A Novel Approach for Estimating Ovarian Cancer Tissue Heterogeneity through the Application of Image Processing Techniques and Artificial Intelligence
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Datasets
- T2-weighted DIXON sequences in the axial plane;
- Diffusion-weighted imaging (DWI) in the axial plane.
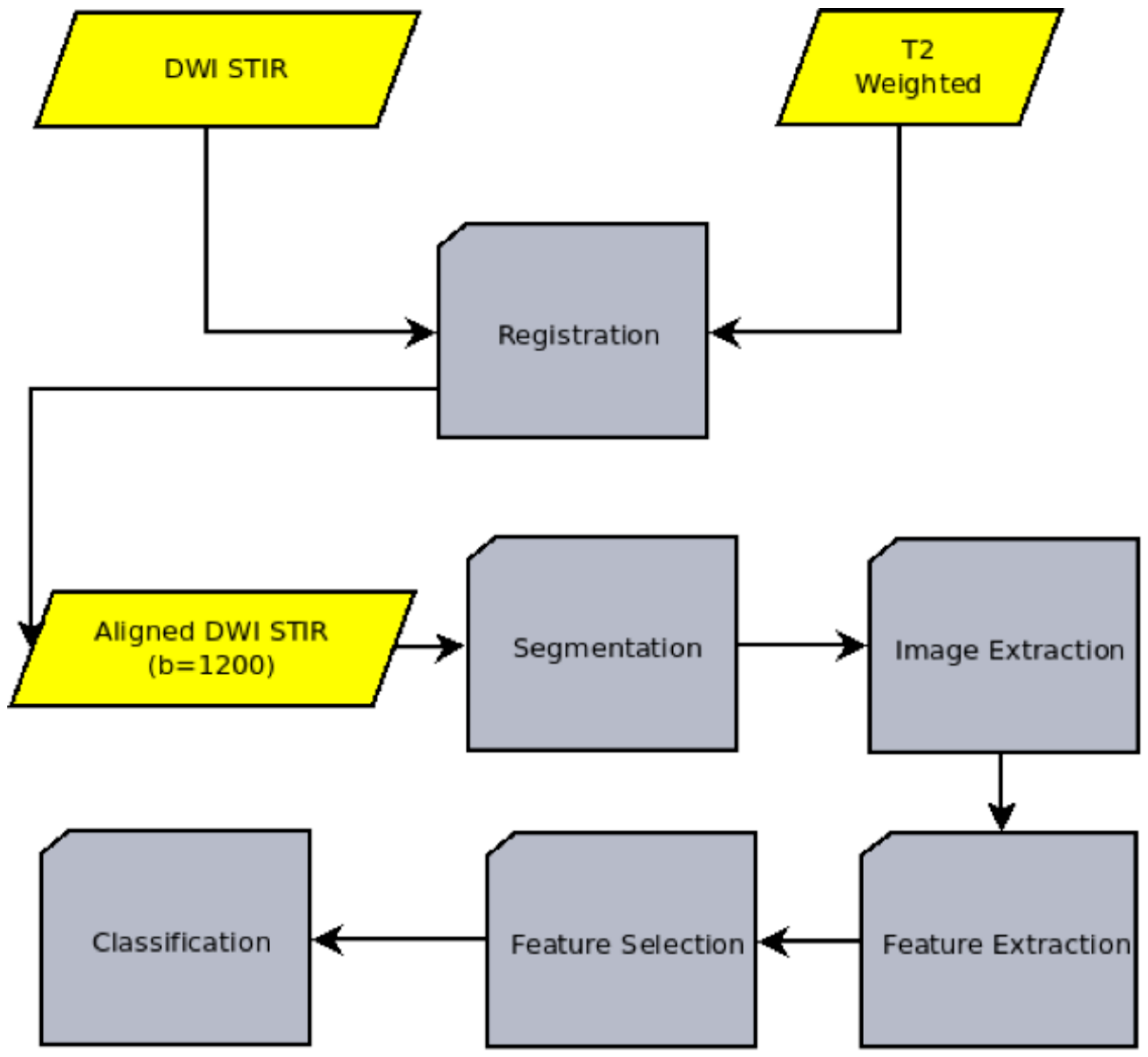
2.2. Algorithm
- Image registration: The process of aligning two or more medical images of the same or different modalities (such as CT, MRI, PET, SPECT, or ultrasound) to analyze them. In our case, the T2-weighted and the DWI images are aligned.
- Image segmentation: The process of dividing an image into multiple regions or segments, each of which corresponds to a different anatomical structure or tissue type. The goal of medical image segmentation is to detect the outline of the cancerous region.
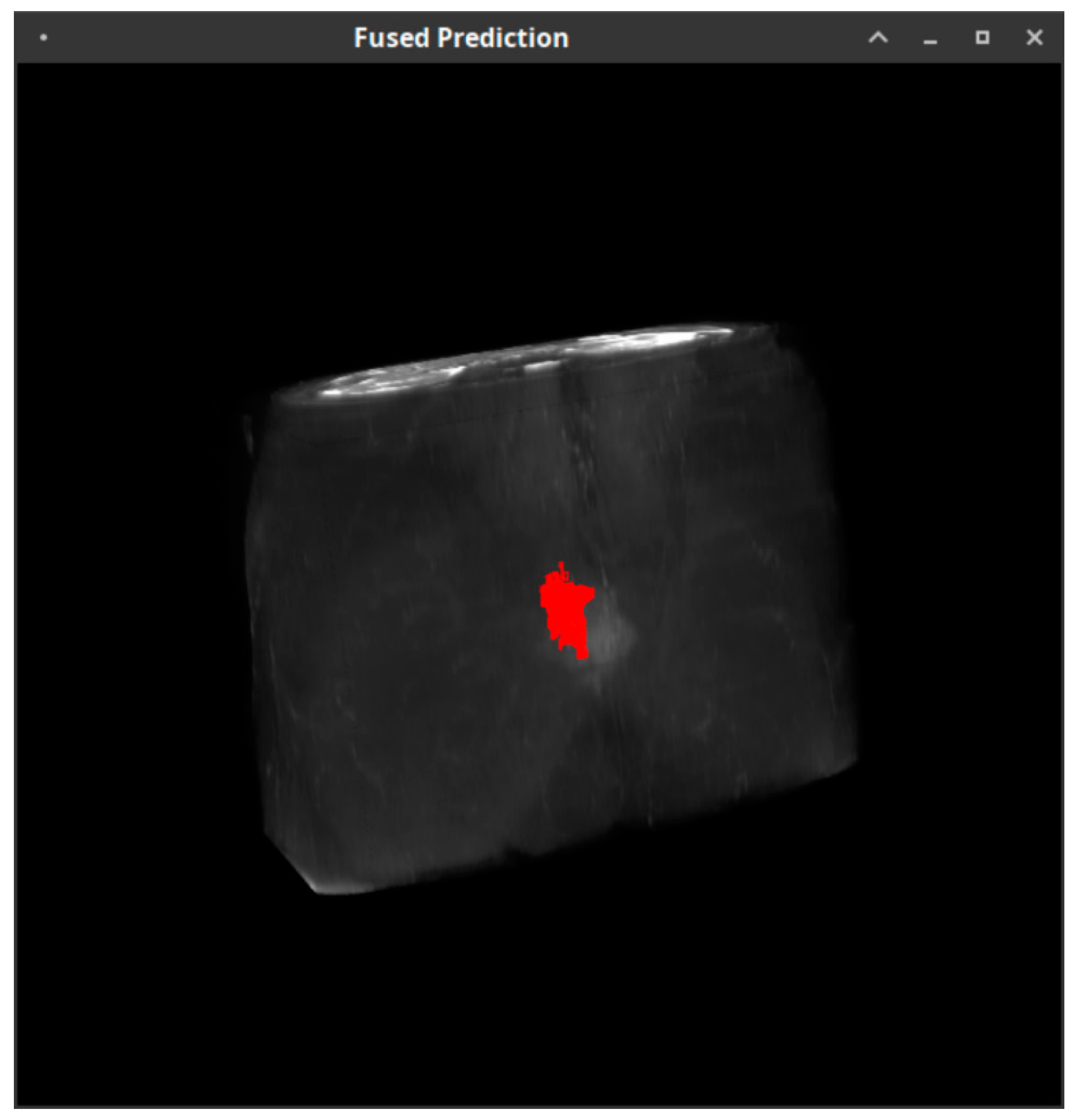
- Image extraction: The process of extracting the cancerous region. The image extraction is needed for both the machine learning pipeline itself and the validation of the whole model through the comparison with the ADC values derived from the DWI sequence.
- Feature extraction: The process of extracting quantitative characteristics, called radiomic features, from the above-mentioned extracted images.
- Feature selection: The process of identifying the subset of the most relevant and useful features from the extracted radiomic features.
- Classification: A machine learning technique that involves training a model to assign a class label to the input data, namely the set of T2-weighted images, on a pixel-by-pixel basis, in an attempt to characterize each region of the data as potentially dangerous or non-dangerous.
2.3. Radiomics
- Wavelet filter: A wavelet filter is a mathematical tool used, in our case, to analyze and process image data signals. It is based on the concept of wavelets, which are small, localized functions that can be used to represent signals at different scales and resolutions. This filter applies a wavelet filter to the input image and yields the decompositions and the approximation.
- Laplacian of Gaussian filter: The particular filter applies a Laplacian of Gaussian filter to the input image and yields a derived image for each sigma value specified. A Laplacian of Gaussian image is obtained by convolving the image with the second derivative (Laplacian) of a Gaussian kernel.
- Square filter: Takes the square of the image intensities and linearly scales them back to the original range. It is used to enhance certain features or attributes of the T2-weighted image, such as sharpness and contrast.
- Square root filter: The square root filter is a type of image processing technique that is used to enhance the contrast and texture of the input image. It takes the square root of the absolute image intensities and scales them back to the original range. The square root filter works by raising each pixel value in the image to the power of 0.5 and has the effect of enhancing the contrast of the image, increasing the visibility of small structures.
- Logarithm filter: The logarithm filter works by applying a logarithmic transformation to the pixel intensity values in the image. This transformation maps the pixel values to a new scale, which can help to stretch out the differences between pixels with relatively small intensity values and those with relatively large intensity values. This can make it easier to distinguish between different tissue types or structures within the image.
- Exponential filter: The exponential filter considers the exponential absolute intensity. It works by applying a weighted average to the pixel values in a local region around a central pixel, with the weighting decreasing exponentially as the distance from the central pixel increases.
- Gradient: The gradient returns the magnitude of the local gradient. The gradient is a measure of the local intensity change or slope of an image. It is calculated by taking the derivative of the intensity values in the image with respect to position. The gradient can provide information about the edges and boundaries of objects in an image, as well as the texture and shape of these objects.
- Local Binary Pattern: This is a simple and efficient method for extracting texture features from images. It works by dividing an image into small cells or neighborhoods and comparing the intensity values of each pixel to the center pixel value. Based on the comparison, the pixel is assigned a binary value (0 or 1). These binary values are then concatenated to form a binary pattern, which is a unique descriptor of the texture in the neighborhood.
- Nineteen features from first-order statistics. First-order statistics are a type of feature that can be extracted from medical images using radiomics techniques. These features describe the overall distribution of intensity values within an image and can provide information about the shape, symmetry and intensity range of the structures in the image. Examples of first-order statistics include the mean, median and standard deviation of the intensity values within an image.
- Shape-based (2D) features. Shape-based features in radiomics involve the measurement and characterization of the shape of structures in the images. These features can include measures of size, shape and spatial relationships, such as the volume, surface area and compactness of a structure. These features are independent of the gray level intensity distribution in the ROI.
- Features from the gray-level cooccurrence matrix describe the second-order joint probability function of an image region. One of these features is autocorrelation. Autocorrelation is a quantitative measure of fineness or coarseness of the texture, which expresses the average intensity difference between a center point and the points of the whole region, indicating the spatial rate of change.
- Sixteen features from the gray-level run-length matrix. This matrix corresponds to the continuous image points that have the same intensity value. One example of such a feature is the run percentage (RP). RP measures the coarseness of the texture by taking the ratio of the number of runs and the number of voxels in a region of interest.
- Sixteen features from gray-level size zone that quantifies gray-level zones in an image. An example of these features is large area emphasis (LAE). LAE is a measure of the distribution of large area size zones, with a greater value indicative of larger size zones and more coarse textures.
- Fourteen features from the gray-level dependence matrix (GLDM) that quantifies gray-level dependencies in an image. An example of these features is dependence non-uniformity (DN). DN measures the similarity of dependence throughout the image, with a lower value indicating more homogeneity among dependencies in the image.
- Five features from the neighboring gray-tone difference matrix, which quantifies the difference between a gray value and the average gray value of its neighbors within a distance δ. An example of these features is coarseness, which has been described above.
2.4. Evaluation Criteria
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.-A.; Fu, L.; Goyeneche, A.; Gao, Z.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chan, W.C.; Ngai, C.H.; Lok, V.; Zhang, L.; Lucero-Prisno, D.E.; Xu, W.; Zheng, Z.-J.; Elcarte, E.; Withers, M.; et al. Worldwide Burden, Risk Factors, and Temporal Trends of Ovarian Cancer: A Global Study. Cancers 2022, 14, 2230. [Google Scholar] [CrossRef] [PubMed]
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.-J.; Bast, R.C.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking Ovarian Cancer II: Reducing Mortality from High-Grade Serous Ovarian Cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef]
- Ramón y Cajal, S.; Sesé, M.; Capdevila, C.; Aasen, T.; de Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical Implications of Intratumor Heterogeneity: Challenges and Opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Zheng, Z.; Yu, T.; Zhao, X.; Gao, X.; Zhao, Y.; Liu, G. Intratumor Heterogeneity: A New Perspective on Colorectal Cancer Research. Cancer Med. 2020, 9, 7637–7645. [Google Scholar] [CrossRef]
- Medeiros, L.R.; Freitas, L.B.; Rosa, D.D.; Silva, F.R.; Silva, L.S.; Birtencourt, L.T.; Edelweiss, M.I.; Rosa, M.I. Accuracy of Magnetic Resonance Imaging in Ovarian Tumor: A Systematic Quantitative Review. Am. J. Obstet. Gynecol. 2011, 204, 67.e1–67.e10. [Google Scholar] [CrossRef]
- Sala, E.; Wakely, S.; Senior, E.; Lomas, D. MRI of Malignant Neoplasms of the Uterine Corpus and Cervix. Am. J. Roentgenol. 2007, 188, 1577–1587. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian Cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Auersperg, N.; Wong, A.S.T.; Choi, K.-C.; Kang, S.K.; Leung, P.C.K. Ovarian Surface Epithelium: Biology, Endocrinology, and Pathology. Endocr. Rev. 2001, 22, 255–288. [Google Scholar] [CrossRef] [PubMed]
- Panayides, A.S.; Amini, A.; Filipovic, N.D.; Sharma, A.; Tsaftaris, S.A.; Young, A.; Foran, D.; Do, N.; Golemati, S.; Kurc, T.; et al. AI in Medical Imaging Informatics: Current Challenges and Future Directions. IEEE J. Biomed. Health Inform. 2020, 24, 1837–1857. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Vargas, H.A.; Wassberg, C.; Fox, J.J.; Wibmer, A.; Goldman, D.A.; Kuk, D.; Gonen, M.; Larson, S.M.; Morris, M.J.; Scher, H.I. Association of Overall Survival with Glycolytic Activity of Castrate-Resistant Prostate Cancer Metastases Response. Radiology 2015, 274, 625. [Google Scholar] [PubMed]
- Vargas, H.A.; Veeraraghavan, H.; Micco, M.; Nougaret, S.; Lakhman, Y.; Meier, A.A.; Sosa, R.; Soslow, R.A.; Levine, D.A.; Weigelt, B.; et al. A Novel Representation of Inter-Site Tumour Heterogeneity from Pre-Treatment Computed Tomography Textures Classifies Ovarian Cancers by Clinical Outcome. Eur. Radiol. 2017, 27, 3991–4001. [Google Scholar] [CrossRef]
- Parekh, V.; Jacobs, M.A. Radiomics: A New Application from Established Techniques. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 207–226. [Google Scholar] [CrossRef]
- Brown, L.G. A Survey of Image Registration Techniques. ACM Comput. Surv. 1992, 24, 325–376. [Google Scholar] [CrossRef]
- Mattes, D.; Haynor, D.R.; Vesselle, H.; Lewellen, T.K.; Eubank, W. PET-CT Image Registration in the Chest Using Free-Form Deformations. IEEE Trans. Med. Imaging 2003, 22, 120–128. [Google Scholar] [CrossRef]
- Rahunathan, S.; Stredney, D.; Schmalbrock, P.; Clymer, B.D. Image Registration Using Rigid Registration and Maximization of Mutual Information. In Proceedings of the 13th Annual Conference on Medicine Meets Virtual Reality, Long Beach, CA, USA, 26–29 January 2005. [Google Scholar]
- Thirion, J.-P. Image Matching as a Diffusion Process: An Analogy with Maxwell’s Demons. Med. Image Anal. 1998, 2, 243–260. [Google Scholar] [CrossRef]
- Savva, A.D.; Economopoulos, T.L.; Matsopoulos, G.K. Geometry-Based vs. Intensity-Based Medical Image Registration: A Comparative Study on 3D CT Data. Comput. Biol. Med. 2016, 69, 120–133. [Google Scholar] [CrossRef]
- Jule, L.T.; Kumar, A.S.; Ramaswamy, K. Micrarray Image Segmentation Using Protracted K-Means Net Algorithm in Enhancement of Accuracy and Robustness. Ann. Rom. Soc. Cell Biol. 2021, 25, 5669–5682. [Google Scholar]
- Taxt, T.; Lundervold, A.; Fuglaas, B.; Lien, H.; Abeler, V. Multispectral Analysis of Uterine Corpus Tumors in Magnetic Resonance Imaging. Magn. Reason. Med. 1992, 23, 55–76. [Google Scholar] [CrossRef]
- Sala, E.; Kataoka, M.Y.; Priest, A.N.; Gill, A.B.; McLean, M.A.; Joubert, I.; Graves, M.J.; Crawford, R.A.F.; Jimenez-Linan, M.; Earl, H.M.; et al. Advanced Ovarian Cancer: Multiparametric MR Imaging Demonstrates Response- and Metastasis-Specific Effects. Radiology 2012, 263, 149–159. [Google Scholar] [CrossRef]
- Nasr, E.; Hamed, I.; Abbas, I.; Khalifa, N.M. Dynamic Contrast Enhanced MRI in Correlation with Diffusion Weighted (DWI) MR for Characterization of Ovarian Masses. Egypt. J. Radiol. Nucl. Med. 2014, 45, 975–985. [Google Scholar] [CrossRef]
- Mansour, S.; Wessam, R.; Raafat, M. Diffusion-Weighted Magnetic Resonance Imaging in the Assessment of Ovarian Masses with Suspicious Features: Strengths and Challenges. Egypt. J. Radiol. Nucl. Med. 2015, 46, 1279–1289. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification Using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine Learning Algorithm Validation with a Limited Sample Size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Zhou, J.; Lu, J.; Gao, C.; Zeng, J.; Zhou, C.; Lai, X.; Cai, W.; Xu, M. Predicting the Response to Neoadjuvant Chemotherapy for Breast Cancer: Wavelet Transforming Radiomics in MRI. BMC Cancer 2020, 20, 100. [Google Scholar] [CrossRef]
- Chaddad, A.; Sabri, S.; Niazi, T.; Abdulkarim, B. Prediction of Survival with Multi-Scale Radiomic Analysis in Glioblastoma Patients. Med. Biol. Eng. Comput. 2018, 56, 2287–2300. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging—“How-to” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Randen, T.; Husoy, J.H. Filtering for Texture Classification: A Comparative Study. IEEE Trans. Pattern Anal. Mach. Intell. 1999, 21, 291–310. [Google Scholar] [CrossRef]
- Aggarwal, N.; Agrawal, R.K. First and Second Order Statistics Features for Classification of Magnetic Resonance Brain Images. J. Signal Inf. Process. 2012, 3, 146–153. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for Multi-Class Classification: An Overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Spelmen, V.S.; Porkodi, R. A Review on Handling Imbalanced Data. In Proceedings of the 2018 International Conference on Current Trends towards Converging Technologies (ICCTCT), Coimbatore, India, 1–3 March 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–11. [Google Scholar]
- Tongman, S.; Chanama, S.; Chanama, M.; Plaimas, K.; Lursinsap, C. Metabolic Pathway Synthesis Based on Predicting Compound Transformable Pairs by Using Neural Classifiers with Imbalanced Data Handling. Expert Syst. Appl. 2017, 88, 45–57. [Google Scholar] [CrossRef]
- “ITK|Insight Toolkit”. Available online: https://itk.org/ (accessed on 25 December 2022).
- “VTK—The Visualization Toolkit”. Available online: https://vtk.org/ (accessed on 25 December 2022).
- “Welcome to Python.org”. Available online: https://www.python.org/ (accessed on 24 May 2022).
- Pattichis, C.S.; Pitris, C.; Liang, J.; Zhang, Y. Guest Editorial on the Special Issue on Integrating Informatics and Technology for Precision Medicine. IEEE J. Biomed. Health Inform. 2019, 23, 12–13. [Google Scholar] [CrossRef]
- Nougaret, S.; McCague, C.; Tibermacine, H.; Vargas, H.A.; Rizzo, S.; Sala, E. Radiomics and Radiogenomics in Ovarian Cancer: A Literature Review. Abdom. Radiol. 2021, 46, 2308–2322. [Google Scholar] [CrossRef]
- Martin-Gonzalez, P.; Crispin-Ortuzar, M.; Rundo, L.; Delgado-Ortet, M.; Reinius, M.; Beer, L.; Woitek, R.; Ursprung, S.; Addley, H.; Brenton, J.D. Integrative Radiogenomics for Virtual Biopsy and Treatment Monitoring in Ovarian Cancer. Insights Imaging 2020, 11, 94. [Google Scholar] [CrossRef]
- Nougaret, S.; Sadowski, E.; Lakhman, Y.; Rousset, P.; Lahaye, M.; Worley, M.; Sgarbura, O.; Shinagare, A.B. The BUMPy Road of Peritoneal Metastases in Ovarian Cancer. Diagn. Interv. Imaging 2022, 103, 448–459. [Google Scholar] [CrossRef]
- Crispin-Ortuzar, M.; Sala, E. Precision Radiogenomics: Fusion Biopsies to Target Tumour Habitats In Vivo. Br. J. Cancer 2021, 125, 778–779. [Google Scholar] [CrossRef]
- Roberts, C.M.; Cardenas, C.; Tedja, R. The Role of Intra-Tumoral Heterogeneity and Its Clinical Relevance in Epithelial Ovarian Cancer Recurrence and Metastasis. Cancers 2019, 11, 1083. [Google Scholar] [CrossRef]
- Shui, L.; Ren, H.; Yang, X.; Li, J.; Chen, Z.; Yi, C.; Zhu, H.; Shui, P. The Era of Radiogenomics in Precision Medicine: An Emerging Approach to Support Diagnosis, Treatment Decisions, and Prognostication in Oncology. Front. Oncol. 2021, 10, 570465. [Google Scholar] [CrossRef] [PubMed]

| Patient | Accuracy | Balanced Accuracy | Sensitivity | Specificity | ||||
|---|---|---|---|---|---|---|---|---|
| SVM | SGD | SVM | SGD | SVM | SGD | SVM | SGD | |
| 1 | 0.55 | 0.78 | 0.69 | 0.55 | 0.44 | 0.97 | 0.94 | 0.12 |
| 2 | 0.99 | 1 | 0.86 | 1 | 0.99 | 1 | 0.73 | 1 |
| 3 | 0.99 | 1 | 0.91 | 0.96 | 0.99 | 1 | 0.81 | 0.92 |
| 4 | 0.98 | 1 | 0.99 | 1 | 0.98 | 1 | 1.00 | 1 |
| 5 | 0.86 | 1 | 0.93 | 1 | 0.86 | 1 | 1.00 | 1 |
| 6 | 0.98 | 1 | 0.50 | 1 | 1.00 | 1 | 0.01 | 1 |
| 7 | 0.63 | 0.68 | 0.58 | 0.77 | 0.67 | 0.59 | 0.49 | 0.94 |
| 8 | 0.99 | 0.98 | 0.99 | 0.99 | 0.99 | 0.98 | 1.00 | 1 |
| 9 | 0.92 | 0.99 | 0.94 | 0.99 | 0.92 | 0.99 | 0.96 | 0.99 |
| 10 | 0.88 | 0.85 | 0.77 | 0.77 | 0.92 | 0.88 | 0.62 | 0.65 |
| 11 | 0.97 | 0.97 | 0.91 | 0.94 | 0.98 | 0.98 | 0.83 | 0.9 |
| 12 | 0.87 | 0.68 | 0.87 | 0.83 | 0.87 | 0.67 | 0.87 | 1 |
| 13 | 0.99 | 0.99 | 0.99 | 0.86 | 0.99 | 0.99 | 1.00 | 0.73 |
| 14 | 0.80 | 0.97 | 0.77 | 0.86 | 0.80 | 0.98 | 0.75 | 0.74 |
| 15 | 0.95 | 0.98 | 0.95 | 0.98 | 0.90 | 0.97 | 1.00 | 0.99 |
| 16 | 0.74 | 0.65 | 0.79 | 0.74 | 0.69 | 0.58 | 0.89 | 0.89 |
| 17 | 0.81 | 0.93 | 0.78 | 0.58 | 0.81 | 0.94 | 0.75 | 0.22 |
| 18 | 0.77 | 0.88 | 0.77 | 0.88 | 0.85 | 0.79 | 0.69 | 0.97 |
| 19 | 0.99 | 0.99 | 0.73 | 0.62 | 0.99 | 0.99 | 0.48 | 0.25 |
| 20 | 0.92 | 0.87 | 0.67 | 0.78 | 0.99 | 0.9 | 0.36 | 0.66 |
| 21 | 0.48 | 0.59 | 0.71 | 0.63 | 0.42 | 0.57 | 1.00 | 0.69 |
| 22 | 0.87 | 0.79 | 0.84 | 0.86 | 0.87 | 0.78 | 0.81 | 0.93 |
| Average | 0.86 | 0.89 | 0.81 | 0.84 | 0.86 | 0.89 | 0.77 | 0.8 |
| ±Std. Dev. | 0.14 | 0.14 | 0.13 | 0.15 | 0.16 | 0.15 | 0.25 | 0.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binas, D.A.; Tzanakakis, P.; Economopoulos, T.L.; Konidari, M.; Bourgioti, C.; Moulopoulos, L.A.; Matsopoulos, G.K. A Novel Approach for Estimating Ovarian Cancer Tissue Heterogeneity through the Application of Image Processing Techniques and Artificial Intelligence. Cancers 2023, 15, 1058. https://doi.org/10.3390/cancers15041058
Binas DA, Tzanakakis P, Economopoulos TL, Konidari M, Bourgioti C, Moulopoulos LA, Matsopoulos GK. A Novel Approach for Estimating Ovarian Cancer Tissue Heterogeneity through the Application of Image Processing Techniques and Artificial Intelligence. Cancers. 2023; 15(4):1058. https://doi.org/10.3390/cancers15041058
Chicago/Turabian StyleBinas, Dimitrios A., Petros Tzanakakis, Theodore L. Economopoulos, Marianna Konidari, Charis Bourgioti, Lia Angela Moulopoulos, and George K. Matsopoulos. 2023. "A Novel Approach for Estimating Ovarian Cancer Tissue Heterogeneity through the Application of Image Processing Techniques and Artificial Intelligence" Cancers 15, no. 4: 1058. https://doi.org/10.3390/cancers15041058
APA StyleBinas, D. A., Tzanakakis, P., Economopoulos, T. L., Konidari, M., Bourgioti, C., Moulopoulos, L. A., & Matsopoulos, G. K. (2023). A Novel Approach for Estimating Ovarian Cancer Tissue Heterogeneity through the Application of Image Processing Techniques and Artificial Intelligence. Cancers, 15(4), 1058. https://doi.org/10.3390/cancers15041058









