Similarities and Differences in the Protein Composition of Cutaneous Melanoma Cells and Their Exosomes Identified by Mass Spectrometry
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Lines and Culture Conditions
2.3. Isolation of Exosomes and Assessment of the Purity of the Exosome Samples
2.4. LC–MS/MS Proteomics
2.5. Analysis of Proteomic Data
2.6. Bioinformatic Analysis
3. Results
3.1. Characterization of CM Exosome Samples
3.2. Identified Proteins of CM Cells and Exosomes and Their Functional Classification
3.3. Functional Similarities and Differences in the Protein Composition of CM Cells and CM-Derived Exosomes
4. Discussion
4.1. CM-Derived Exosomes Are Enriched in Proteins with Functional Implications in Cancer
4.2. Clinical Relevance of Proteins Identified in CM-Derived Exosomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CM | Cutaneous Melanoma |
| EV | Extracellular Vesicles |
| LC–MS/MS | Liquid Chromatography and Mass Spectrometry |
| NTA | Nanoparticle Tracking Analysis |
Appendix A

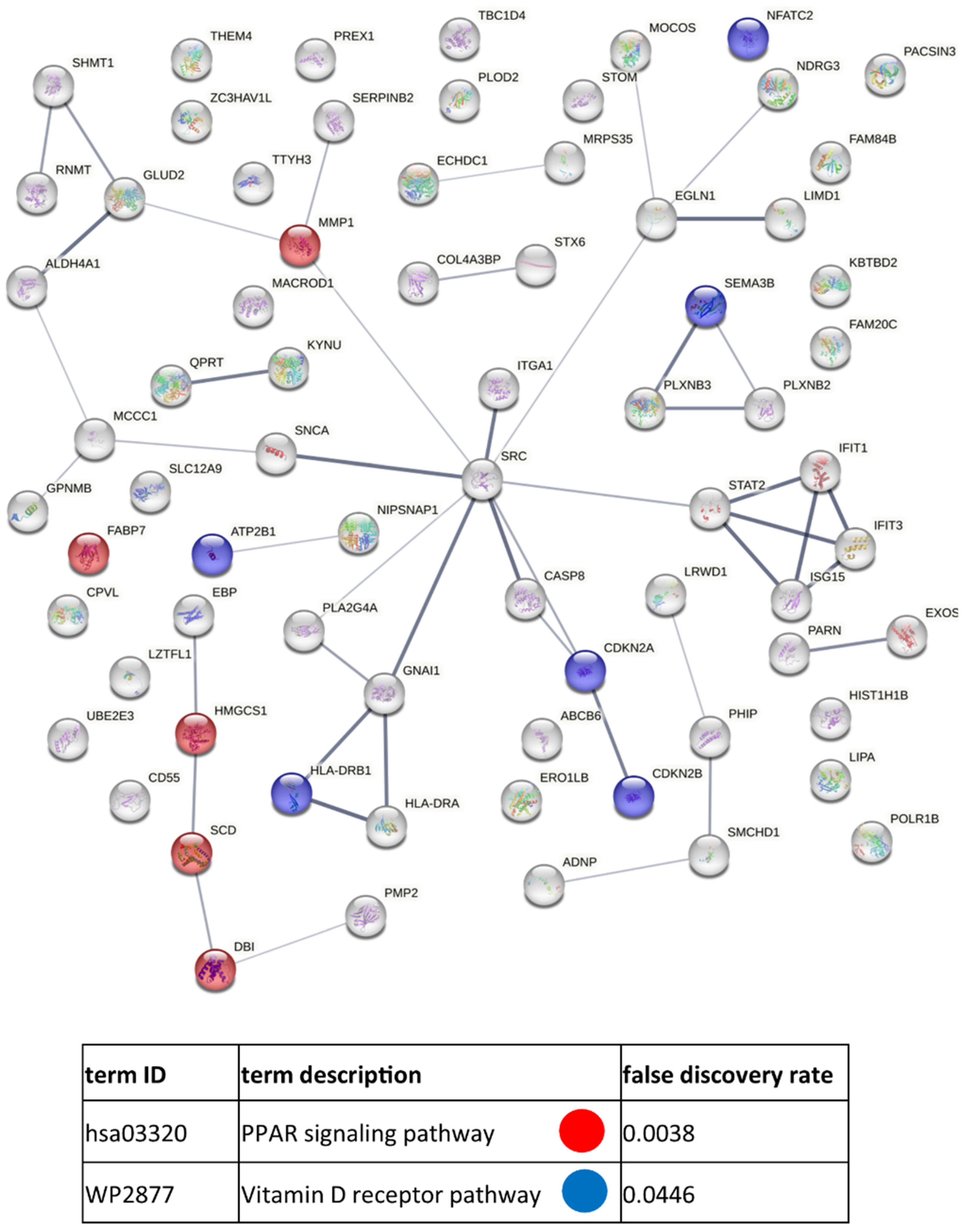



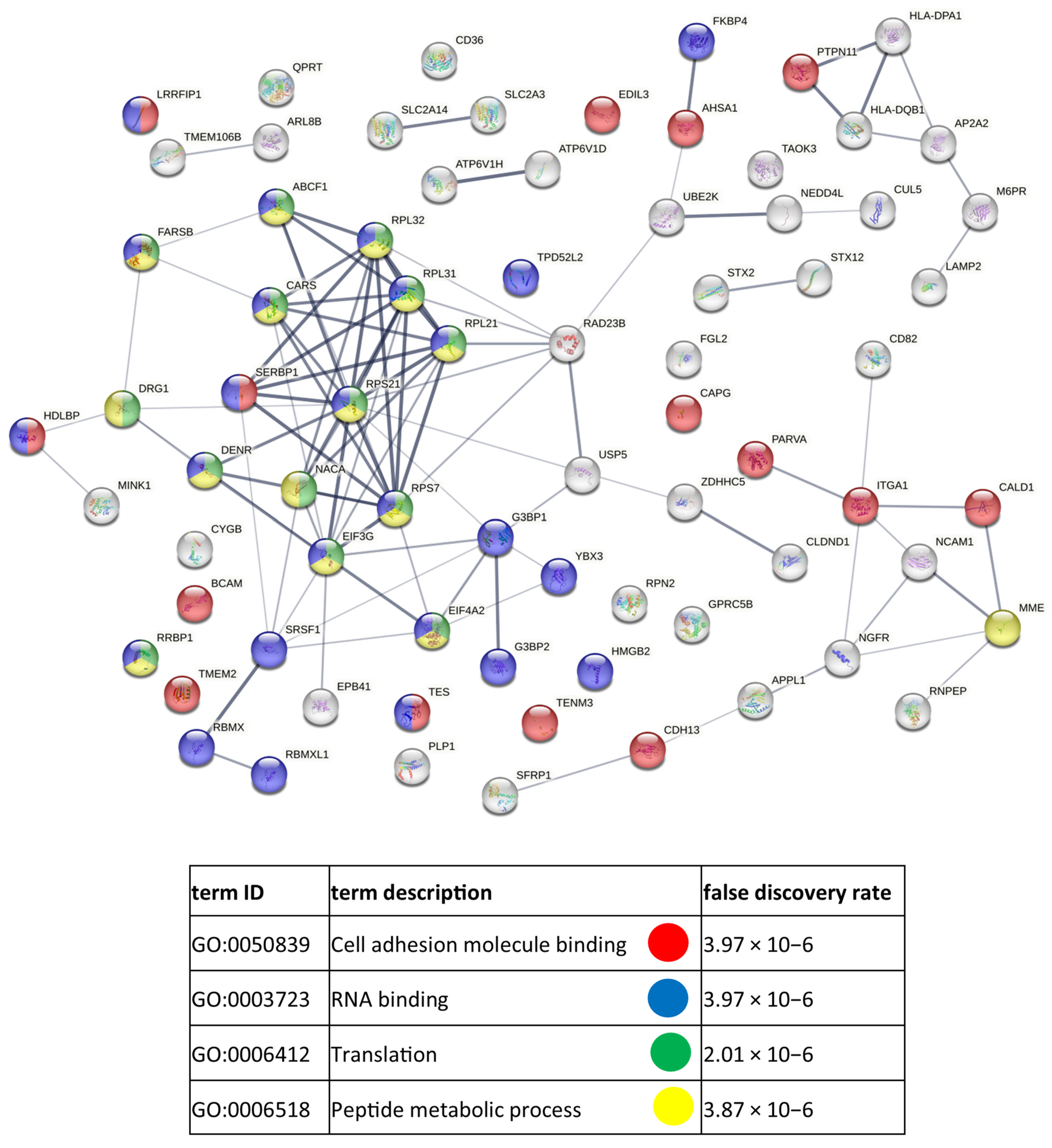


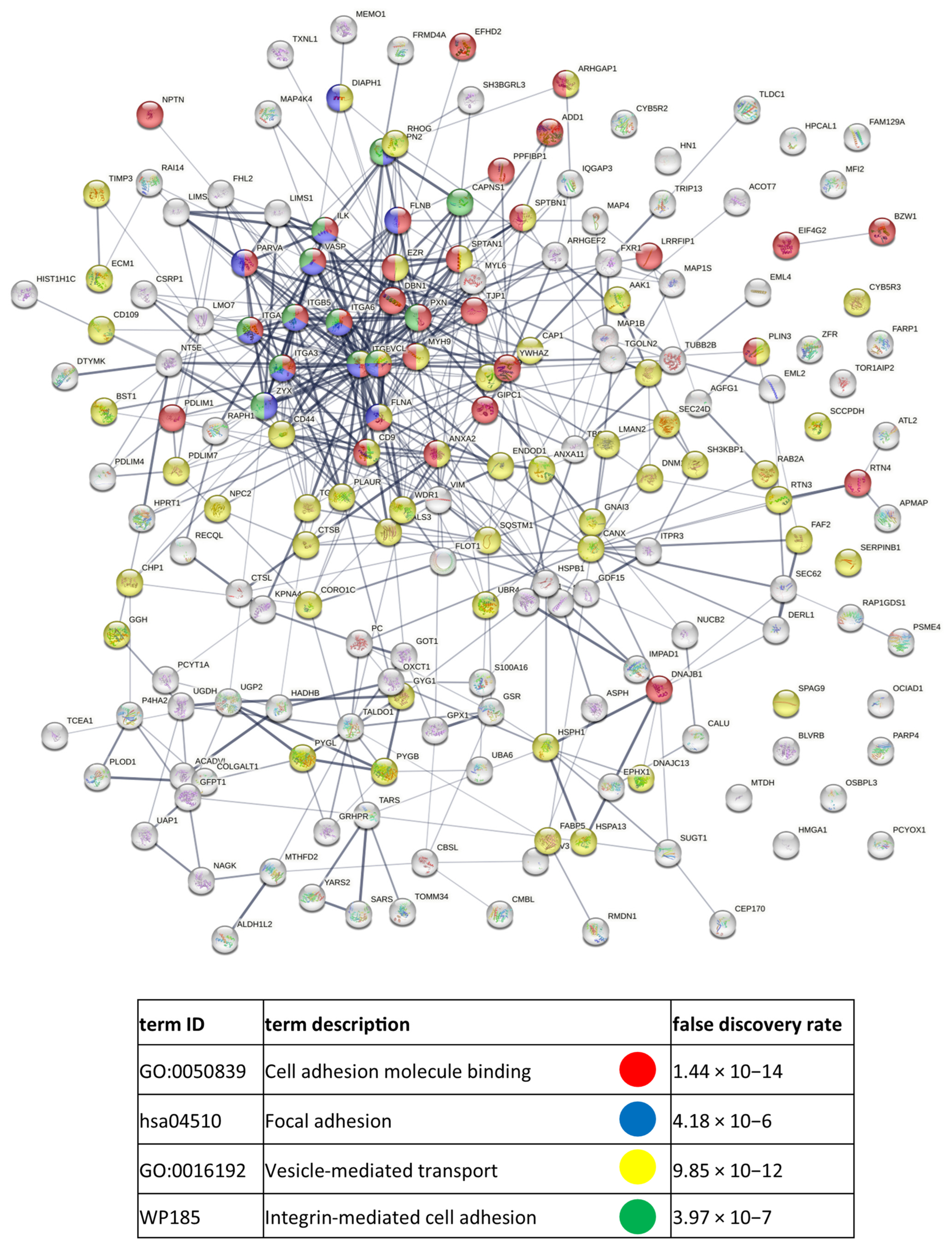
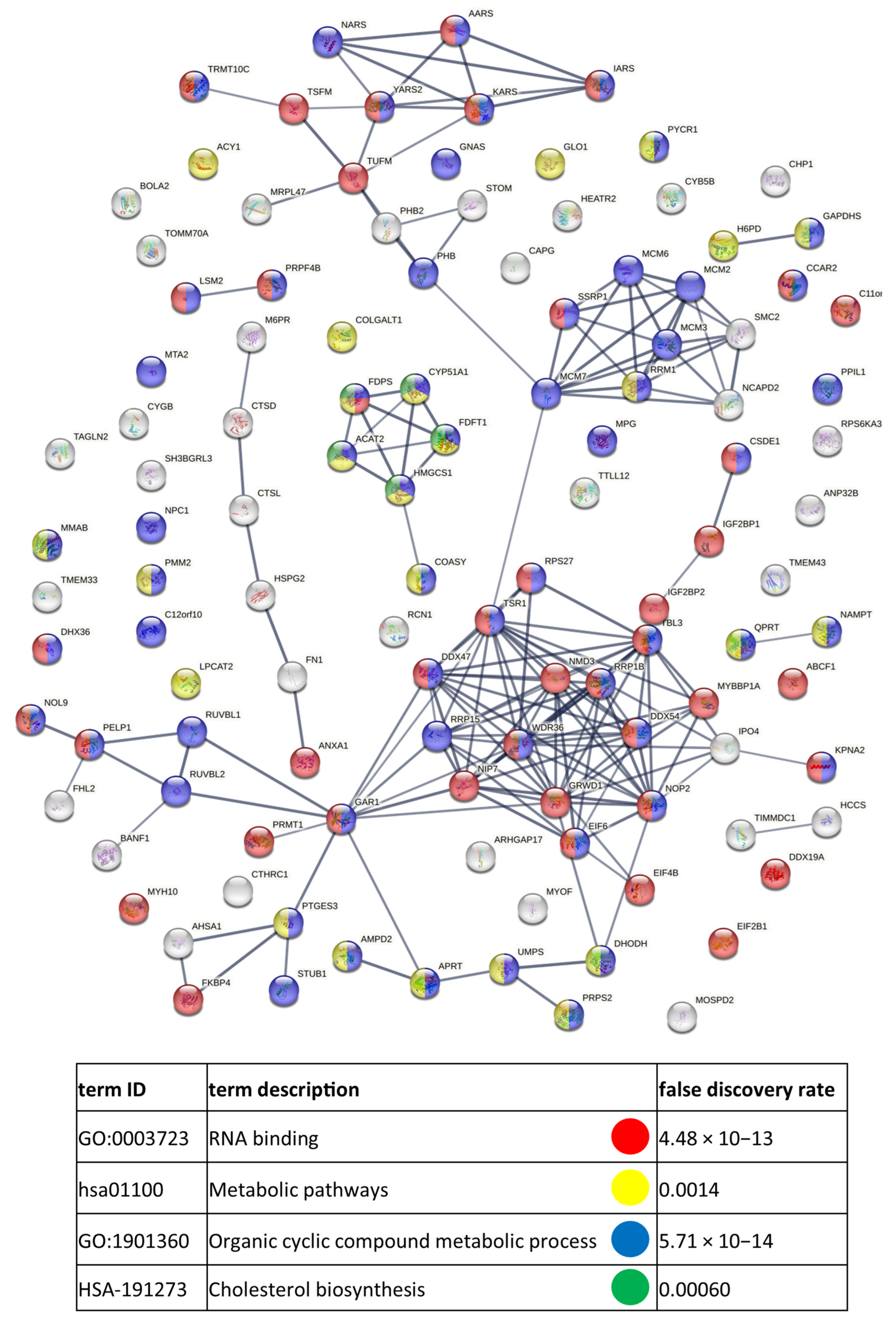
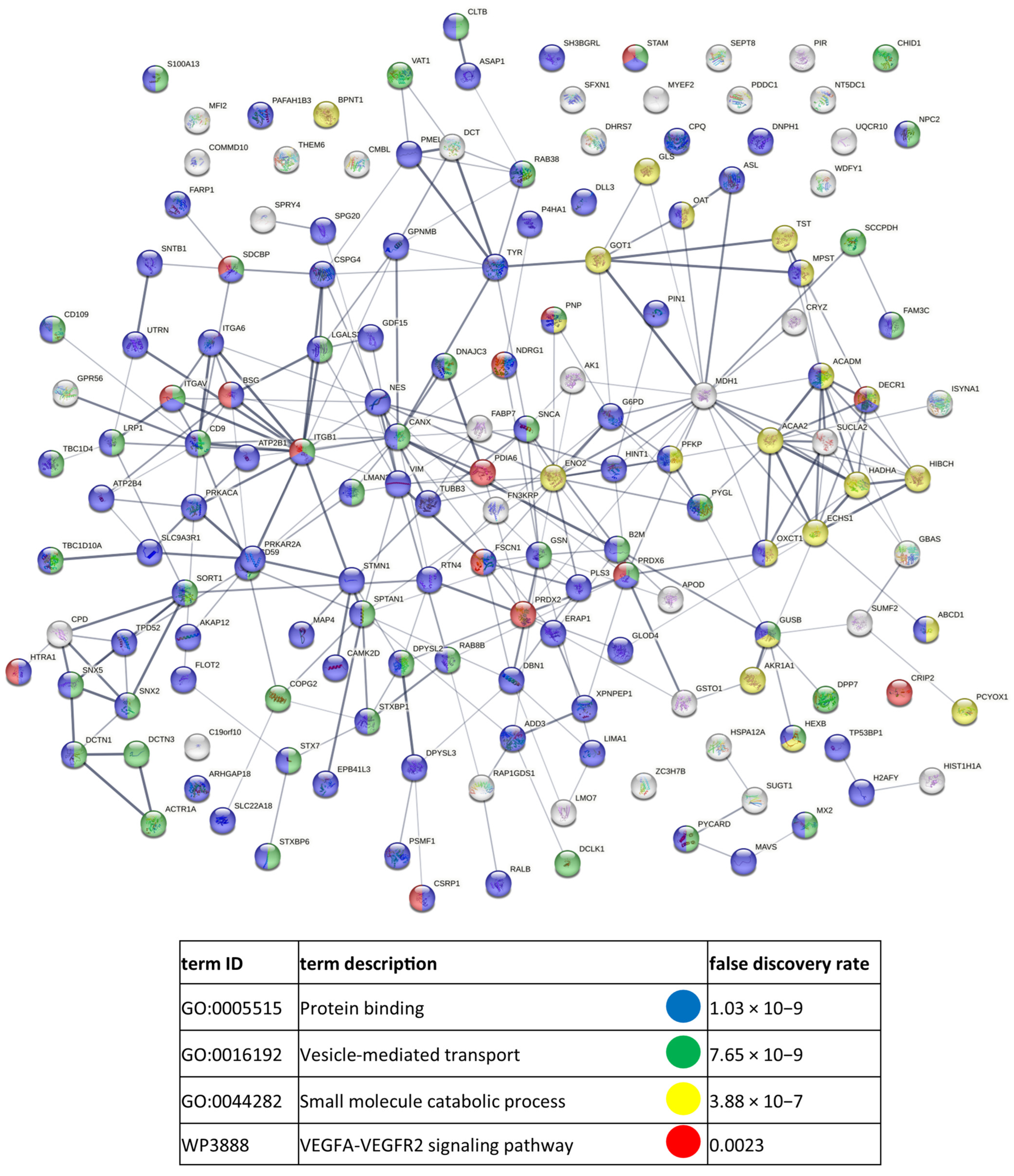
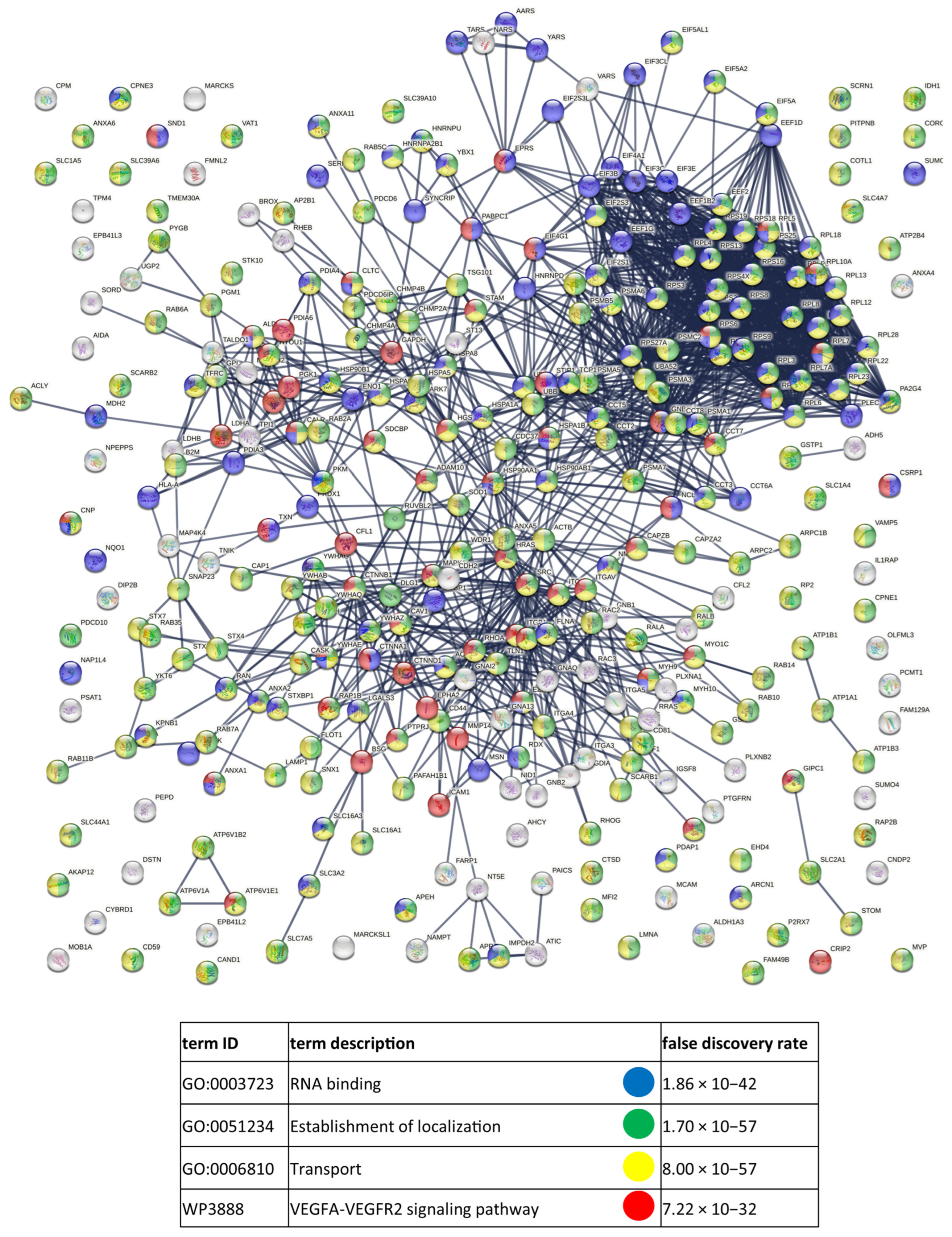

References
- Karimkhani, C.; Green, A.C.; Nijsten, T.; Weinstock, M.A.; Dellavalle, R.P.; Naghavi, M.; Fitzmaurice, C. The global burden of melanoma: Results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017, 177, 134–140. [Google Scholar] [CrossRef]
- Sung, H.; Ferllay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wróbel, S.; Przybyło, M.; Stępień, E. The clinical trial landscape for melanoma therapies. J. Clin. Med. 2019, 8, E368. [Google Scholar] [CrossRef]
- Sabag, N.; Yakobson, A.; Retchkiman, M.; Silberstein, E. Novel biomarkers and therapeutic targets for melanoma. Int. J. Mol. Sci. 2022, 23, 11656. [Google Scholar] [CrossRef]
- Switzer, B.; Puzanov, I.; Skitzki, J.J.; Hamad, L.; Ernstoff, M.S. Managing metastatic melanoma in 2022: A clinical review. JCO Oncol. Pract. 2022, 18, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Mancini, S.J.C.; Balabanian, K.; Corre, I.; Gavard, J.; Lazennec, G.; Le Bousse-Kerdilès, M.-C.; Louache, F.; Maguer-Satta, V.; Mazure, N.M.; Mechta-Grigoriou, F.; et al. Deciphering tumor niches: Lessons from solid and hematological malignancies. Front. Immunol. 2021, 12, 766275. [Google Scholar] [CrossRef]
- Murgai, M.; Giles, A.; Kaplan, R. Physiological, tumor, and metastatic niche: Opportunities and challenges for targeting the tumor microenvirinment. Crit. Rev. Oncog. 2015, 20, 301–314. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Clancy, J.; D’Souza-Schorey, C. Extracellular vesicles in cancer: Purpose and promise. Cancer J. 2018, 24, 65–69. [Google Scholar] [CrossRef]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Dong, Q.; Liu, X.; Cheng, K.; Sheng, J.; Kong, J.; Liu, T. Pre-metastatic niche formation in different organs induced by tumor extracellular vesicles. Front. Cell Dev. Biol. 2021, 9, 733627. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Surman, M.; Stępień, E.; Przybyło, M. Melanome-derived extracellular vesicles: Focus on their proteome. Proteomes 2019, 7, 21. [Google Scholar] [CrossRef]
- Lattman, E.; Levesque, M.P. The role of extracellular vesicles in melanoma progression. Cancers 2022, 14, 3086. [Google Scholar] [CrossRef]
- Surman, M.; Hoja-Łukowicz, D.; Szwed, S.; Kędracka-Krok, S.; Jankowska, U.; Kurtyka, M.; Drożdż, A.; Lityńska, A.; Stępień, E.; Przybyło, M. An insight into the proteome of uveal melanoma-derived ectosomes reveals the presence of potentially useful biomarkers. Int. J. Mol. Sci. 2019, 20, 3789. [Google Scholar] [CrossRef]
- Herlyn, M.; Balaban, G.; Bennicelli, J.; Guerry, D.; Halaban, R.; Herlyn, D.; Elder, D.E.; Maul, G.G.; Steplewski, Z.; Nowell, P.C. Primary melanoma cells of the vertical growth phase: Similarities to metastatic cells. J. Natl. Cancer Inst. 1985, 74, 283–289. [Google Scholar]
- Cornil, I.; Theodorescu, D.; Man, S.; Herlyn, M.; Jambrosic, J.; Kerbel, R.S. Fibroblast cell interactions with human melanoma cells affect tumor cell growth as a function of tumor progression. Proc. Natl. Acad. Sci. USA 1991, 88, 6028–6032. [Google Scholar] [CrossRef]
- Juhasz, I.; Albelda, S.M.; Elder, D.E.; Murphy, G.F.; Adachi, K.; Herlyn, D.; Valyi-Nagy, I.T.; Herlyn, M. Growth and invasion of human melanomas in human skin grafted to immunodeficient mice. Am. J. Pathol. 1993, 143, 528–537. [Google Scholar]
- Drożdż, A.; Kamińska, A.; Surman, M.; Gonet-Surówka, A.; Jach, R.; Huras, H.; Przybyło, M.; Stępień, E.Ł. Low-Vacuum Filtration as an Alternative Extracellular Vesicle Concentration Method: A Comparison with Ultracentrifugation and Differential Centrifugation. Pharmaceutics. 2020, 12, 872. [Google Scholar] [CrossRef] [PubMed]
- Surman, M.; Hoja-Łukowicz, D.; Szwed, S.; Drożdż, A.; Stępień, E.; Przybyło, M. Human melanoma-derived ectosomes are enriched with specific glycan epitopes. Life Sci. 2018, 207, 395–411. [Google Scholar] [CrossRef]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Mercapide, J.; Anzanello, F.; Pope, R.M.; Lorico, A. Biochemical and biological characterization of exosomes containing prominin-1/CD133. Mol. Cancer 2013, 12, 62. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Wang, S.; Zhao, R.C. Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol. Cell Biochem. 2013, 383, 13–20. [Google Scholar] [CrossRef]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef]
- Liao, C.F.; Lin, S.H.; Chen, H.C.; Tai, C.J.; Chang, C.C.; Li, L.T.; Yeh, C.M.; Yeh, K.T.; Chen, Y.C.; Hsu, T.H.; et al. CSE1L, a novel microvesicle membrane protein, mediates Ras-triggered microvesicle generation and metastasis of tumor cells. Mol. Med. 2012, 18, 1269–1280. [Google Scholar] [CrossRef]
- Hatanaka, M.; Higashi, Y.; Fukushige, T.; Baba, N.; Kawai, K.; Hashiguchi, T.; Su, J.; Zeng, W.; Chen, X.; Kanekura, T. Cleaved CD147 shed from the surface of malignant melanoma cells activates MMP2 produced by fibroblasts. Anticancer Res. 2014, 34, 7091–7096. [Google Scholar]
- Clancy, J.W.; Sedgwick, A.; Rosse, C.; Muralidharan-Chari, V.; Raposo, G.; Method, M.; Chavrier, P.; D’Souza-Schorey, C. Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun. 2015, 6, 691. [Google Scholar] [CrossRef]
- Zhao, X.P.; Wang, M.; Song, Y.; Song, K.; Yan, T.L.; Wang, L.; Liu, K.; Shang, Z.J. Membrane microvesicles as mediators for melanoma-fibroblasts communication: Roles of the VCAM-1/VLA-4 axis and the ERK1/2 signal pathway. Cancer Lett. 2015, 360, 125–133. [Google Scholar] [CrossRef]
- Surman, M.; Kędracka-Krok, S.; Hoja-Łukowicz, D.; Jankowska, U.; Drożdż, A.; Stępień, E.Ł.; Przybyło, M. Mass spectrometry-based proteomic characterization of cutaneous melanoma ectosomes reveals the presence of cancer-related molecules. Int. J. Mol. Sci. 2020, 21, 2934. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a prometastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Kim, J.; Afshari, A.; Sengupta, R.; Sebastiano, V.; Gupta, A.; Kim, Y.H.; Reproducibility Project: Cancer Biology. Replication study: Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. elife 2018, 7, e39944. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Ducoux-Petit, M.; Denat, L.; Soldan, V.; Dauvillier, S.; Balor, S.; Burlet-Schiltz, O.; Larue, L.; Muller, C.; et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Cell Melanoma Res. 2015, 28, 464–475. [Google Scholar] [CrossRef]
- Hao, S.; Ye, Z.; Li, F.; Meng, Q.; Qureshi, M.; Yang, J.; Xiang, J. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp. Oncol. 2006, 28, 126–131. [Google Scholar] [PubMed]
- Mannavola, F.; Tucci, M.; Felici, C.; Passarelli, A.; D’Oronzo, S.; Silvestris, F. Tumor-derived exosomes promote the in vitro osteotropism of melanoma cells by activating the SDF-1/CXCR4/CXCR7 axis. J. Transl. Med. 2019, 17, 230. [Google Scholar] [CrossRef]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef]
- Wang, P.; Wu, Y.; Chen, W.; Zhang, M.; Qin, J. Malignant melanoma-derived exosomes induce endothelial damage and glial activation on a human BBB chip model. Biosensors 2022, 12, 89. [Google Scholar] [CrossRef]
- Xiao, D.; Barry, S.; Kmetz, D.; Egger, M.; Pan, J.; Rai, S.N.; Qu, J.; McMasters, K.M.; Hao, H. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016, 376, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L.; Pan, H.; Lanza, G.M.; Wickline, S.A. Paracrine induction of endothelium by tumor exosomes. Lab. Investig. 2009, 89, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, E.J.; Bergenfelz, C.; von Bulow, V.; Serifler, F.; Carlemalm, E.; Jonsson, G.; Andersson, T.; Leandersson, K. WNT5A induces release of exosomes containing pro-angiogenic and immunosuppressive factors from malignant melanoma cells. Mol. Cancer 2014, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Biagioni, A.; Laurenzana, A.; Menicacci, B.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Guasti, D.; Paoli, P.; Serratì, S.; Mocali, A.; et al. uPAR-expressing melanoma exosomes promote angiogenesis by VE-Cadherin, EGFR and uPAR overexpression and rise of ERK1,2 signaling in endothelial cells. Cell. Mol. Life Sci. 2021, 78, 3057–3072. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37. [Google Scholar] [CrossRef]
- Leary, N.; Walser, S.; He, Y.; Cousin, N.; Pereira, P.; Gallo, A.; Collado-Diaz, V.; Halin, C.; Garcia-Silva, S.; Peinado, H.; et al. Melanoma-derived extracellular vesicles mediate lymphatic remodelling and impair tumour immunity in draining lymph nodes. J. Extracell. Vesicles 2022, 11, e12197. [Google Scholar] [CrossRef]
- García-Silva, S.; Benito-Martín, A.; Nogués, L.; Hernández-Barranco, A.; Mazariegos, M.S.; Santos, V.; Hergueta-Redondo, M.; Ximénez-Embún, P.; Kataru, R.P.; Lopez, A.A.; et al. Melanoma-derived small extracellular vesicles induce lymphangiogenesis and metastasis through an NGFR-dependent mechanism. Nat. Cancer 2021, 2, 1387–1405. [Google Scholar] [CrossRef]
- Strnadová, K.; Pfeiferová, L.; Přikryl, P.; Dvořánková, B.; Vlčák, E.; Frýdlová, J.; Vokurka, M.; Novotný, J.; Šáchová, J.; Hradilová, M.; et al. Exosomes produced by melanoma cells significantly influence the biological properties of normal and cancer-associated fibroblasts. Histochem. Cell Biol. 2022, 157, 153–172. [Google Scholar] [CrossRef]
- Bardi, G.T.; Smith, M.A.; Hood, J.L. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine 2018, 105, 63–72. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Yang, L.; Jiang, Y.; Qian, Q. TIM-3 shuttled by MV3 cells-secreted exosomes inhibits CD4+ T cell immune function and induces macrophage M2 polarization to promote the growth and metastasis of melanoma cells. Transl. Oncol. 2022, 18, 101334. [Google Scholar] [CrossRef]
- Bland, C.L.; Byrne-Hoffman, C.N.; Fernandez, A.; Rellick, S.L.; Deng, W.O. Exosomes derived from B16F0 melanoma cells alter the transcriptome of cytotoxic T cells that impacts mitochondrial respiration. FEBS J. 2018, 285, 1033–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhong, W.; Wang, B.; Yang, J.; Yang, J.; Yu, Z.; Qin, Z.; Shi, A.; Xu, W.; Zheng, C.; et al. ICAM-1-mediated adhesion is a prerequisite for exosome-induced T cell suppression. Dev. Cell 2022, 57, 329–343.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, Y.; Wang, W.; Zhang, Y.; Chen, Z.; Hao, C.; Zhang, J.P. Melanoma-released exosomes directly activate the mitochondrial apoptotic pathway of CD4(+) T cells through their microRNA cargo. Exp. Cell Res. 2018, 371, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liu, C.; Su, K.; Wang, J.; Liu, Y.; Zhang, L.; Li, C.; Cong, Y.; Kimberly, R.; Grizzle, W.E.; et al. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J. Immunol. 2007, 178, 6867–6875. [Google Scholar] [CrossRef]
- Boussadia, Z.; Lamberti, J.; Mattei, F.; Pizzi, E.; Puglisi, R.; Zanetti, C.; Pasquini, L.; Fratini, F.; Fantozzi, L.; Felicetti, F.; et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res. 2018, 37, 245. [Google Scholar] [CrossRef]
- Kruijff, S.; Hoekstra, H.J. The current status of S-100B as a biomarker in melanoma. Eur. J. Surg. Oncol. 2012, 38, 281–285. [Google Scholar] [CrossRef]
- Barak, V.; Leibovici, V.; Peretz, T.; Kalichman, I.; Lotem, M.; Merims, S. Assessing response to new treatments and prognosis in melanoma patients, by the biomarker S-100β. Anticancer Res. 2015, 35, 6755–6760. [Google Scholar]
- Jurisic, V.; Radenkovic, S.; Konjevic, G. The Actual Role of LDH as Tumor Marker, Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 115–124. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extr. Ves. 2013, 2, 20424. [Google Scholar] [CrossRef]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabro, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef]
- Garcia-Silva, S.; Benito-Martin, A.; Sanchez-Redondo, S.; Hernandez-Barranco, A.; Ximenez-Embun, P.; Nogues, L.; Mazariegos, M.S.; Brinkmann, K.; López, A.A.; Meyer, L.; et al. Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF (V600E) mutation. J. Exp. Med. 2019, 216, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Marconcini, R.; Pasquini, G.; Rofi, E.; Vivaldi, C.; Bloise, F.; Restante, G.; Arrigoni, E.; Caparello, C.; Bianco, M.G.; et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br. J. Cancer 2018, 118, 820–824. [Google Scholar] [CrossRef] [PubMed]
- Serratì, S.; Guida, M.; Di Fonte, R.; De Summa, S.; Strippoli, S.; Iacobazzi, R.M.; Quarta, A.; De Risi, I.; Guida, G.; Paradiso, A.; et al. Circulating extracellular vesicles expressing PD1 and PD-L1 predict response and mediate resistance to checkpoint inhibitors immunotherapy in metastatic melanoma. Mol. Cancer 2022, 21, 20. [Google Scholar] [CrossRef] [PubMed]
- Turiello, R.; Capone, M.; Morretta, E.; Monti, M.C.; Madonna, G.; Azzaro, R.; Del Gaudio, P.; Simeone, E.; Sorrentino, A.; Ascierto, P.A.; et al. Exosomal CD73 from serum of patients with melanoma suppresses lymphocyte functions and is associated with therapy resistance to anti-PD-1 agents. J. Immunother. Cancer 2022, 10, e004043. [Google Scholar] [CrossRef] [PubMed]
- Pietrowska, M.; Zebrowska, A.; Gawin, M.; Marczak, L.; Sharma, P.; Mondal, S.; Mika, J.; Polańska, J.; Ferrone, S.; Kirkwood, J.M.; et al. Proteomic profile of melanoma cell-derived small extracellular vesicles in patients’ plasma: A potential correlate of melanoma progression. J. Extracell. Vesicles 2021, 10, e12063. [Google Scholar] [CrossRef] [PubMed]
- Boudhraa, Z.; Rondepierre, F.; Ouchchane, L.; Kintossou, R.; Trzeciakiewicz, A.; Franck, F.; Kanitakis, J.; Labeille, B.; Joubert-Zakeyh, J.; Bouchon, B.; et al. Annexin A1 in primary tumors promotes melanoma dissemination. Clin. Exp. Metastasis 2014, 31, 749–760. [Google Scholar] [CrossRef]
- Shin, J.; Song, I.S.; Pak, J.H.; Jang, S.W. Upregulation of annexin A1 expression by butyrate in human melanoma cells induces invasion by inhibiting E-cadherin expression. Tumour Biol. 2016, 37, 14577–14584. [Google Scholar] [CrossRef]
- Finger, E.C.; Castellini, L.; Rankin, E.B.; Vilalta, M.; Krieg, A.J.; Jiang, D.; Banh, A.; Zundel, W.; Powell, M.B.; Giaccia, A.J. Hypoxic induction of AKAP12 variant 2 shifts PKA-mediated protein phosphorylation to enhance migration and metastasis of melanoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4441–4446. [Google Scholar] [CrossRef]
- Fukunaga-Kalabis, M.; Martinez, G.; Nguyen, T.K.; Kim, D.; Santiago-Walker, A.; Roesch, A.; Herlyn, M. Tenascin-C promotes melanoma progression by maintaining the ABCB5-positive side population. Oncogene 2010, 29, 6115–6124. [Google Scholar] [CrossRef]
- Hu, W.; Jin, L.; Jiang, C.C.; Long, G.V.; Scolyer, R.A.; Wu, Q.; Zhang, X.D.; Mei, Y.; Wu, M. AEBP1 upregulation confers acquired resistance to BRAF (V600E) inhibition in melanoma. Cell Death Dis. 2013, 4, e914. [Google Scholar] [CrossRef]
- D’Aguanno, S.; Valentini, E.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Spagnuolo, M.; Buglioni, S.; Ercolani, C.; Falcone, I.; De Dominici, M.; et al. Semaphorin 5A drives melanoma progression: Role of Bcl-2, miR-204 and c-Myb. J. Exp. Clin Cancer Res. 2018, 37, 278. [Google Scholar] [CrossRef] [PubMed]
- Komina, A.V.; Palkina, N.V.; Aksenenko, M.B.; Lavrentev, S.N.; Moshev, A.V.; Savchenko, A.A.; Averchuk, A.S.; Rybnikov, Y.A.; Ruksha, T.G. Semaphorin-5A downregulation is associated with enhanced migration and invasion of BRAF-positive melanoma cells under vemurafenib treatment in melanomas with heterogeneous BRAF status. Melanoma Res. 2019, 29, 544–548. [Google Scholar] [CrossRef] [PubMed]


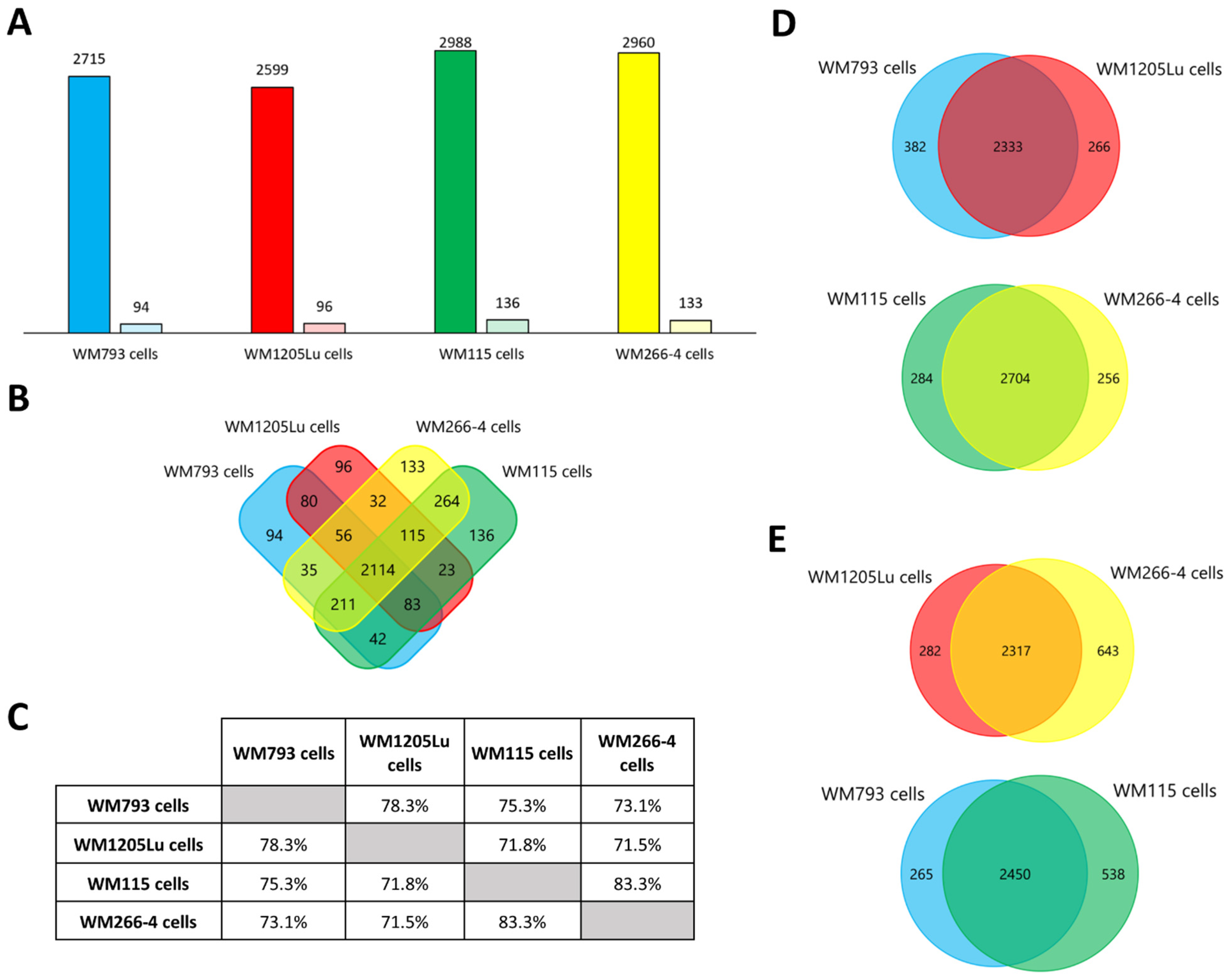
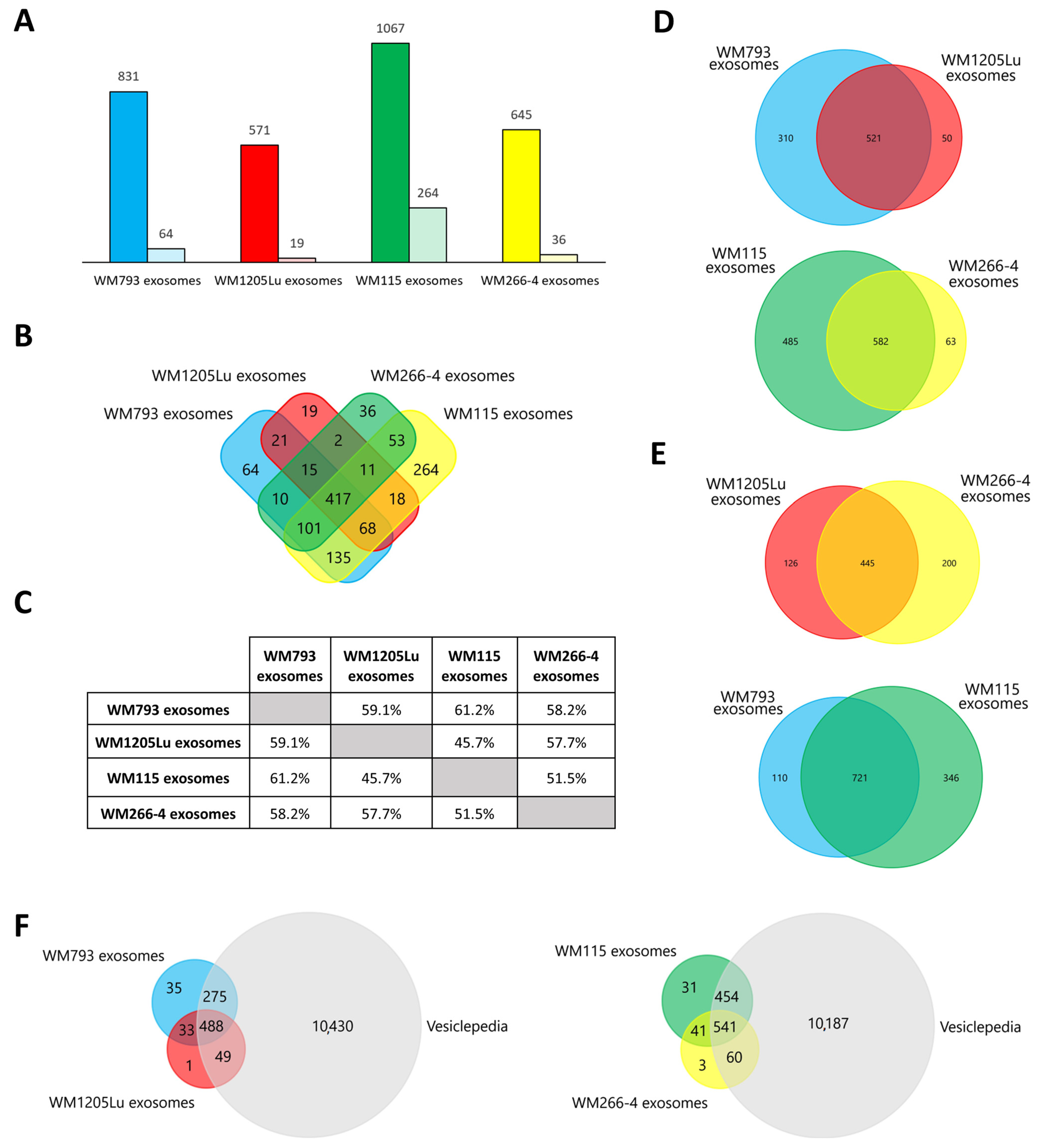
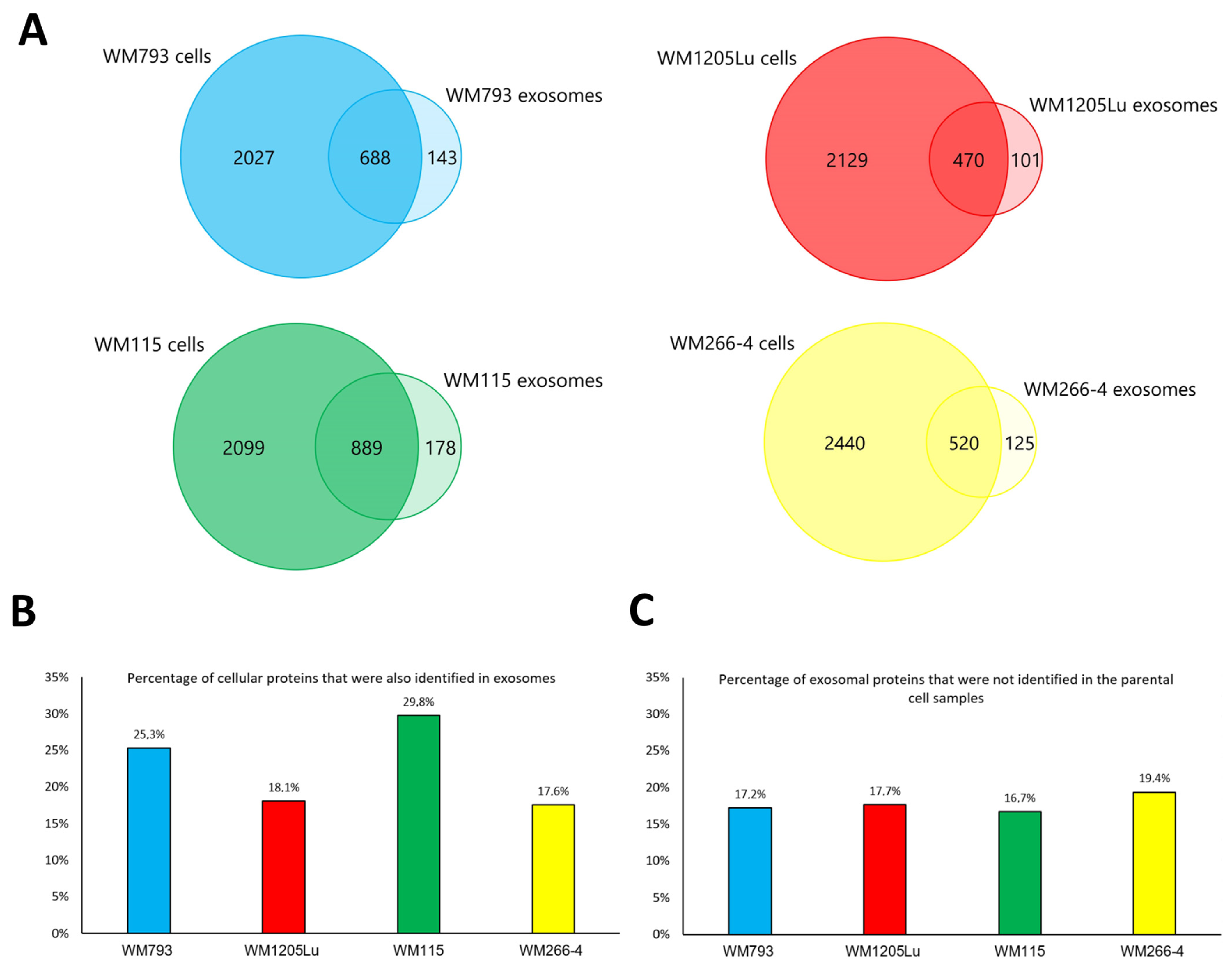
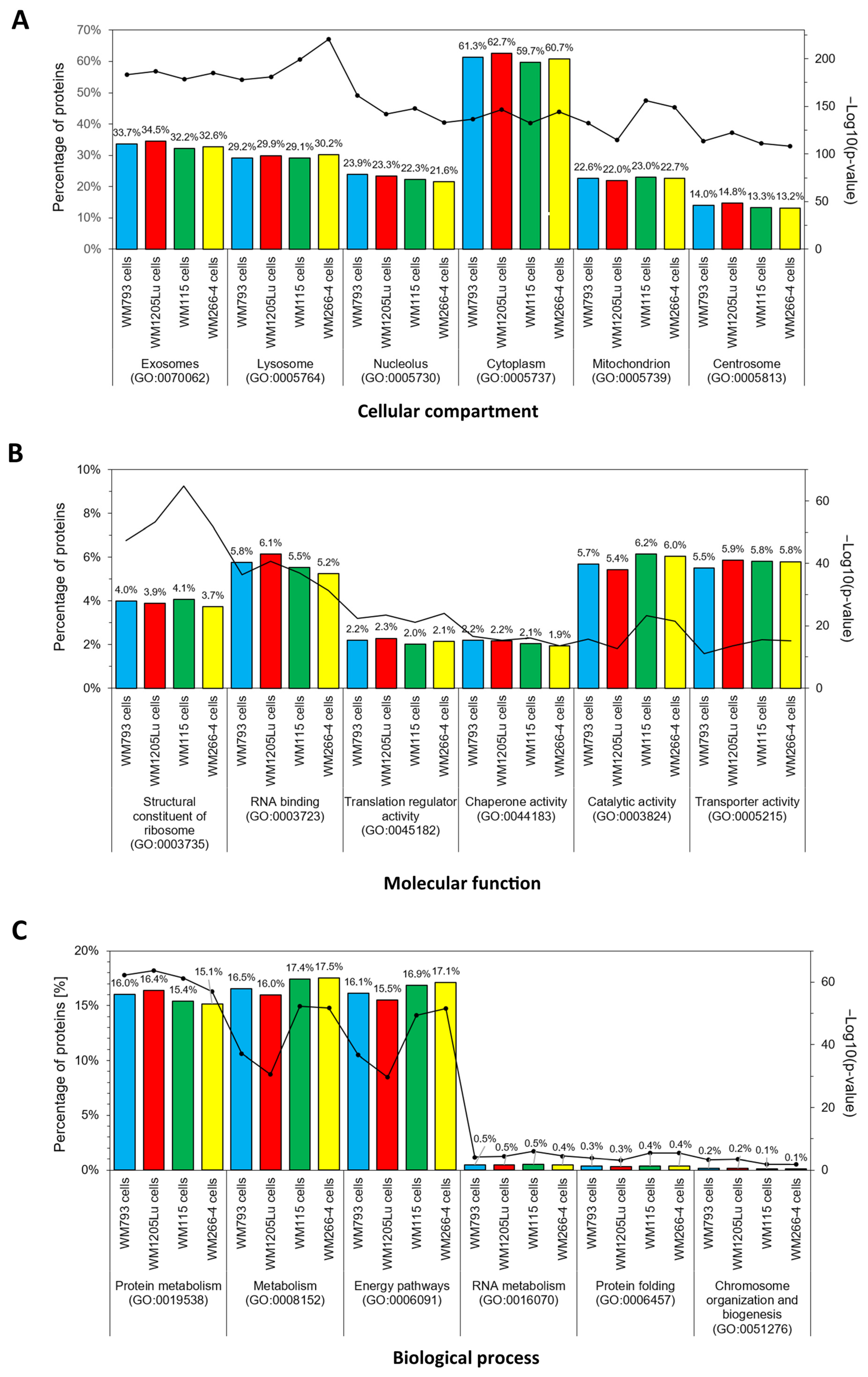
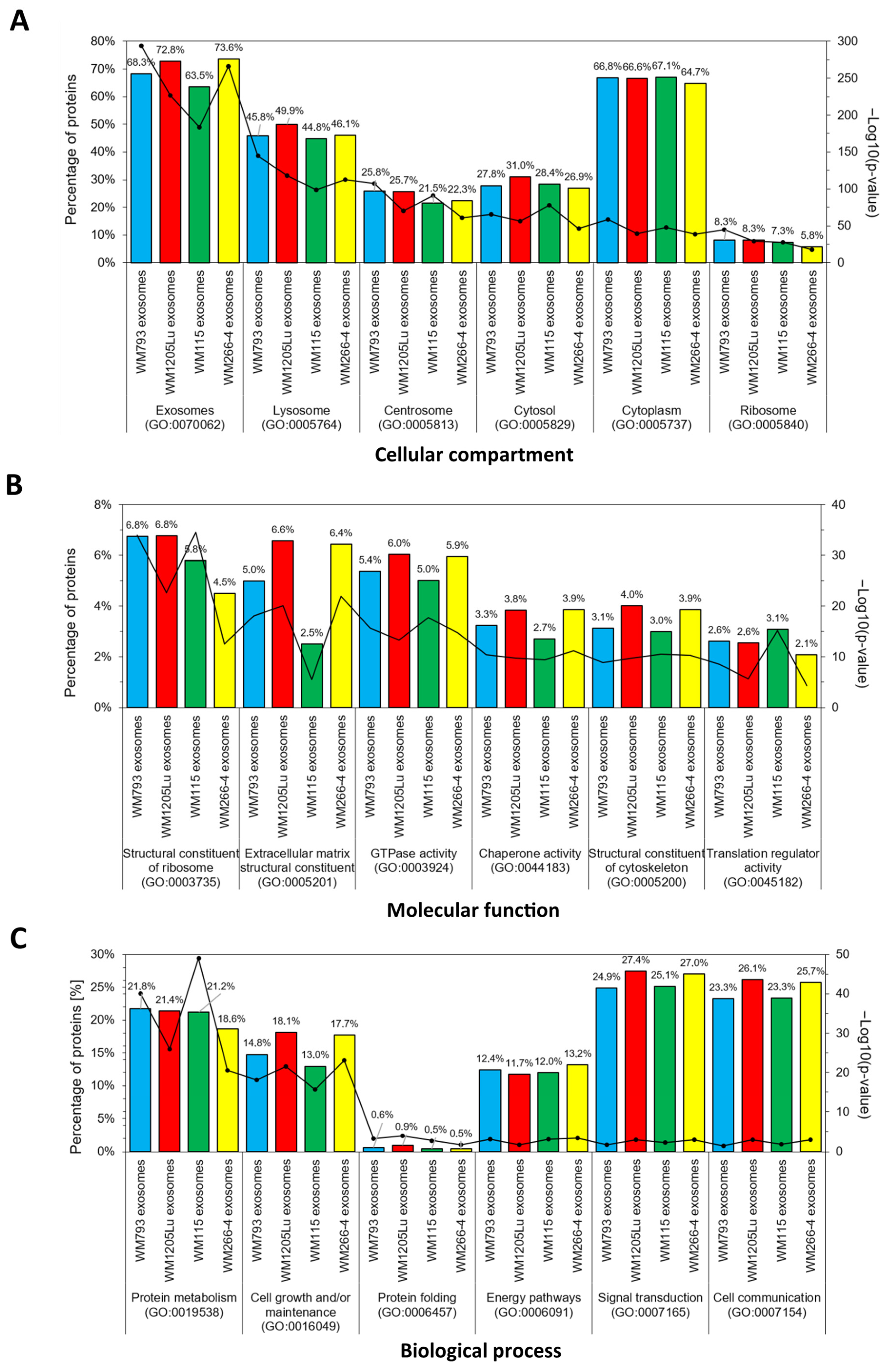
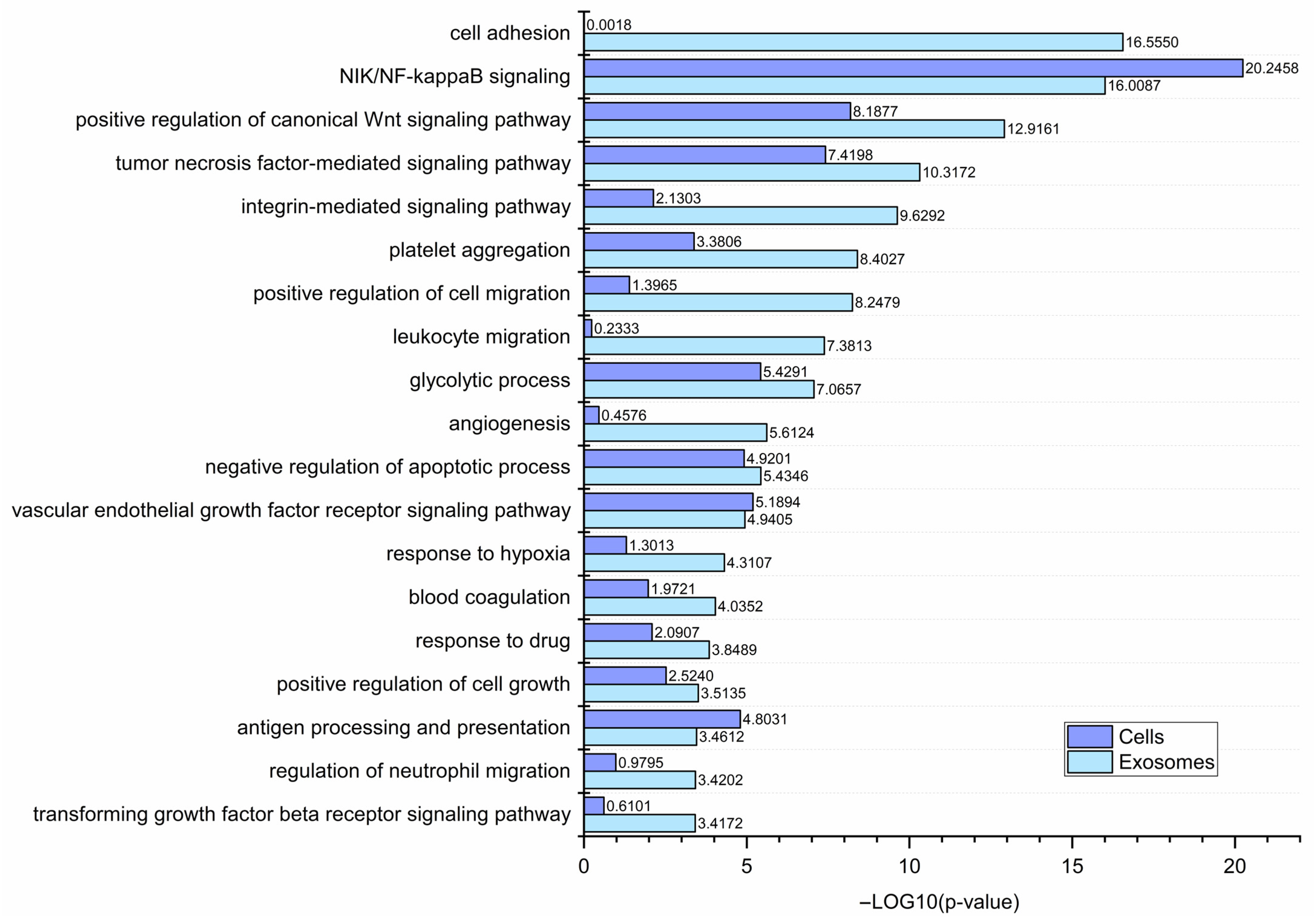
| Present Only in WM793 Cells | Present Only in WM1205Lu Cells | |
|---|---|---|
| Structural maintenance of chromosomes flexible hinge domain-containing protein 1 Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 Acyl-CoA desaturase Interferon-induced protein with tetratricopeptide repeats 3 Plexin-B2 Fatty acid-binding protein, brain mRNA cap guanine-N7 methyltransferase Syntaxin-6 TBC1 domain family member 4 Poly(A)-specific ribonuclease PARN E3 ubiquitin-protein ligase HERC2 Adenosine deaminase HLA class II histocompatibility antigen, DR alpha chain HLA class II histocompatibility antigen, DRB1-15 beta chain Interstitial collagenase 22 kDa interstitial collagenase 27 kDa interstitial collagenase Plasminogen activator inhibitor 2 Ubiquitin-like protein ISG15 Acyl-CoA-binding protein Complement decay-accelerating factor Interferon-induced protein with tetratricopeptide repeats 1 Proto-oncogene tyrosine-protein kinase Src Histone H1.5 Plasma membrane calcium-transporting ATPase 1 Erythrocyte band 7 integral membrane protein Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial Serine hydroxymethyltransferase, cytosolic Alpha-synuclein Lysosomal acid lipase/cholesteryl ester hydrolase Cyclin-dependent kinase inhibitor 2A Cyclin-dependent kinase 4 inhibitor B Cytosolic phospholipase A2 Phospholipase A2 Lysophospholipase Glutamate dehydrogenase 2, mitochondrial Signal transducer and activator of transcription 2 Integrin alpha-1 Guanine nucleotide-binding protein G(i) subunit alpha-1 HLA class II histocompatibility antigen, DR beta 3 chain | Hydroxymethylglutaryl-CoA synthase, cytoplasmic Semaphorin-3B Nuclear factor of activated T-cells, cytoplasmic 2 Caspase-8 Caspase-8 subunit p18 Caspase-8 subunit p10 Transmembrane glycoprotein NMB 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase Nicotinate-nucleotide pyrophosphorylase [carboxylating] Kynureninase Acyl-coenzyme A thioesterase THEM4 ERO1-like protein beta Extracellular serine/threonine protein kinase FAM20C Kelch repeat and BTB domain-containing protein 2 Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein PH-interacting protein Ubiquitin-conjugating enzyme E2 E3 Molybdenum cofactor sulfurase Zinc finger CCCH-type antiviral protein 1-like Protein FAM84B Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial Protein NipSnap homolog 1 O-acetyl-ADP-ribose deacetylase MACROD1 Solute carrier family 12 member 9 Protein tweety homolog 3 Egl nine homolog 1 Activity-dependent neuroprotector homeobox protein Probable serine carboxypeptidase CPVL DNA-directed RNA polymerase I subunit RPA2 ATP-binding cassette sub-family B member 6, mitochondrial Exosome complex component RRP41 Leucine zipper transcription factor-like protein 1 Ethylmalonyl-CoA decarboxylase Leucine-rich repeat and WD repeat-containing protein 1 LIM domain-containing protein 1 Protein NDRG3 Protein kinase C and casein kinase substrate in neurons protein 3 Collagen type IV alpha-3-binding protein 28S ribosomal protein S35, mitochondrial | Citron Rho-interacting kinase Neuropilin-1 Triple functional domain protein Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 3 Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 2 Sorbin and SH3 domain-containing protein 2 Tissue-type plasminogen activator;Tissue-type plasminogen activator chain A;Tissue-type plasminogen activator chain B Alpha-crystallin B chain Apolipoprotein D Neuromodulin Protein S100-A1 Thromboxane-A synthase CAP-Gly domain-containing linker protein 1 Carbonic anhydrase-related protein AP-1 complex subunit sigma-2 TNF receptor-associated factor 1 Growth factor receptor-bound protein 10 Calcyphosin Dihydropyrimidinase-related protein 1 Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2 Glycerol-3-phosphate acyltransferase 3 HCLS1-binding protein 3 Uncharacterized protein FLJ45252 Fermitin family homolog 3 5-nucleotidase domain-containing protein 3 Protein AHNAK2 Actin filament-associated protein 1-like 2 EH domain-binding protein 1 E3 ubiquitin-protein ligase DTX3L PDZ domain-containing protein GIPC3 Ubiquitin carboxyl-terminal hydrolase 47 Sorting nexin-18 Protein FAM107B Synaptic vesicle membrane protein VAT-1 homolog-like T-complex protein 11-like protein 1 EH domain-containing protein 2 Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1 Absent in melanoma 1 protein |
| Present Only in WM115 Cells | Present Only in WM266-4 Cells | |
|---|---|---|
| Serine protease 23 HLA class II histocompatibility antigen, DR alpha chain HLA class II histocompatibility antigen, DQ beta 1 chain HLA class II histocompatibility antigen, DR beta 4 chain Nidogen-1 Platelet glycoprotein 4 HLA class II histocompatibility antigen, DP alpha 1 chain HLA class II histocompatibility antigen, DM beta chain Nicotinamide N-methyltransferase High mobility group protein HMGI-C Methylosome subunit pICln Ubiquitin-conjugating enzyme E2 G1 Ubiquitin-conjugating enzyme E2 G1, N-terminally processed HLA class II histocompatibility antigen, DR beta 3 chain Mitotic spindle assembly checkpoint protein MAD2A E3 ubiquitin-protein ligase TRIP12 Cysteine and glycine-rich protein 2 Mitochondrial 10-formyltetrahydrofolate dehydrogenase G antigen 13 G antigen 2D G antigen 2A G antigen 2B/2C G antigen 12J G antigen 12H G antigen 12B/C/D/E Mitochondrial Rho GTPase 2 Cap-specific mRNA (nucleoside-2-O-)-methyltransferase 1 Protein SAAL1 PITH domain-containing protein 1 Probable serine carboxypeptidase CPVL 39S ribosomal protein L17, mitochondrial | Shootin-1 Monocarboxylate transporter 4 Syndecan-3 Heat shock 70 kDa protein 4L Cholinesterase Annexin A3 Insulin-like growth factor-binding protein 2 Protein S100-A1 Caspase-1 Caspase-1 subunit p20 Caspase-1 subunit p10 Neural cell adhesion molecule L1 Sulfotransferase 1A1 Sulfotransferase 1A2 H(+)/Cl(−) exchange transporter 3 Serine beta-lactamase-like protein LACTB, mitochondrial Collagen alpha-1 (XIV) chain Selenium-binding protein 1 Inactive tyrosine-protein kinase 7 AP2-associated protein kinase 1 NADH-cytochrome b5 reductase 2 Nucleoredoxin Uncharacterized protein FLJ45252 Myosin-14 MICAL-like protein 1 Probable aminopeptidase NPEPL1 PRKC apoptosis WT1 regulator protein Ras-related protein Rab-34 Leucine-rich repeat-containing protein C10orf11 Pleckstrin homology domain-containing family A member 2 Ras-related protein Rab-18 Coatomer subunit zeta-2 Calcium-dependent secretion activator 1 Integral membrane protein 2B BRI2, membrane form BRI2 intracellular domain BRI2C, soluble form | Bri23 peptide FH1/FH2 domain-containing protein 1 Sulfide:quinone oxidoreductase, mitochondrial |
| Present Only in WM793 Exosomes | Present Only in WM1205Lu Exosomes | |
|---|---|---|
| Na(+)/H(+) exchange regulatory cofactor NHE-RF1 Neurosecretory protein VGF Neuroendocrine regulatory peptide-1 Neuroendocrine regulatory peptide-2 Antimicrobial peptide VGF [554–577] Fatty acid-binding protein, brain Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 Heterogeneous nuclear ribonucleoprotein R EGF-like repeat and discoidin I-like domain-containing protein 3 HLA class II histocompatibility antigen, DRB1-15 beta chain Serotransferrin Lupus La protein Beta-hexosaminidase subunit alpha Fumarate hydratase, mitochondrial U1 small nuclear ribonucleoprotein 70 kDa Heterogeneous nuclear ribonucleoprotein A1 Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed Heterogeneous nuclear ribonucleoprotein A1-like 2 Poly [ADP-ribose] polymerase 1 Hyaluronan and proteoglycan link protein 1 X-ray repair cross-complementing protein 5 Aminopeptidase N Follistatin Protachykinin-1 Substance P Neurokinin A Neuropeptide K Neuropeptide gamma C-terminal-flanking peptide Lamin-B1 Splicing factor, proline- and glutamine-rich Myelin protein P0 | Sulfate transporter Palmitoyl-protein thioesterase 1 Hepatoma-derived growth factor 26S protease regulatory subunit 8 HLA class II histocompatibility antigen, DR beta 3 chain Protein SET; Protein SETSIP RNA-binding protein EWS Prolow-density lipoprotein receptor-related protein 1 Low-density lipoprotein receptor-related protein 1 85 kDa subunit Low-density lipoprotein receptor-related protein 1 515 kDa subunit Low-density lipoprotein receptor-related protein 1 intracellular domain ATP-dependent RNA helicase A EGF-containing fibulin-like extracellular matrix protein 1 Sorting nexin-1 CD166 antigen Collagen alpha-1(XIX) chain Polypeptide N-acetylgalactosaminyltransferase 5 Extracellular serine/threonine protein kinase FAM20C Minor histocompatibility antigen H13 Proteasome subunit beta type-7 Acetyl-CoA acetyltransferase, cytosolic Probable serine carboxypeptidase CPVL Protein kinase C and casein kinase substrate in neurons protein 3 Angiopoietin-related protein 2 Basic leucine zipper and W2 domain-containing protein 2 | Fermitin family homolog 3 Ras GTPase-activating-like protein IQGAP3 |
| Present Only in WM115 Exosomes | Present Only in WM266-4 Exosomes | ||
|---|---|---|---|
| Nascent polypeptide-associated complex subunit alpha, muscle-specific form Nascent polypeptide-associated complex subunit alpha Tumor protein D54 Density-regulated protein EGF-like repeat and discoidin I-like domain-containing protein 3 Eukaryotic translation initiation factor 3 subunit G AP-2 complex subunit alpha-2 Activator of 90 kDa heat shock protein ATPase homolog 1 HLA class II histocompatibility antigen, DQ beta 1 chain Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 Tumor necrosis factor receptor superfamily member 16 Neprilysin Solute carrier family 2, facilitated glucose transporter member 3 Solute carrier family 2, facilitated glucose transporter member 14 Protein 4.1 Lysosome-associated membrane glycoprotein 2 Neural cell adhesion molecule 1 HLA class II histocompatibility antigen, DR beta 4 chain Platelet glycoprotein 4 Y-box-binding protein 3 HLA class II histocompatibility antigen, DP alpha 1 chain Cation-dependent mannose-6-phosphate receptor High mobility group protein B2 | Syntaxin-2 RNA-binding motif protein, X chromosome RNA-binding motif protein, X chromosome, N-terminally processed RNA binding motif protein, X-linked-like-1 Macrophage-capping protein Ubiquitin carboxyl-terminal hydrolase 5 60S ribosomal protein L21 Cysteine--tRNA ligase, cytoplasmic Basal cell adhesion molecule UV excision repair protein RAD23 homolog B Cadherin-13 Integrin alpha-1 Myelin proteolipid protein Ubiquitin-conjugating enzyme E2 K 40S ribosomal protein S7 60S ribosomal protein L31 60S ribosomal protein L32 40S ribosomal protein S21 Vigilin Peptidyl-prolyl cis-trans isomerase FKBP4 Peptidyl-prolyl cis-trans isomerase FKBP4, N-terminally processed Caldesmon Tyrosine-protein phosphatase non-receptor type 11 Serine/arginine-rich splicing factor 1 Ras GTPase-activating protein-binding protein 1 Eukaryotic initiation factor 4A-II;Eukaryotic initiation factor 4A-II, N-terminally processed Fibroleukin | Nicotinate-nucleotide pyrophosphorylase [carboxylating] Leucine-rich repeat flightless-interacting protein 1 Syntaxin-12 Secreted frizzled-related protein 1 Misshapen-like kinase 1 Plasminogen activator inhibitor 1 RNA-binding protein ATP-binding cassette sub-family F member 1 Cytoglobin Cullin-5 E3 ubiquitin-protein ligase NEDD4-like Palmitoyltransferase ZDHHC5 Serine/threonine-protein kinase TAO3 Aminopeptidase B Phenylalanine--tRNA ligase beta subunit Transmembrane protein 106B Alpha-parvin ADP-ribosylation factor-like protein 8B Claudin domain-containing protein 1 G-protein coupled receptor family C group 5 member B Teneurin-3 Ribosome-binding protein 1 Testin Transmembrane protein 2 V-type proton ATPase subunit H DCC-interacting protein 13-alpha Ras GTPase-activating protein-binding protein 2 Developmentally-regulated GTP-binding protein 1 V-type proton ATPase subunit D Nicotinate-nucleotide CD82 antigen | Laminin subunit alpha-5 Fatty acid-binding protein, brain Protocadherin-7 Metalloproteinase inhibitor 1 Collagen alpha-2(VI) chain Versican core protein Peptidyl-glycine alpha-amidating monooxygenase;Peptidylglycine alpha-hydroxylating monooxygenase Peptidyl-alpha-hydroxyglycine alpha-amidating lyase Biglycan Fibulin-1 Laminin subunit alpha-2 Thrombospondin-2 Fibrillin-1 Pigment epithelium-derived factor Low-density lipoprotein receptor-related protein 2 Collagen alpha-1(XIV) chain Selenium-binding protein 1 Receptor-type tyrosine-protein phosphatase S Integrin alpha-7;Integrin alpha-7 heavy chain Integrin alpha-7 light chain;Integrin alpha-7 70 kDa form SPARC-like protein 1 Collagen alpha-1(XIX) chain Insulin-like growth factor-binding protein 7 Glutaminyl-peptide cyclotransferase FRAS1-related extracellular matrix protein 2 Polypeptide N-acetylgalactosaminyltransferase 5 Xylosyltransferase 1 Latent-transforming growth factor beta-binding protein 4 Brevican core protein Collagen alpha-1(XII) chain EMILIN-2 Fibroblast growth factor-binding protein 2 Glyoxalase domain-containing protein 4 Angiopoietin-related protein 2 |
| Upregulated in WM793 Cells | Upregulated in WM1205Lu Cells | ||
| Proteins | Fold Change | Proteins | Fold Change |
| Unconventional myosin-Ib | 16.750 | UDP-N-acetylhexosamine pyrophosphorylase | 22.502 |
| Sulfate transporter | 7.4312 | Tight junction protein ZO-1 | 19.663 |
| Inter-alpha-trypsin inhibitor heavy chain H5 | 7.259 | Protein Niban | 18.700 |
| Death-inducer obliterator 1 | 6.965 | Ras GTPase-activating-like protein IQGAP3 | 11.358 |
| Protein TANC1 | 6.833 | Epididymal secretory protein E1 | 9.054 |
| Acetyl-CoA acetyltransferase, cytosolic | 6.364 | Integrin alpha-2 | 8.955 |
| Signal transducer and activator of transcription 3 | 5.631 | Growth/differentiation factor 15 | 8.3022 |
| Elongation factor 1-alpha 2 | 5.212 | Protein-glutamine gamma-glutamyltransferase 2 | 8.283 |
| Superoxide dismutase [Mn], mitochondrial | 4.863 | PDZ and LIM domain protein 7 | 7.297 |
| Farnesyl pyrophosphate synthase | 4.605 | Melanotransferrin | 6.855 |
| Upregulated in WM115 Cells | Upregulated in WM266-4 Cells | ||
| Proteins | Fold Change | Proteins | Fold Change |
| Macrophage-capping protein | 526.300 | Serine/threonine-protein kinase DCLK1 | 28.759 |
| Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | 37.595 | Melanotransferrin | 10.494 |
| Cytoglobin | 7.445 | LIM domain only protein 7 | 9.851 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | 7.287 | CD109 antigen | 7.568 |
| Fibronectin | 5.340 | Fatty acid-binding protein, brain | 6.679 |
| Condensin complex subunit 1 | 4.970 | Protein S100-A13 | 5.543 |
| Structural maintenance of chromosomes protein 2 | 4.576 | 3-ketoacyl-CoA thiolase, mitochondrial | 5.407 |
| Cathepsin L1 | 4.179 | Ras-related protein Rab-8B | 3.668 |
| Myoferlin | 3.939 | Band 4.1-like protein 3 | 3.623 |
| Squalene synthase | 3.245 | Integrin alpha-6 | 3.529 |
| Upregulated in WM115 Exosomes | Upregulated in WM266-4 Exosomes | ||
| Proteins | Fold Change | Proteins | Fold Change |
| Plexin-A1 | 7.044 | Matrilin-2 | 35.037 |
| Transferrin receptor protein 1 | 6.849 | Collagen alpha-1(VI) chain | 25.929 |
| Annexin A1 | 6.160 | Extracellular matrix protein 1 | 23.545 |
| Tyrosine--tRNA ligase, cytoplasmic | 6.085 | C-type lectin domain family 11 member A | 17.627 |
| 60S ribosomal protein L5 | 5.851 | Adipocyte enhancer-binding protein 1 | 17.042 |
| 60S ribosomal protein L7a | 5.456 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 16.935 |
| Erythrocyte band 7 integral membrane protein | 5.422 | Semaphorin-5A | 14.349 |
| 60S ribosomal protein L7 | 5.390 | Vitamin K-dependent protein S | 14.241 |
| Sodium/potassium-transporting ATPase subunit beta-1 | 5.368 | Tenascin | 13.403 |
| A-kinase anchor protein 12 | 5.131 | Agrin | 11.801 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Surman, M.; Jankowska, U.; Wilczak, M.; Przybyło, M. Similarities and Differences in the Protein Composition of Cutaneous Melanoma Cells and Their Exosomes Identified by Mass Spectrometry. Cancers 2023, 15, 1097. https://doi.org/10.3390/cancers15041097
Surman M, Jankowska U, Wilczak M, Przybyło M. Similarities and Differences in the Protein Composition of Cutaneous Melanoma Cells and Their Exosomes Identified by Mass Spectrometry. Cancers. 2023; 15(4):1097. https://doi.org/10.3390/cancers15041097
Chicago/Turabian StyleSurman, Magdalena, Urszula Jankowska, Magdalena Wilczak, and Małgorzata Przybyło. 2023. "Similarities and Differences in the Protein Composition of Cutaneous Melanoma Cells and Their Exosomes Identified by Mass Spectrometry" Cancers 15, no. 4: 1097. https://doi.org/10.3390/cancers15041097
APA StyleSurman, M., Jankowska, U., Wilczak, M., & Przybyło, M. (2023). Similarities and Differences in the Protein Composition of Cutaneous Melanoma Cells and Their Exosomes Identified by Mass Spectrometry. Cancers, 15(4), 1097. https://doi.org/10.3390/cancers15041097






