The KRAS-Mutant Consensus Molecular Subtype 3 Reveals an Immunosuppressive Tumor Microenvironment in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
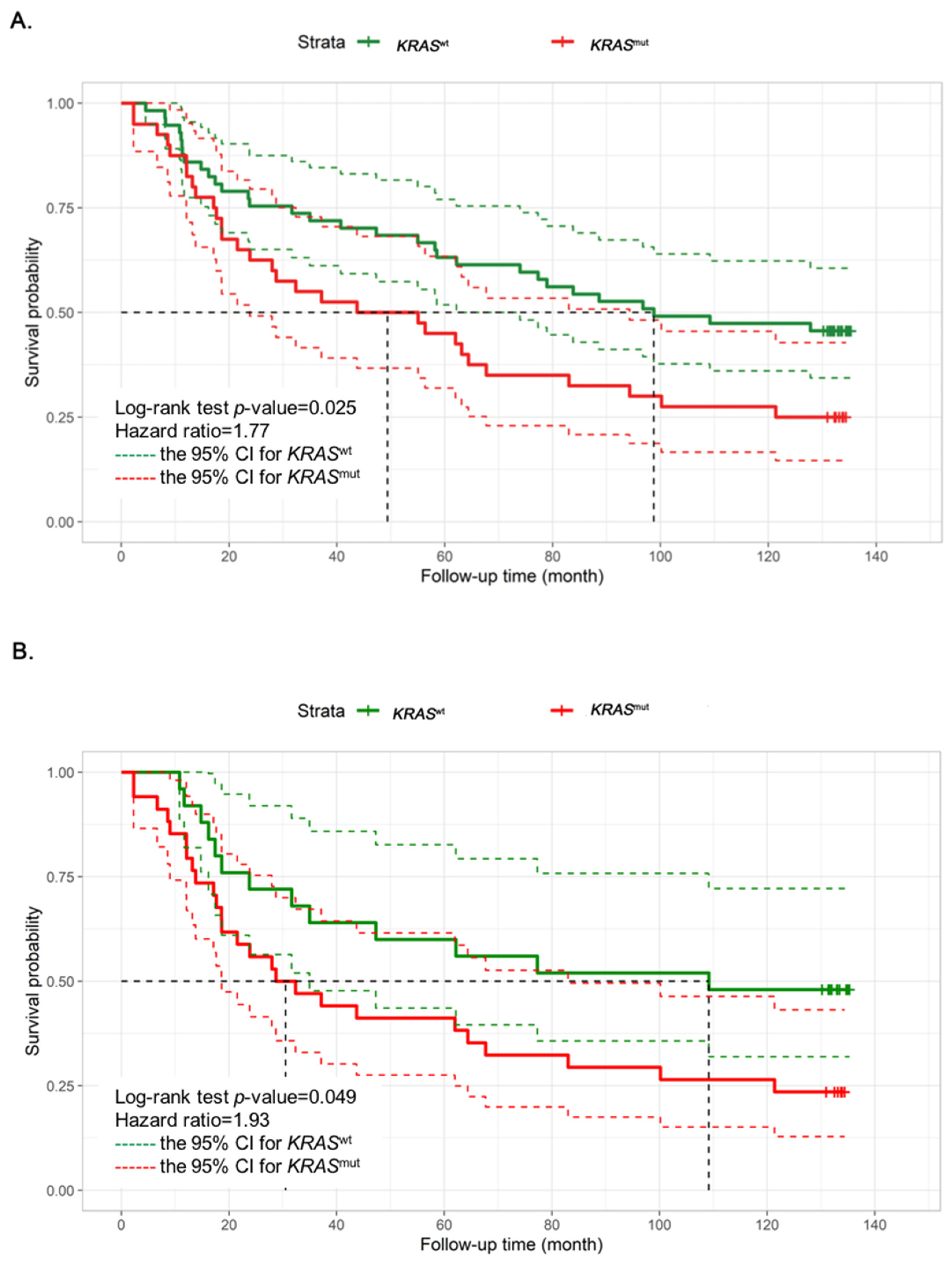
2.2. Survival, Clinical and Statistical Analysis
2.3. DNA Isolation
2.4. RNA Isolation
2.5. KRAS Mutation Screening
2.6. KRAS-Related Gene Expression Profiling Using NanoString Platform
2.7. Pathway Analysis
2.8. CMS Classification Using the NanoString Platform
2.9. Digital Spatial Profiling (DSP)
3. Results
3.1. CRC Patients with KRASmut Show a Shorter Overall Survival That Is Mainly Correlated with Their Transcriptomic Profile
3.2. DEG of KRASmut Tumors Enrich in Immune Signature and TGFβ Pathways
3.3. KRASmut CMS3 Classified Tumors Show a Distinct Immune Suppression of the Gene Expression Pattern in the TME
3.4. The upregulation of CD40 in the TME of KRASmut
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Schmitt, M.; Greten, F.R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 2021, 21, 653–667. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar]
- Phipps, A.I.; Buchanan, D.D.; Makar, K.W.; Win, A.K.; Baron, J.A.; Lindor, N.M.; Potter, J.D.; Newcomb, P.A. KRAS-mutation status in relation to colorectal cancer survival: The joint impact of correlated tumour markers. Br. J. Cancer 2013, 108, 1757–1764. [Google Scholar]
- Conlin, A.; Smith, G.; Carey, F.A.; Wolf, C.R.; Steele, R.J. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 2005, 54, 1283–1286. [Google Scholar] [CrossRef]
- Nash, G.M.; Gimbel, M.; Shia, J.; Nathanson, D.R.; Ndubuisi, M.I.; Zeng, Z.S.; Kemeny, N.; Paty, P.B. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann. Surg. Oncol. 2010, 17, 572–578. [Google Scholar]
- Arrington, A.K.; Heinrich, E.L.; Lee, W.; Duldulao, M.; Patel, S.; Sanchez, J.; Garcia-Aguilar, J.; Kim, J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int. J. Mol. Sci. 2012, 13, 12153–12168. [Google Scholar]
- Van Krieken, J.H.; Jung, A.; Kirchner, T.; Carneiro, F.; Seruca, R.; Bosman, F.T.; Quirke, P.; Flejou, J.F.; Plato Hansen, T.; de Hertogh, G.; et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: Proposal for an European quality assurance program. Virchows Arch. 2008, 453, 417–431. [Google Scholar]
- Ryan, M.B.; Coker, O.; Sorokin, A.; Fella, K.; Barnes, H.; Wong, E.; Kanikarla, P.; Gao, F.; Zhang, Y.; Zhou, L.; et al. KRAS(G12C)-independent feedback activation of wild-type RAS constrains KRAS(G12C) inhibitor efficacy. Cell Rep. 2022, 39, 110993. [Google Scholar]
- Hamarsheh, S.; Gross, O.; Brummer, T.; Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 2020, 11, 5439. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar]
- Lal, N.; White, B.S.; Goussous, G.; Pickles, O.; Mason, M.J.; Beggs, A.D.; Taniere, P.; Willcox, B.E.; Guinney, J.; Middleton, G.W. KRAS Mutation and Consensus Molecular Subtypes 2 and 3 Are Independently Associated with Reduced Immune Infiltration and Reactivity in Colorectal Cancer. Clin. Cancer Res. 2018, 24, 224–233. [Google Scholar] [CrossRef]
- Lal, N.; Beggs, A.D.; Willcox, B.E.; Middleton, G.W. An immunogenomic stratification of colorectal cancer: Implications for development of targeted immunotherapy. Oncoimmunology 2015, 4, e976052. [Google Scholar] [CrossRef]
- Becht, E.; de Reynies, A.; Giraldo, N.A.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautes-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066. [Google Scholar] [CrossRef]
- Boutin, A.T.; Liao, W.T.; Wang, M.; Hwang, S.S.; Karpinets, T.V.; Cheung, H.; Chu, G.C.; Jiang, S.; Hu, J.; Chang, K.; et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017, 31, 370–382. [Google Scholar]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Canellas, A.; Hernando-Momblona, X.; et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Zhang, B.; Halder, S.K.; Kashikar, N.D.; Cho, Y.J.; Datta, A.; Gorden, D.L.; Datta, P.K. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology 2010, 138, 969–980 e961–963. [Google Scholar]
- Inamoto, S.; Itatani, Y.; Yamamoto, T.; Minamiguchi, S.; Hirai, H.; Iwamoto, M.; Hasegawa, S.; Taketo, M.M.; Sakai, Y.; Kawada, K. Loss of SMAD4 Promotes Colorectal Cancer Progression by Accumulation of Myeloid-Derived Suppressor Cells through the CCL15-CCR1 Chemokine Axis. Clin. Cancer Res. 2016, 22, 492–501. [Google Scholar] [CrossRef]
- Gonzalez, D.M.; Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014, 7, re8. [Google Scholar] [CrossRef]
- Kim, S.T.; Park, K.H.; Shin, S.W.; Kim, Y.H. Dose KRAS Mutation Status Affect on the Effect of VEGF Therapy in Metastatic Colon Cancer Patients? Cancer Res. Treat. 2014, 46, 48–54. [Google Scholar]
- De Cuba, E.M.; Snaebjornsson, P.; Heideman, D.A.; van Grieken, N.C.; Bosch, L.J.; Fijneman, R.J.; Belt, E.; Bril, H.; Stockmann, H.B.; Hooijberg, E.; et al. Prognostic value of BRAF and KRAS mutation status in stage II and III microsatellite instable colon cancers. Int. J. Cancer 2016, 138, 1139–1145. [Google Scholar]
- Soldevilla, B.; Carretero-Puche, C.; Gomez-Lopez, G.; Al-Shahrour, F.; Riesco, M.C.; Gil-Calderon, B.; Alvarez-Vallina, L.; Espinosa-Olarte, P.; Gomez-Esteves, G.; Rubio-Cuesta, B.; et al. The correlation between immune subtypes and consensus molecular subtypes in colorectal cancer identifies novel tumour microenvironment profiles, with prognostic and therapeutic implications. Eur. J. Cancer 2019, 123, 118–129. [Google Scholar]
- Ge, P.; Wang, W.; Li, L.; Zhang, G.; Gao, Z.; Tang, Z.; Dang, X.; Wu, Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed. Pharmacother. 2019, 118, 109228. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar]
- Gao, F.; Wang, W.; Tan, M.; Zhu, L.; Zhang, Y.; Fessler, E.; Vermeulen, L.; Wang, X. DeepCC: A novel deep learning-based framework for cancer molecular subtype classification. Oncogenesis 2019, 8, 44. [Google Scholar] [CrossRef]
- Bourn, J.R.; Ruiz-Torres, S.J.; Hunt, B.G.; Benight, N.M.; Waltz, S.E. Tumor cell intrinsic RON signaling suppresses innate immune responses in breast cancer through inhibition of IRAK4 signaling. Cancer Lett. 2021, 503, 75–90. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Hirata, E.; Sahai, E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026781. [Google Scholar] [CrossRef]
- Yang, L.; Pang, Y.; Moses, H.L. TGF-beta and immune cells: An important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010, 31, 220–227. [Google Scholar]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-beta Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef]
- Yamada, N.; Kuranaga, Y.; Kumazaki, M.; Shinohara, H.; Taniguchi, K.; Akao, Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-beta1-mediated suppression. Oncotarget 2016, 7, 27033–27043. [Google Scholar]
- Kitamura, T.; Kometani, K.; Hashida, H.; Matsunaga, A.; Miyoshi, H.; Hosogi, H.; Aoki, M.; Oshima, M.; Hattori, M.; Takabayashi, A.; et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat. Genet. 2007, 39, 467–475. [Google Scholar]
- Zhu, Z.; Aref, A.R.; Cohoon, T.J.; Barbie, T.U.; Imamura, Y.; Yang, S.; Moody, S.E.; Shen, R.R.; Schinzel, A.C.; Thai, T.C.; et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014, 4, 452–465. [Google Scholar] [CrossRef]
- Hawinkels, L.J.; Paauwe, M.; Verspaget, H.W.; Wiercinska, E.; van der Zon, J.M.; van der Ploeg, K.; Koelink, P.J.; Lindeman, J.H.; Mesker, W.; ten Dijke, P.; et al. Interaction with colon cancer cells hyperactivates TGF-beta signaling in cancer-associated fibroblasts. Oncogene 2014, 33, 97–107. [Google Scholar] [CrossRef]
- Calon, A.; Espinet, E.; Palomo-Ponce, S.; Tauriello, D.V.; Iglesias, M.; Cespedes, M.V.; Sevillano, M.; Nadal, C.; Jung, P.; Zhang, X.H.; et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell 2012, 22, 571–584. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar]
- West, N.R.; McCuaig, S.; Franchini, F.; Powrie, F. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 2015, 15, 615–629. [Google Scholar]
- Nagasaki, T.; Hara, M.; Nakanishi, H.; Takahashi, H.; Sato, M.; Takeyama, H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: Anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br. J. Cancer 2014, 110, 469–478. [Google Scholar] [CrossRef]
- Tacconi, C.; Ungaro, F.; Correale, C.; Arena, V.; Massimino, L.; Detmar, M.; Spinelli, A.; Carvello, M.; Mazzone, M.; Oliveira, A.I.; et al. Activation of the VEGFC/VEGFR3 Pathway Induces Tumor Immune Escape in Colorectal Cancer. Cancer Res. 2019, 79, 4196–4210. [Google Scholar] [CrossRef]
- Lim, E.J.; Kang, J.H.; Kim, Y.J.; Kim, S.; Lee, S.J. ICAM-1 promotes cancer progression by regulating SRC activity as an adapter protein in colorectal cancer. Cell Death Dis. 2022, 13, 417. [Google Scholar] [CrossRef]
- Velez-Delgado, A.; Donahue, K.L.; Brown, K.L.; Du, W.; Irizarry-Negron, V.; Menjivar, R.E.; Lasse Opsahl, E.L.; Steele, N.G.; The, S.; Lazarus, J.; et al. Extrinsic KRAS Signaling Shapes the Pancreatic Microenvironment Through Fibroblast Reprogramming. Cell Mol. Gastroenterol. Hepatol. 2022, 13, 1673–1699. [Google Scholar] [CrossRef]
- Huang, C.Y.; Chiang, S.F.; Ke, T.W.; Chen, T.W.; You, Y.S.; Chen, W.T.; Chao, K.S.C. Clinical significance of programmed death 1 ligand-1 (CD274/PD-L1) and intra-tumoral CD8+ T-cell infiltration in stage II-III colorectal cancer. Sci. Rep. 2018, 8, 15658. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar]
- Lee, J.; Li, L.; Gretz, N.; Gebert, J.; Dihlmann, S. Absent in Melanoma 2 (AIM2) is an important mediator of interferon-dependent and -independent HLA-DRA and HLA-DRB gene expression in colorectal cancers. Oncogene 2012, 31, 1242–1253. [Google Scholar] [CrossRef]
- Walsh, M.D.; Dent, O.F.; Young, J.P.; Wright, C.M.; Barker, M.A.; Leggett, B.A.; Bokey, L.; Chapuis, P.H.; Jass, J.R.; Macdonald, G.A. HLA-DR expression is associated with better prognosis in sporadic Australian clinicopathological Stage C colorectal cancers. Int. J. Cancer 2009, 125, 1231–1237. [Google Scholar]
- Kitamura, T.; Qian, B.Z.; Pollard, J.W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 2015, 15, 73–86. [Google Scholar] [CrossRef]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef]
- Engblom, C.; Pfirschke, C.; Pittet, M.J. The role of myeloid cells in cancer therapies. Nat. Rev. Cancer 2016, 16, 447–462. [Google Scholar] [CrossRef]
- Ogawa, R.; Yamamoto, T.; Hirai, H.; Hanada, K.; Kiyasu, Y.; Nishikawa, G.; Mizuno, R.; Inamoto, S.; Itatani, Y.; Sakai, Y.; et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tumor-Associated Neutrophils via the CXCL1/8-CXCR2 Axis. Clin. Cancer Res. 2019, 25, 2887–2899. [Google Scholar]
- Liu, C.; Yao, Z.; Wang, J.; Zhang, W.; Yang, Y.; Zhang, Y.; Qu, X.; Zhu, Y.; Zou, J.; Peng, S.; et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ. 2020, 27, 1765–1781. [Google Scholar]
- Siddiqa, A.; Sims-Mourtada, J.C.; Guzman-Rojas, L.; Rangel, R.; Guret, C.; Madrid-Marina, V.; Sun, Y.; Martinez-Valdez, H. Regulation of CD40 and CD40 ligand by the AT-hook transcription factor AKNA. Nature 2001, 410, 383–387. [Google Scholar]
- Kashyap, A.S.; Schmittnaegel, M.; Rigamonti, N.; Pais-Ferreira, D.; Mueller, P.; Buchi, M.; Ooi, C.H.; Kreuzaler, M.; Hirschmann, P.; Guichard, A.; et al. Optimized antiangiogenic reprogramming of the tumor microenvironment potentiates CD40 immunotherapy. Proc. Natl. Acad. Sci. USA 2020, 117, 541–551. [Google Scholar]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]



| Characteristics | Total N (%), n = 97 (100) | KRASwt N (%), n = 57 (58.76) | KRASmut N (%), n = 40 (41.24) | p-Value |
|---|---|---|---|---|
| Mutation status | ||||
| Exon 2 codon 12 | - | 28 (70) | ||
| Exon 2 codon 13 | - | 8 (20) | ||
| Exon 3 codon 61 | - | 4 (10) | ||
| Mean age | 64.86 | 63.01 | 67.48 | 0.101 |
| Sex | 0.459 | |||
| Male | 48 | 30 (62.5) | 18 (37.5) | |
| Female | 49 | 27 (55.1) | 22 (44.9) | |
| Tumor site | 0.381 | |||
| Rectum | 38 | 18 (50.0) | 18 (50.0) | |
| Left side colon | 43 | 28 (65.1) | 15 (34.9) | |
| Right side colon | 18 | 11 (61.1) | 7 (38.9) | |
| Stage | 0.109 | |||
| I | 13 | 11 (84.6) | 2 (15.4) | |
| II | 31 | 16 (51.6) | 15 (48.4) | |
| III | 33 | 21 (63.6) | 12 (36.4) | |
| IV | 20 | 9 (45.0) | 11 (55.0) | |
| Metastatic site | 1.000 | |||
| Liver | 16 | 8 (50.0) | 8 (50.0) | |
| Lung | 1 | - | 1 (100) | |
| Liver and lung | 2 | - | 2 (100) | |
| Others | 1 | 1 (100) | - | |
| Pathological grade | 0.126 | |||
| Moderately differentiated | 86 | 53 (61.6) | 33 (38.4) | |
| Well differentiated | 9 | 3 (33.8) | 6 (66.7) | |
| Poorly differentiated | 2 | 1(50.0) | 1(50.0) | |
| Adjuvant chemotherapy | 0.662 | |||
| Yes | 57 | 29 (50.9) | 28 (49.1) | |
| No | 40 | 23 (50.9) | 17 (49.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanjak, P.; Chaiboonchoe, A.; Suwatthanarak, T.; Acharayothin, O.; Thanormjit, K.; Chanthercrob, J.; Suwatthanarak, T.; Wannasuphaphol, B.; Chumchuen, K.; Suktitipat, B.; et al. The KRAS-Mutant Consensus Molecular Subtype 3 Reveals an Immunosuppressive Tumor Microenvironment in Colorectal Cancer. Cancers 2023, 15, 1098. https://doi.org/10.3390/cancers15041098
Tanjak P, Chaiboonchoe A, Suwatthanarak T, Acharayothin O, Thanormjit K, Chanthercrob J, Suwatthanarak T, Wannasuphaphol B, Chumchuen K, Suktitipat B, et al. The KRAS-Mutant Consensus Molecular Subtype 3 Reveals an Immunosuppressive Tumor Microenvironment in Colorectal Cancer. Cancers. 2023; 15(4):1098. https://doi.org/10.3390/cancers15041098
Chicago/Turabian StyleTanjak, Pariyada, Amphun Chaiboonchoe, Tharathorn Suwatthanarak, Onchira Acharayothin, Kullanist Thanormjit, Jantappapa Chanthercrob, Thanawat Suwatthanarak, Bundit Wannasuphaphol, Kemmapon Chumchuen, Bhoom Suktitipat, and et al. 2023. "The KRAS-Mutant Consensus Molecular Subtype 3 Reveals an Immunosuppressive Tumor Microenvironment in Colorectal Cancer" Cancers 15, no. 4: 1098. https://doi.org/10.3390/cancers15041098
APA StyleTanjak, P., Chaiboonchoe, A., Suwatthanarak, T., Acharayothin, O., Thanormjit, K., Chanthercrob, J., Suwatthanarak, T., Wannasuphaphol, B., Chumchuen, K., Suktitipat, B., Sampattavanich, S., Korphaisarn, K., Pongpaibul, A., Poungvarin, N., Grove, H., Riansuwan, W., Trakarnsanga, A., Methasate, A., Pithukpakorn, M., & Chinswangwatanakul, V. (2023). The KRAS-Mutant Consensus Molecular Subtype 3 Reveals an Immunosuppressive Tumor Microenvironment in Colorectal Cancer. Cancers, 15(4), 1098. https://doi.org/10.3390/cancers15041098







