehman, A.; Iqbal, M.; Saba, T. Skin cancer detection from der-moscopic images using deep learning and fuzzy k-means clustering. Microsc. Res. Tech. 2022, 85, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Anas, M.; Gupta, K.; Ahmad, S. Skin cancer classification using K-means clustering. Int. J. Tech. Res. Appl. 2017, 5, 62–65. [Google Scholar]
- Hossain, M.S.; Muhammad, G.; Alhamid, M.F.; Song, B.; Al-Mutib, K. Audio-Visual Emotion Recognition Using Big Data Towards 5G. Mob. Networks Appl. 2016, 21, 753–763. [Google Scholar] [CrossRef]
- Khan, M.Q.; Hussain, A.; Rehman, S.U.; Khan, U.; Maqsood, M.; Mehmood, K.; Khan, M.A. Classification of Melanoma and Nevus in Digital Images for Diagnosis of Skin Cancer; IEEE: Washington, DC, USA, 2019; Volume 7, pp. 90132–90144. [Google Scholar]
- Janney, B.; Roslin, E. Analysis of Skin Cancer using K-Means Clustering and Hybrid Classification Model. Indian J. Public Health Res. Dev. 2019, 10, 1371–1378. [Google Scholar] [CrossRef]
- Murugan, A.; Nair, S.H.; Kumar, K.P.S. Detection of Skin Cancer Using SVM, Random Forest and kNN Classifiers. J. Med. Syst. 2019, 43, 269. [Google Scholar] [CrossRef]
- Luu, N.T.; Le, T.-H.; Phan, Q.-H.; Pham, T.-T. Characterization of Mueller matrix elements for classifying human skin cancer utilizing random forest algorithm. J. Biomed. Opt. 2021, 26, 075001. [Google Scholar] [CrossRef] [PubMed]
- Nandhini, S.; Sofiyan, M.A.; Kumar, S.; Afridi, A. Skin cancer classification using random forest. Int. J. Manag. Humanit. 2019, 4, 39–42. [Google Scholar] [CrossRef]
- Dhivyaa, C.R.; Sangeetha, K.; Balamurugan, M.; Amaran, S.; Vetriselvi, T.; Johnpaul, P. Skin lesion classification using decision trees and random forest algorithms. J. Ambient. Intell. Humaniz. Comput. 2020, 1–13. [Google Scholar] [CrossRef]
- Melbin, K.; Raj, Y. Integration of modified ABCD features and support vector machine for skin lesion types classi-fication. Multimed. Tools Appl. 2021, 80, 8909–8929. [Google Scholar] [CrossRef]
- Alsaeed, A.A.D. On the development of a skin cancer computer aided diagnosis system using support vector machine. Biosci. Biotechnol. Res. Commun. 2019, 12, 297–308. [Google Scholar]
- Neela, A.G. Implementation of support vector machine for identification of skin cancer. Int. J. Eng. Manuf. 2019, 9, 42–52. [Google Scholar]
- Arora, G.; Dubey, A.K.; Jaffery, Z.A.; Rocha, A. Bag of feature and support vector machine based early diagnosis of skin cancer. Neural Comput. Appl. 2020, 34, 8385–8392. [Google Scholar] [CrossRef]
- Poovizhi, S.; Tr, G.B. An Efficient Skin Cancer Diagnostic System Using Bendlet Transform and Support Vector Machine. An. Acad. Bras. Ciências 2020, 92. [Google Scholar] [CrossRef]
- Schaefer, G.; Krawczyk, B.; Celebi, M.E.; Iyatomi, H. An ensemble classification approach for melanoma diagnosis. Memetic Comput. 2014, 6, 233–240. [Google Scholar] [CrossRef]
- Rahman, Z.; Hossain, M.S.; Islam, M.R.; Hasan, M.M.; Hridhee, R.A. An approach for multiclass skin lesion clas-sification based on ensemble learning. Inform. Med. Unlocked 2021, 25, 100659. [Google Scholar] [CrossRef]
- Divya, D.; Ganeshbabu, T.R. Fitness adaptive deer hunting-based region growing and recurrent neural network for melanoma skin cancer detection. Int. J. Imaging Syst. Technol. 2020, 30, 731–752. [Google Scholar] [CrossRef]
- Ahmad, B.; Usama, M.; Ahmad, T.; Khatoon, S.; Alam, C.M. An ensemble model of convolution and recurrent neural network for skin disease classification. Int. J. Imaging Syst. Technol. 2021, 32, 218–229. [Google Scholar] [CrossRef]
- Patil, R.S.; Biradar, N. Automated mammogram breast cancer detection using the optimized combination of con-volutional and recurrent neural network. Evol. Intell. 2021, 14, 1459–1474. [Google Scholar] [CrossRef]
- Alom, M.Z.; Aspiras, T.; Taha, T.M.; Asari, V.K. Skin cancer segmentation and classification with NABLA-N and inception recurrent residual convolutional networks. arXiv 2019, arXiv:1904.11126. [Google Scholar]
- Toğaçar, M.; Cömert, Z.; Ergen, B. Intelligent skin cancer detection applying autoencoder, MobileNetV2 and spiking neural networks. Chaos Solitons Fractals 2021, 144, 110714. [Google Scholar] [CrossRef]
- Diame, Z.E.; ElBery, M.; Salem MA, M.; Roushdy, M.I. Experimental Comparative Study on Autoencoder Per-formance for Aided Melanoma Skin Disease Recognition. Int. J. Intell. Comput. Inf. Sci. 2022, 22, 88–97. [Google Scholar]
- Majji, R.; Prakash, P.G.O.; Cristin, R.; Parthasarathy, G. Social bat optimisation dependent deep stacked auto-encoder for skin cancer detection. IET Image Process. 2020, 14, 4122–4131. [Google Scholar] [CrossRef]
- Diame, Z.E.; Al-Berry, M.N.; Salem, M.A.-M.; Roushdy, M. Autoencoder Performance Analysis of Skin Lesion Detection. J. Southwest Jiaotong Univ. 2021, 56, 937–947. [Google Scholar] [CrossRef]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Wu, X.; Wang, H.-Y.; Shi, P.; Sun, R.; Wang, X.; Luo, Z.; Zeng, F.; Lebowitz, M.S.; Lin, W.-Y.; Lu, J.-J.; et al. Long short-term memory model—A deep learning approach for medical data with irregularity in cancer predication with tumor markers. Comput. Biol. Med. 2022, 144, 105362. [Google Scholar] [CrossRef]
- Elashiri, M.A.; Rajesh, A.; Pandey, S.N.; Shukla, S.K.; Urooj, S.; Lay-Ekuakille, A. Ensemble of weighted deep concatenated features for the skin disease classification model using modified long short term memory. Biomed. Signal Process. Control. 2022, 76, 103729. [Google Scholar] [CrossRef]
- Liao, J.; Liu, L.; Duan, H.; Huang, Y.; Zhou, L.; Chen, L.; Wang, C. Using a Convolutional Neural Network and Convolutional Long Short-term Memory to Automatically Detect Aneurysms on 2D Digital Subtraction Angiography Images: Framework Development and Validation. JMIR Public Health Surveill. 2022, 10, e28880. [Google Scholar] [CrossRef] [PubMed]
- Mazoure, B.; Mazoure, A.; Bédard, J.; Makarenkov, V. DUNEScan: A web server for uncertainty estimation in skin cancer detection with deep neural networks. Sci. Rep. 2022, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Khan, M.A.; Sharif, M.; Akram, T.; Kadry, S.; Hsu, C.-H. A two-stream deep neural network-based intelligent system for complex skin cancer types classification. Int. J. Intell. Syst. 2022, 37, 10621–10649. [Google Scholar] [CrossRef]
- Han, S.S.; Park, I.; Chang, S.E.; Lim, W.; Kim, M.S.; Park, G.H.; Chae, J.B.; Huh, C.H.; Na, J.-I. Augmented Intelligence Dermatology: Deep Neural Networks Empower Medical Professionals in Diagnosing Skin Cancer and Predicting Treatment Options for 134 Skin Disorders. J. Investig. Dermatol. 2020, 140, 1753–1761. [Google Scholar] [CrossRef]
- Wan, J.-J.; Chen, B.-L.; Kong, Y.-X.; Ma, X.-G.; Yu, Y.-T. An Early Intestinal Cancer Prediction Algorithm Based on Deep Belief Network. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Al-Antari, M.A.; Al-Masni, M.; Park, S.-U.; Park, J.; Metwally, M.K.; Kadah, Y.M.; Han, S.-M.; Kim, T.-S. An Automatic Computer-Aided Diagnosis System for Breast Cancer in Digital Mammograms via Deep Belief Network. J. Med. Biol. Eng. 2017, 38, 443–456. [Google Scholar] [CrossRef]
- Farhi, L.; Kazmi, S.M.; Imam, H.; Alqahtani, M.; Rehman, F.U. Dermoscopic Image Classification Using Deep Belief Learning Network Architecture. Wirel. Commun. Mob. Comput. 2022, 2022, 2415726. [Google Scholar] [CrossRef]
- Dorj, U.-O.; Lee, K.-K.; Choi, J.-Y.; Lee, M. The skin cancer classification using deep convolutional neural network. Multimedia Tools Appl. 2018, 77, 9909–9924. [Google Scholar] [CrossRef]
- Refianti, R.; Mutiara, A.B.; Priyandini, R.P. Classification of melanoma skin cancer using convolutional neural network. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 409–417. [Google Scholar] [CrossRef]
- Höhn, J.; Hekler, A.; Krieghoff-Henning, E.; Kather, J.N.; Utikal, J.S.; Meier, F.; Brinker, T.J. Integrating patient data into skin cancer classification using convolutional neural networks: Systematic review. J. Med. Internet Res. 2021, 23, e20708. [Google Scholar] [CrossRef]
- Han, S.S.; Moon, I.J.; Lim, W.; Suh, I.S.; Lee, S.Y.; Na, J.-I.; Kim, S.H.; Chang, S.E. Keratinocytic Skin Cancer Detection on the Face Using Region-Based Convolutional Neural Network. JAMA Dermatol. 2020, 156, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.S.; Tembhurne, J.V.; Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimedia Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Li, Y.; Fauteux, F.; Zou, J.; Nantel, A.; Pan, Y. Personalized prediction of genes with tumor-causing somatic mutations based on multi-modal deep Boltzmann machine. Neurocomputing 2019, 324, 51–62. [Google Scholar] [CrossRef]
- Peter Soosai Anandaraj, A.; Gomathy, V.; Amali Angel Punitha, A.; Abitha Kumari, D.; Sheeba Rani, S.; Sureshkumar, S. Internet of Medical Things (IoMT) Enabled Skin Lesion Detection and Classification Using Optimal Segmentation and Restricted Boltzmann Machines. In Cognitive Internet of Medical Things for Smart Healthcare; Springer: Cham, Switzerland, 2021; pp. 195–209. [Google Scholar]
- Usmani, U.A.; Watada, J.; Jaafar, J.; Aziz, I.A.; Roy, A. A Reinforcement Learning Algorithm for Automated Detection of Skin Lesions. Appl. Sci. 2021, 11, 9367. [Google Scholar] [CrossRef]
- Wang, S.; Hamian, M. Skin Cancer Detection Based on Extreme Learning Machine and a Developed Version of Thermal Exchange Optimization. Comput. Intell. Neurosci. 2021, 2021, 9528664. [Google Scholar] [CrossRef] [PubMed]
- Sayed, G.I.; Soliman, M.M.; Hassanien, A.E. A novel melanoma prediction model for imbalanced data using optimized SqueezeNet by bald eagle search optimization. Comput. Biol. Med. 2021, 136, 104712. [Google Scholar] [CrossRef]
- Afza, F.; Sharif, M.; Khan, M.A.; Tariq, U.; Yong, H.-S.; Cha, J. Multiclass Skin Lesion Classification Using Hybrid Deep Features Selection and Extreme Learning Machine. Sensors 2022, 22, 799. [Google Scholar] [CrossRef]
- Khan, I.U.; Aslam, N.; Anwar, T.; Aljameel, S.S.; Ullah, M.; Khan, R.; Rehman, A.; Akhtar, N. Remote Diagnosis and Triaging Model for Skin Cancer Using EfficientNet and Extreme Gradient Boosting. Complexity 2021, 2021, 5591614. [Google Scholar] [CrossRef]
- Alabdulkareem, A. Artificial intelligence and dermatologists: Friends or foes? J. Dermatol. Dermatol. Surg. 2019, 23, 57. [Google Scholar] [CrossRef]
- Shen, C.; Li, C.; Xu, F.; Wang, Z.; Shen, X.; Gao, J.; Ko, R.; Jing, Y.; Tang, X.; Yu, R.; et al. Web-based study on Chinese dermatologists’ attitudes towards artificial intelligence. Ann. Transl. Med. 2020, 8, 698. [Google Scholar] [CrossRef] [PubMed]
- Maron, R.C.; Utikal, J.S.; Hekler, A.; Hauschild, A.; Sattler, E.; Sondermann, W.; Haferkamp, S.; Schilling, B.; Heppt, M.V.; Jansen, P.; et al. Artificial Intelligence and Its Effect on Dermatologists’ Accuracy in Dermoscopic Melanoma Image Classification: Web-Based Survey Study. J. Med. Internet Res. 2020, 22, e18091. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.A.; Pérez-Chada, L.M.; Creadore, A.; Li, S.J.; Lo, K.; Manjaly, P.; Mostaghimi, A. Patient perspectives on the use of artificial intelligence for skin cancer screening: A qualitative study. JAMA Dermatol. 2020, 156, 501–512. [Google Scholar] [CrossRef]
- Sreelatha, T.; Subramanyam, M.V.; Prasad, M.N.G. A Survey work on Early Detection methods of Melanoma Skin Cancer. Res. J. Pharm. Technol. 2019, 12, 2589. [Google Scholar] [CrossRef]
- di Ruffano, L.F.; Dinnes, J.; Deeks, J.J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Takwoingi, Y.; Godfrey, K.; O’Sullivan, C.; Matin, R.N.; et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 12, CD013189. [Google Scholar] [CrossRef]
- Tan, T.Y.; Zhang, L.; Lim, C.P. Intelligent skin cancer diagnosis using improved particle swarm optimization and deep learning models. Appl. Soft Comput. 2019, 84, 105725. [Google Scholar] [CrossRef]
- Gerke, S.; Minssen, T.; Cohen, G. Ethical and legal challenges of artificial intelligence-driven healthcare. In Artificial Intelligence in Healthcare; Academic Press: Cambridge, MA, USA, 2020; pp. 295–336. [Google Scholar]
- Rigby, M.J. Ethical Dimensions of Using Artificial Intelligence in Health Care. AMA J. Ethic- 2019, 21, E121–E124. [Google Scholar] [CrossRef]
- Da Silva, M.; Horsley, T.; Singh, D.; Da Silva, E.; Ly, V.; Thomas, B.; Daniel, R.C.; Chagal-Feferkorn, K.A.; Iantomasi, S.; White, K.; et al. Legal concerns in health-related artificial intelligence: A scoping review protocol. Syst. Rev. 2022, 11, 1–8. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef] [PubMed]
- Lobl, M.B.; Clarey, D.; Higgins, S.; Sutton, A.; Hansen, L.; Wysong, A. Targeted next-generation sequencing of matched localized and metastatic primary high-risk SCCs identifies driver and co-occurring mutations and novel therapeutic targets. J. Dermatol. Sci. 2020, 99, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, R.N.; Robitaille, A.; Dutta, S.; Cuenin, C.; Santare, D.; Skenders, G.; Leja, M.; Fischer, N.; Giuliano, A.R.; Rollison, D.E.; et al. Generation of a novel next-generation sequencing-based method for the isolation of new human papillomavirus types. Virology 2018, 520, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kadampur, M.A.; Al Riyaee, S. Skin cancer detection: Applying a deep learning based model driven architecture in the cloud for classifying dermal cell images. Inform. Med. Unlocked 2019, 18, 100282. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Sharif, M.; Kadry, S.; Nam, Y. Computer Decision Support System for Skin Cancer Localization and Classification. Comput. Mater. Contin. 2021, 68, 1041–1064. [Google Scholar] [CrossRef]
- Sharif, M.I.; Khan, M.A.; Alhussein, M.; Aurangzeb, K.; Raza, M. A decision support system for multimodal brain tumor classification using deep learning. Complex Intell. Syst. 2021, 8, 3007–3020. [Google Scholar] [CrossRef]
- Abdar, M.; Acharya, U.R.; Sarrafzadegan, N.; Makarenkov, V. NE-nu-SVC: A New Nested Ensemble Clinical Decision Support System for Effective Diagnosis of Coronary Artery Disease. IEEE Access 2019, 7, 167605–167620. [Google Scholar] [CrossRef]
- Ray, P.P.; Dash, D.; De, D. A Systematic Review of Wearable Systems for Cancer Detection: Current State and Challenges. J. Med. Syst. 2017, 41, 180. [Google Scholar] [CrossRef]
- Gupta, A.K.; Bharadwaj, M.; Mehrotra, R. Skin Cancer Concerns in People of Color: Risk Factors and Prevention. Asian Pac. J. Cancer Prev. 2016, 17, 5257–5264. [Google Scholar] [CrossRef]
- Sun, M.D.; Kentley, J.; Wilson, B.W.; Soyer, H.P.; Curiel-Lewandrowski, C.N.; Rotemberg, V.; ISIC Technique Working Group. Digital skin imaging applications, part I: Assessment of image acquisition technique features. Ski. Res. Technol. 2022, 28, 623–632. [Google Scholar] [CrossRef]
- Barata, C.; Marques, J.S.; Celebi, M.E. Improving dermoscopy image analysis using color constancy. In 2014 IEEE International Conference on Image Processing (ICIP); IEEE: Washington, DC, USA, 2014; pp. 3527–3531. [Google Scholar]
- Salvi, M.; Branciforti, F.; Veronese, F.; Zavattaro, E.; Tarantino, V.; Savoia, P.; Meiburger, K.M. DermoCC-GAN: A new approach for standardizing dermatological images using generative adversarial networks. Comput. Methods Programs Biomed. 2022, 225, 107040. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.; Holman, D.M.; Maguire-Eisen, M. Ultraviolet Radiation Exposure and Its Impact on Skin Cancer Risk. Semin. Oncol. Nurs. 2016, 32, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Wolner, Z.J.; Yélamos, O.; Liopyris, K.; Rogers, T.; Marchetti, M.A.; Marghoob, A.A. Enhancing Skin Cancer Diagnosis with Dermoscopy. Dermatol. Clin. 2017, 35, 417–437. [Google Scholar] [CrossRef] [PubMed]
- A Comparison of Polarised and Nonpolarised Dermoscopy|DermNet. Available online: https://dermnetnz.org/topics/a-comparison-of-polarised-and-nonpolarised-dermoscopy (accessed on 30 January 2023).
- Hone, N.L.; Grandhi, R.; Ingraffea, A.A. Basal Cell Carcinoma on the Sole: An Easily Missed Cancer. Case Rep. Dermatol. 2016, 8, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Pala, P.; Bergler-Czop, B.S.; Gwiżdż, J. Teledermatology: Idea, benefits and risks of modern age–a systematic review based on melanoma. Adv. Dermatol. Allergol. Postępy Dermatol. I Alergol. 2020, 37, 159–167. [Google Scholar] [CrossRef]
- Veronese, F.; Branciforti, F.; Zavattaro, E.; Tarantino, V.; Romano, V.; Meiburger, K.; Salvi, M.; Seoni, S.; Savoia, P. The Role in Teledermoscopy of an Inexpensive and Easy-to-Use Smartphone Device for the Classification of Three Types of Skin Lesions Using Convolutional Neural Networks. Diagnostics 2021, 11, 451. [Google Scholar] [CrossRef]
- Skin Cancer ISIC. Available online: https://www.kaggle.com/datasets/nodoubttome/skin-cancer9-classesisic (accessed on 30 November 2022).
- Dermatology Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Dermatology?ref=datanews.io (accessed on 30 November 2022).
- The HAM10000 Dataset, a Large Collection of Multi-Source Dermatoscopic Images of Common Pigmented Skin Lesions. Available online: https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/DBW86T (accessed on 30 November 2022).
- Hussain, S.F.; Qaisar, S.M. Epileptic seizure classification using level-crossing EEG sampling and en-semble of sub-problems classifier. Expert Syst. Appl. 2022, 191, 116356. [Google Scholar] [CrossRef]
- Qaisar, S.M.; Alsharif, F.; Subasi, A.; Bensenouci, A. Appliance Identification Based on Smart Meter Data and Event-Driven Processing in the 5G Framework. Procedia Comput. Sci. 2021, 182, 103–108. [Google Scholar] [CrossRef]
- Qaisar, S.M.; Khan, S.I.; Dallet, D.; Tadeusiewicz, R.; Pławiak, P. Signal-piloted processing metaheuristic optimization and wavelet decomposition based elucidation of arrhythmia for mobile healthcare. Biocybern. Biomed. Eng. 2022, 42, 681–694. [Google Scholar] [CrossRef]


| Reference | Year | One-Phrase Summary | Machine Learning Models in Skin Cancer Diagnosis | Deep Learning in Skin Cancer Diagnosis | Open Challenges in Skin Cancer Diagnosis | Future Directions for Skin Cancer Diagnosis |
|---|---|---|---|---|---|---|
| Our review | - | A comprehensive survey on machine learning and deep learning techniques used to diagnose skin cancer | H | H | H | H |
| [11] | 2022 | A review on cancer diagnosis using Artificial Intelligence | H | H | M | N |
| [12] | 2022 | A research article on the recent advancements in cancer diagnosis using machine learning and deep learning techniques | H | H | L | M |
| [6] | 2021 | A review of machine learning and its applications in the field of skin cancer | H | L | M | H |
| [7] | 2021 | A minireview on deep learning and its use in cancer diagnosis and prognosis prediction | N | H | M | H |
| [10] | 2021 | A review on skin disease diagnosis with deep learning | N | H | N | H |
| [14] | 2021 | A review on skin cancer classification via convolution neural networks | N | M | M | N |
| [15] | 2021 | A survey on deep learning techniques for skin lesion analysis and melanoma cancer detection | N | H | M | N |
| [9] | 2020 | A review article on Artificial-Intelligence-based methods for diagnosis of skin cancer | M | M | H | N |
| [13] | 2020 | A review on malignant melanoma classification using deep learning | N | H | M | H |
| [16] | 2020 | A survey in cancer detection using machine learning | H | N | H | H |
| [8] | 2019 | A bibliographic review on cancer diagnosis using deep learning | N | H | M | N |
| Search Term | Set of Keywords |
|---|---|
| Skin | skin cancer, skin disease, skin cancer diagnosis, skin cancer detection, skin lesion |
| Cancer | cancer type, cancer diagnosis |
| Deep | deep learning, deep neural networks |
| Melanoma | melanoma skin cancer, melanoma cancer |
| Machine | machine learning |
| Machine learning techniques | artificial neural network, naïve Bayes, decision tree, k-nearest neighbors, k-means clustering, random forest, support vector machines, ensemble learning |
| Deep learning techniques | recurrent neural networks, deep autoencoders, long short-term memory, deep neural network, deep belief network, deep convolutional neural network, deep Boltzmann machine, deep reinforcement learning, extreme learning machine |
| Reference | Creator and Year of Dataset | Skin Cancer Categories | Dataset Used | Dataset Size | Type of Data | Details About the Dataset |
|---|---|---|---|---|---|---|
| [132] | International Skin Imaging Collaboration, 2020 | Actinic keratosis, basal cell carcinoma, dermatofibroma, melanoma, nevus, seborrheic keratosis, squamous cell carcinoma, vascular lesion | ISIC | 2357 images | Dermoscopic images | Contains images of malignant and benign oncological diseases. Melanoma and mole images are slightly dominant in the dataset |
| [133] | Nilsel Ilter, H. Altay Guvenir, 1998 | Melanoma and non-melanoma | DermIS, DermQuest | 72 images in DermIS and 274 images in DermQuest | Not reported | Contains lesion images. They are subject to various artifacts such as drastic shadow effect and differing illumination. |
| [134] | Tschandl, P., 2018 | Actinic keratoses and intraepithelial carcinoma, basal cell carcinoma, benign keratosis-like lesions, dermatofibroma, melanoma, melanocytic nevi, and vascular lesions | HAM10000 | 10015 images | Dermoscopic images | More than half of lesion images are validated through histopathology. Remaining images are confirmed through expert consensus or in-vivo confocal microscopy. |
| [35] | Dongtan Sacred Heart Hospital, Hallym University, and Sanggye Paik Hospital, Inje University, 2016 | Basal cell carcinoma | Hallym | 152 images | Dermoscopic images | Country of origin is South Korea and a total of 106 members participated in the creation of this dataset |
| [35] | Department of Dermatology at Asan Medical Center, 2017 | Basal cell carcinoma, squamous cell carcinoma, intraepithelial carcinoma, and melanoma | Asan Dataset | 17125 images and 1276 test images | Clinical images | While the thumbnails were available for free downloading, the full-size images required external permission and it came at a cost of US $200 or £145. |
| [34] | Mitko Veta et al., 2016 | Not reported | TUPAC 2016 Dataset | 500 training and 321 test images | Whole slide images | Images to predict tumor proliferation scores from whole slide images. |
| Reference | Skin Cancer Category | Machine Learning Model | Description of Approach Used | Dataset | Key Contribution | Limitations | Performance Evaluation Metrics and Results |
|---|---|---|---|---|---|---|---|
| [38] | Non-melanoma skin cancer | Artificial neural network | 12 neurons in each layer, inputs normalized to fall between 0 and 1, sigmoid activation function | National Health Interview Survey Dataset (NHIS 2016) | Multi-parametrized artificial neural network | Model does not include ultraviolet radiation exposure and family history data while making predictions | AUC is area under ROC curve. Training AUC—0.8058, validation AUC—0.8099 |
| [44] | Skin disease detection and segmentation | Naïve Bayes classifier | Skin lesion segmentation using a dynamic graph cut algorithm followed by a naïve bayes classifier for skin disease classification | ISIC 2017 | Flexible group minimizing for alike functions, making them decipherable in polynomial time | Cannot differentiate between certain colors | Diagnostic accuracy–72.7%, sensitivity–91.7%, specificity-70.1% |
| [47] | Non-melanoma skin cancer | Decision tree | Cox regression analysis to identify variables that enter the decision tree analysis | Oregon Procurement Transplant Network STAR 2016 | Confirms importance of known risk factors and also identifies new variables establishing risk of getting non melanoma skin cancer | Model building and validation sets were not from independent cohorts | Cumulative incidence rate highest risk group: 7.4%, intermediate risk group: 3.1–5.5%, lowest risk group: 0.8% |
| [54] | Skin lesion | K-nearest neighbor classifier | Firefly with k-nearest neighbor algorithm to predict and classify skin cancer using threshold-based segmentation | - | Recognize skin cancer without performing unnecessary skin biopsies | Image pre-processing and segmentation is heavily dependent on threshold values | False predictive value: 0.0, false negative rate: 11.11%, sensitivity: 88.89%, specificity: 100% |
| [56] | Melanoma skin cancer | K-means clustering | Region-based convolutional neural networks along with fuzzy k-means clustering. | ISIC 2016, ISIC 2017, PH2 | Fully automated skin lesion segmentation at its earliest stage | Model is heavily reliant on successful segmentation from the R-CNN stage | Sensitivity: 90%, specificity: 97.1%, accuracy: 95.4% |
| [61] | Melanoma skin Cancer | Random forest | Watershed segmentation used for feature extraction and then classified with random forest | ISIC | Section lesions on skin with increased precision | Same classification can be carried out with higher accuracy using a simple vector machine | Accuracy: 74.32%, sensitivity: 76.85%, specificity: 71.79% |
| [66] | Melanoma skin cancer | Simple vector machine | Extracted features such as texture, color, shape are inputs to the SVM classifier for skin lesion classification | University Medical Center Groningen (UMCG) database | Computer Aided Diagnosis support system for image acquisition, pre-processing, segmentation, extraction, classification, and result viewing | No support for hair removal and image cropping techniques, classification model can be improved further | Confusion matrix: [3,7,62,64], where [true positive, true negative, false positive, false negative] sensitivity: 90%, specificity: 96% |
| [71] | Multi-class skin cancer | Ensemble learning | Weighted average ensemble learning based model using 5 deep learning models | Human Against Machine (HAM10000), ISIC 2019 | Significantly improved result as compared to models individually and existing systems | Trained over a highly imbalanced dataset leading to compromised results while testing and validation | Confusion matrix, ROC-AUC score |
| Reference | Skin Cancer Category | Deep Learning Model | Description of Approach Used | Dataset | Key Contribution | Limitations | Performance Evaluation Metrics and Results |
|---|---|---|---|---|---|---|---|
| [72] | Melanoma skin cancer | Recurrent neural networks | Classification phases uses modified deep learning algorithm by coalescing optimization concepts from RNNs | PH2 | Superior to existing algorithms in terms of optimal segmentation and classification for melanoma skin cancer | Heavy dependence on parameters for segmentation and classification | Algorithmic analysis including specificity: 0.94915, sensitivity: 0.83051, precision: 0.89091, F1-score: 0.85965, etc. |
| [76] | Skin cancer detection | Autoencoders | Dataset is reconstructed using autoencoder model, reconstruction and spiking networks contribute to enhanced performance | ISIC | Feature sets obtained from convolution model are suitable for merging | Model extracts many unnecessary and irrelevant features | Specificity: 0.9332, sensitivity: 0.9372, precision: 0.9450, F1-score: 0.9411, accuracy: 0.9354 |
| [81] | Skin cancer diagnosis | Long short-term memory model | Tumor marker data values were used to train and test an LSTM model | Two independent medical centers | LSTM model demonstrates superiority while dealing with irregular data and can be used when time intervals between tests vary | Inability to analyze irregular tumor marker data for cancer screening | Time-to-cancer diagnosis in different risk groups, risk stratification |
| [87] | Binary classification, multi-class skin cancer diagnosis | Deep neural network | CNN architectures trained on large datasets and evaluated against algorithm-assisted clinicians’ results | Edinburgh and SNU datasets | Model serves as an ancillary tool to enhance diagnostic accuracy of clinicians | Outcome of algorithm is significantly affected by composition of input images; performance is sub-optimal if input image quality is low | Improvement in sensitivity and specificity by 12.1% and 1.1%, respectively |
| [88] | Malignant tumor detection | Deep belief network | Analyze patient data from deep learning perspective, merged with patient attributes and case reports to construct an expert system helping to predict the probability of early cancer | Jiangsu Provincial Hospital of Traditional Chinese Medicine | Relatively effective dimensional reduction and noise cancellation technique, reduces missed clinician diagnoses during endoscopy and treatment | Medium runtime in comparison to other deep learning methods | Accuracy: 0.8148, precision: 0.8571, recall: 0.6, F1 score: 0.7059 |
| [91] | Melanoma, carcinoma, keratosis | Deep convolutional neural network | Classifies skin cancer using ECOC SVM and deep CNN, images are cropped to reduce noise | Pretrained on ImageNet, Internet Images for fine-tuning | Multi-class skin cancer classification using fine-tuned pretrained ImageNet model | Model does not extend to ABCD (asymmetry, border, color, diameter) rule | Accuracy: 0.942, specificity: 0.9074, sensitivity: 0.9783 |
| [96] | Tumor causing somatic mutations | Deep Boltzmann machine | Multi-modal deep Boltzmann machine approach for prediction of somatic mutation genes that undergo malignant transformation, model learns relation between germline and mutation profiles using data | - | Genome-based diagnostic test to monitor for the presence of cancer-driving mutations | Sample size of is limited, Whole Exome Sequencing (WES) data displayed at gene level | Average accuracy: 0.7176, p-value |
| [99] | Melanoma skin cancer | Extreme learning machine | After pre-processing, Otsu method is employed to segment region of interest, subsequently, feature extraction is applied to mine important characteristics, deep belief network is used to categorize and classify | ISIC for training, SIIM-ISIC melanoma for validation | Optimized Pipeline feature designed for efficient detection of melanoma from images, DBN uses Thermal Exchange Optimization Algorithm as new meta-heuristic method | Computationally very intensive and time consuming | Accuracy: 0.9265, specificity: 0.8970, sensitivity: 0.9118, PPV: 0.8676, NPV: 0.9412 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melarkode, N.; Srinivasan, K.; Qaisar, S.M.; Plawiak, P. AI-Powered Diagnosis of Skin Cancer: A Contemporary Review, Open Challenges and Future Research Directions. Cancers 2023, 15, 1183. https://doi.org/10.3390/cancers15041183
Melarkode N, Srinivasan K, Qaisar SM, Plawiak P. AI-Powered Diagnosis of Skin Cancer: A Contemporary Review, Open Challenges and Future Research Directions. Cancers. 2023; 15(4):1183. https://doi.org/10.3390/cancers15041183
Chicago/Turabian StyleMelarkode, Navneet, Kathiravan Srinivasan, Saeed Mian Qaisar, and Pawel Plawiak. 2023. "AI-Powered Diagnosis of Skin Cancer: A Contemporary Review, Open Challenges and Future Research Directions" Cancers 15, no. 4: 1183. https://doi.org/10.3390/cancers15041183
APA StyleMelarkode, N., Srinivasan, K., Qaisar, S. M., & Plawiak, P. (2023). AI-Powered Diagnosis of Skin Cancer: A Contemporary Review, Open Challenges and Future Research Directions. Cancers, 15(4), 1183. https://doi.org/10.3390/cancers15041183






