CXCR3 Expression Is Associated with Advanced Tumor Stage and Grade Influencing Survival after Surgery of Localised Renal Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Clinical Characteristics
2.2. Tumor Samples and Preparation
2.3. Multiplex Immunofluorescence (IF) Staining
2.4. Single-Cell RNA-Seq Analysis
2.5. Statistical Analysis
3. Results
3.1. Patient and Tumor Characteristics
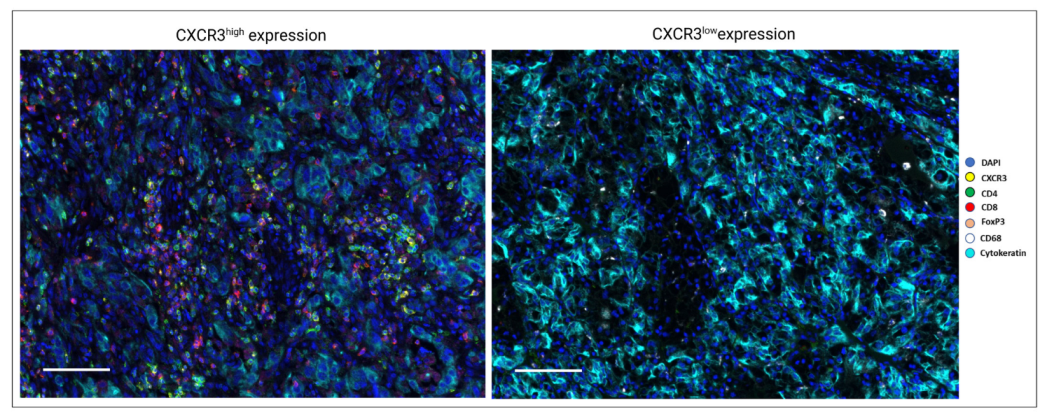
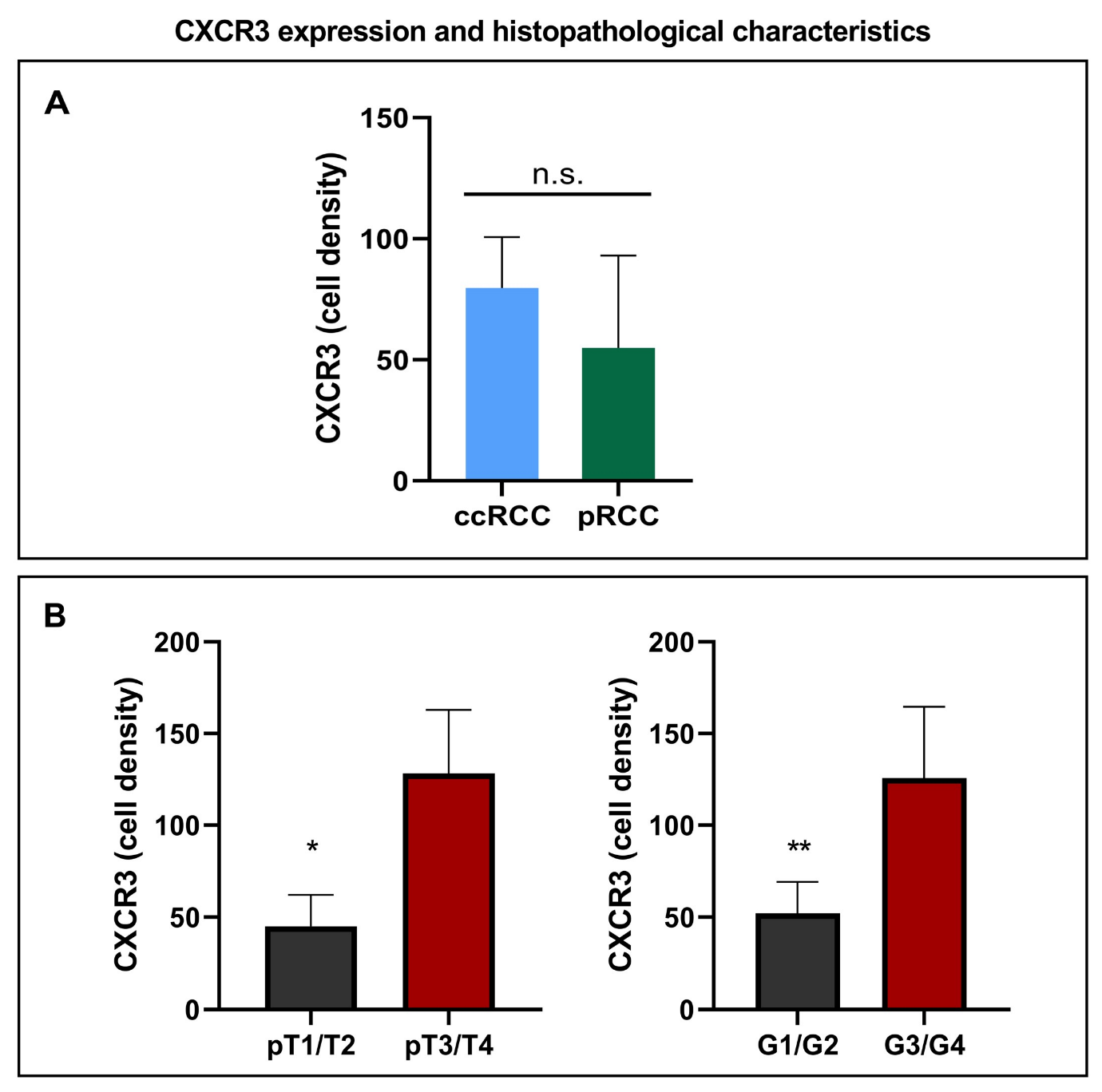
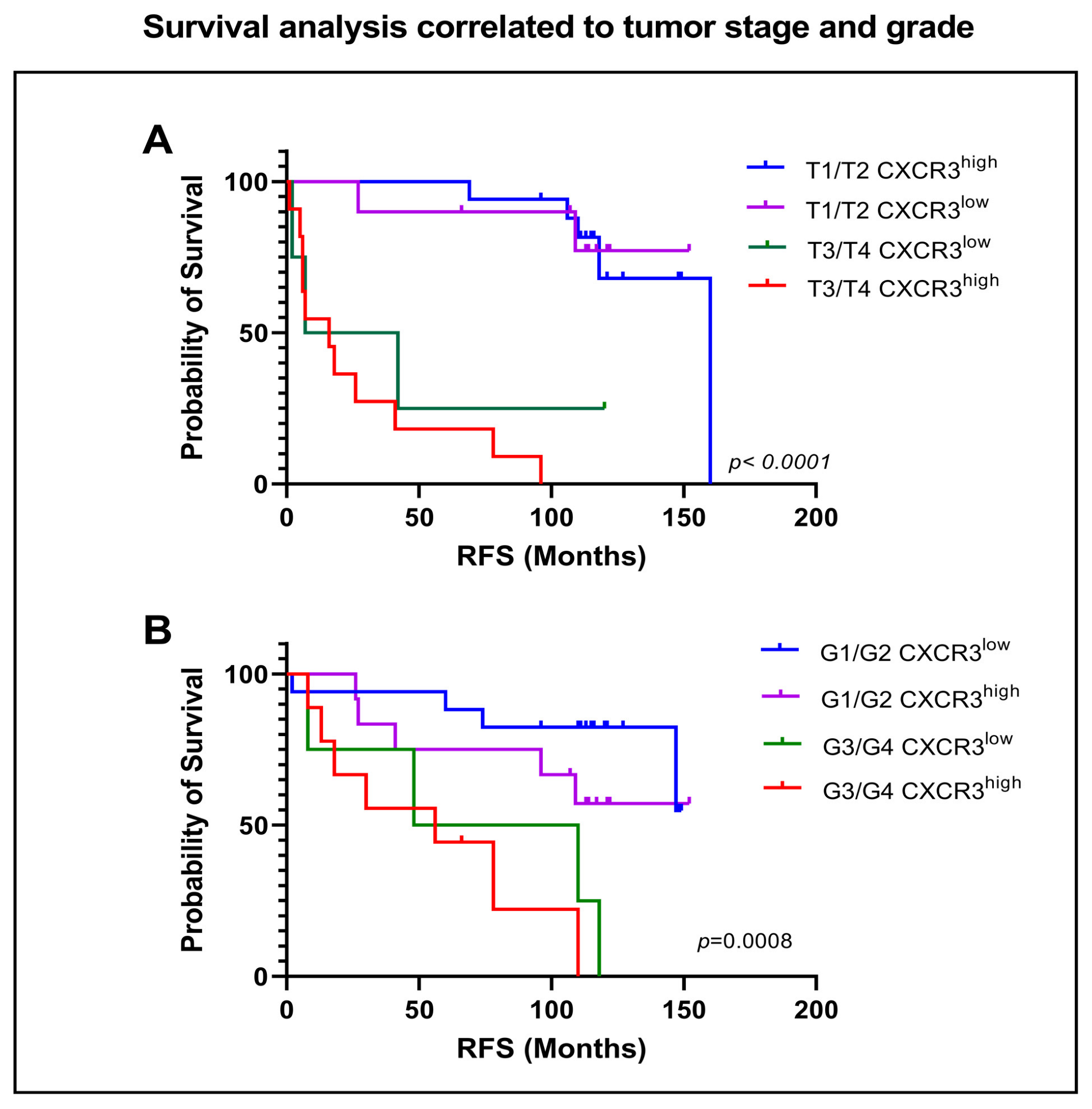
3.2. CXCR3 Expression and Histopathological Characteristics
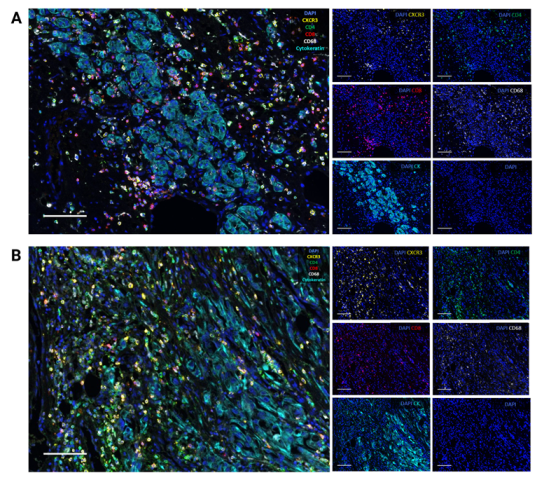
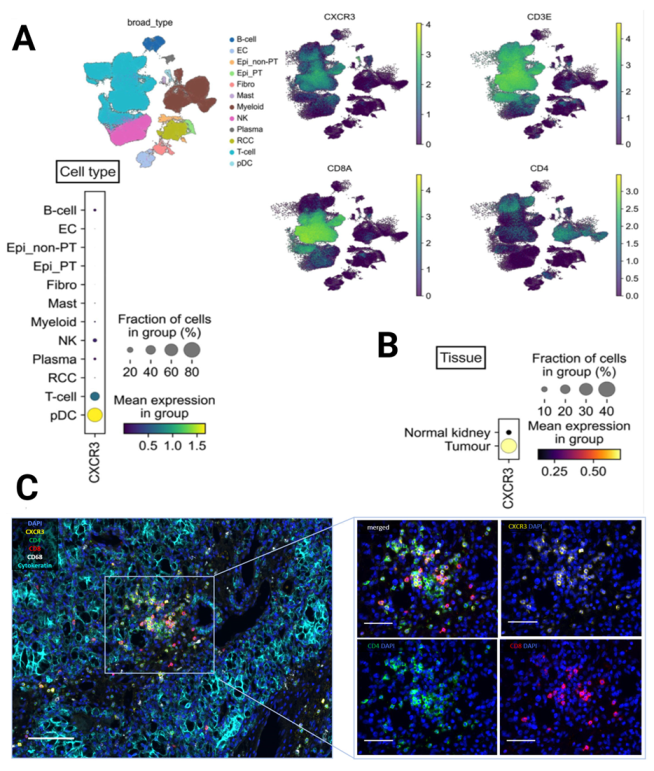
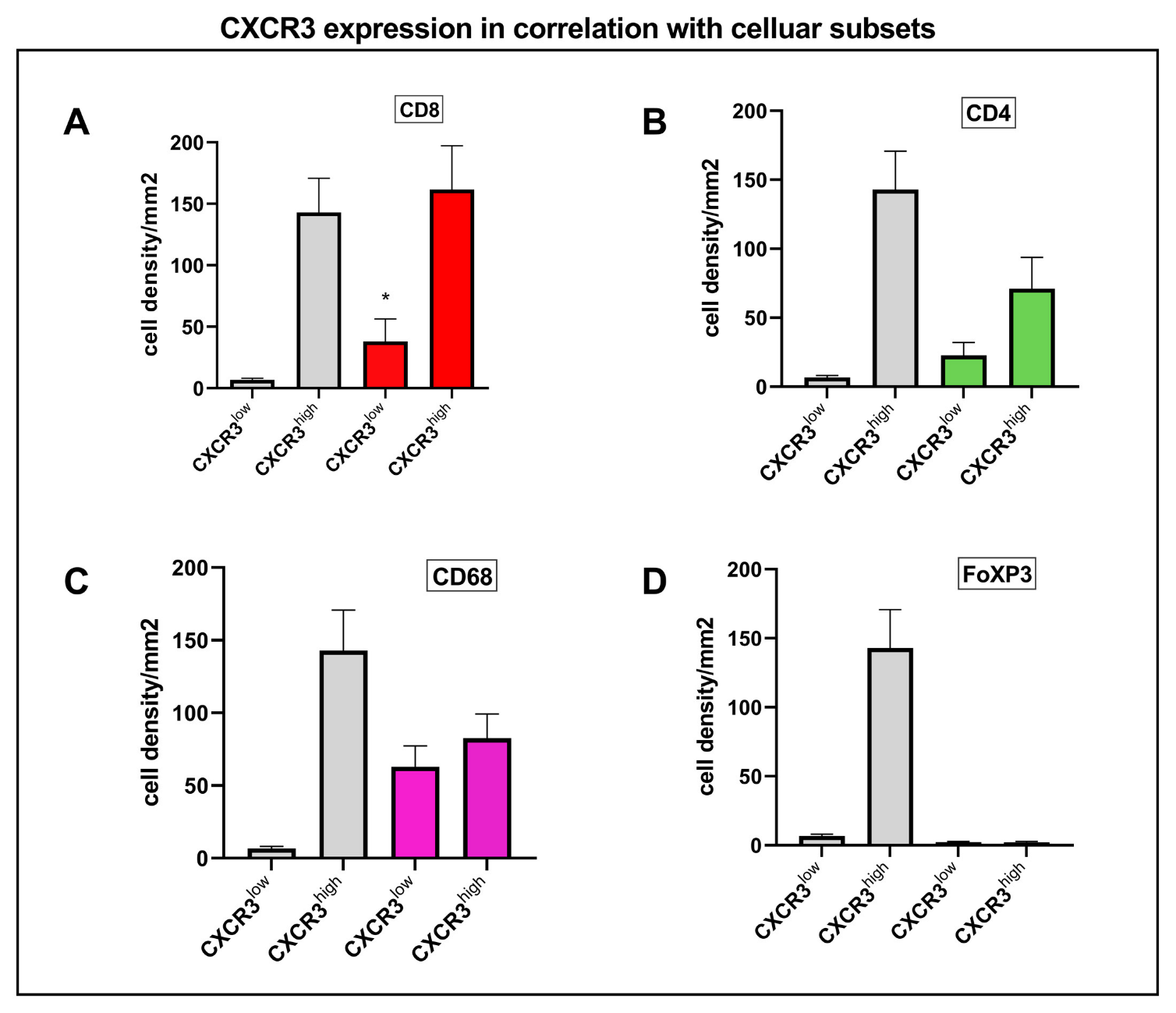
3.3. CXCR3 Is Mainly Expressed on CD8+ and CD4+ T Cells According to Single-Cell RNA-Seq and Multiplex Immunofluorescence
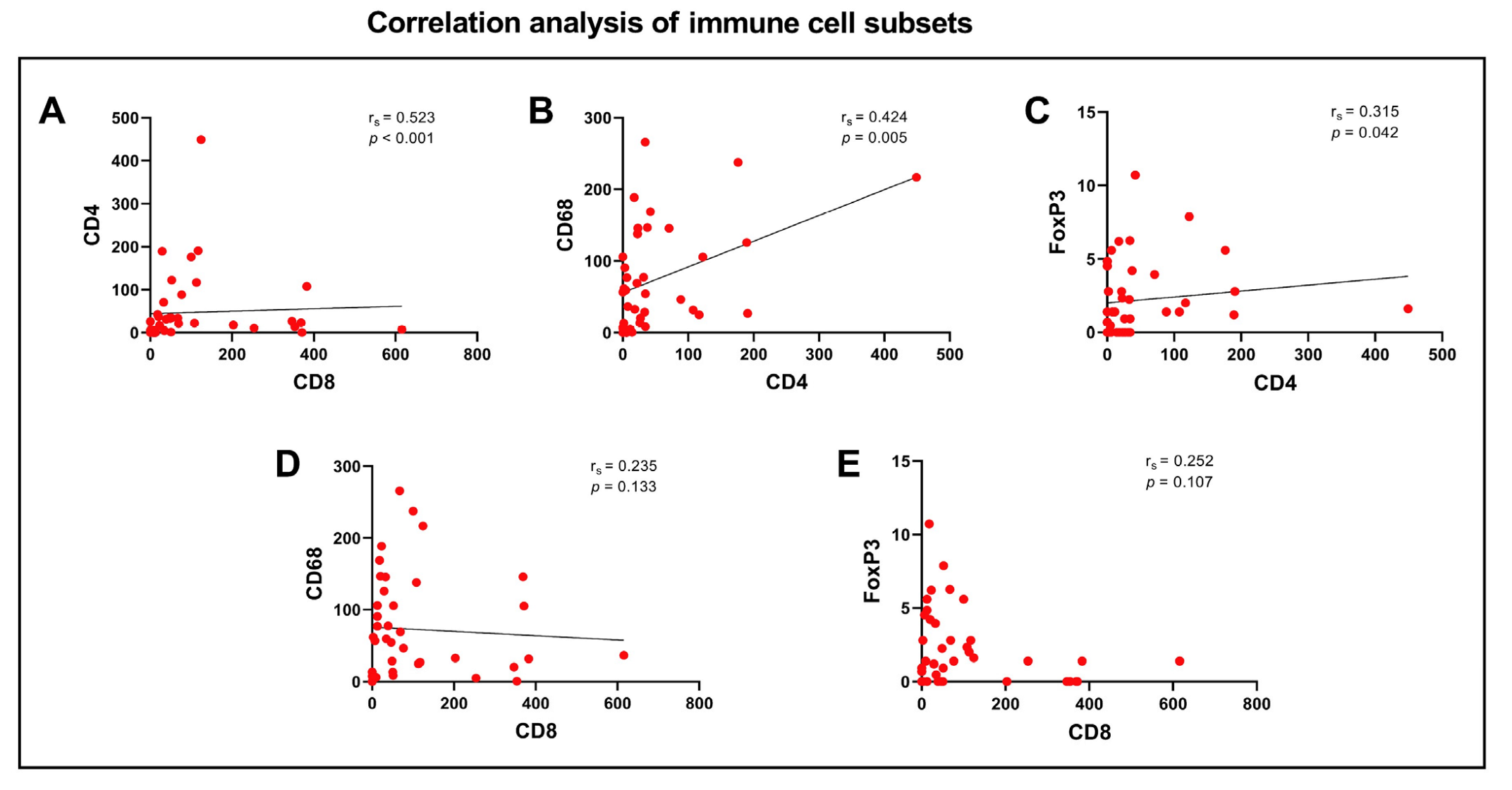
3.4. Correlation Analysis of CXCR3 and Immune Cell Subsets
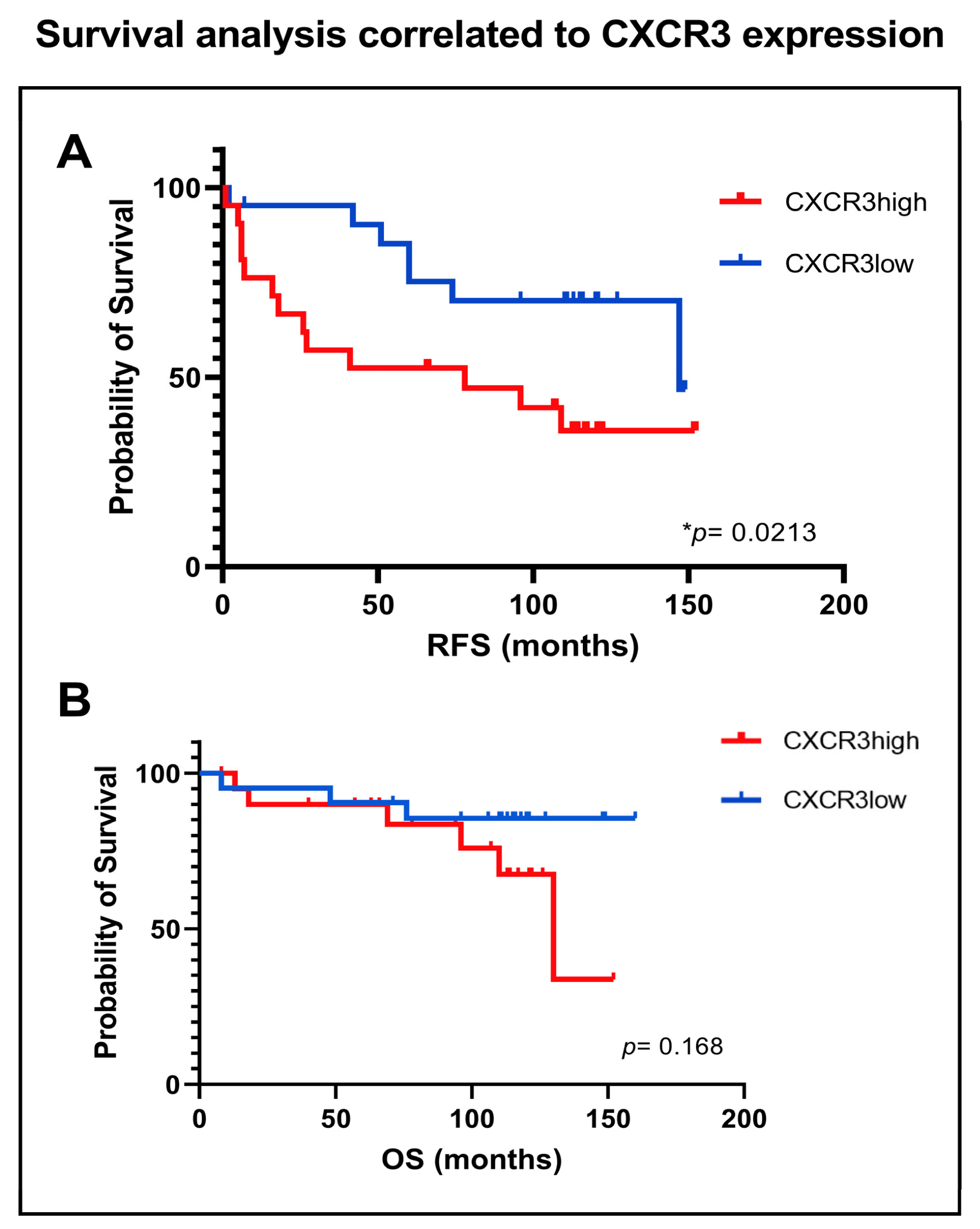
3.5. Influence of CXCR3 Expression on Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bedke, J.; Capitanio, U.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur. Urol. 2022, 82, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Scelo, G.; Larose, T.L. Epidemiology and Risk Factors for Kidney Cancer. J. Clin. Oncol. 2018, 36, JCO2018791905. [Google Scholar] [CrossRef] [PubMed]
- Muglia, V.F.; Prando, A. Renal cell carcinoma: Histological classification and correlation with imaging findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef]
- Decastro, G.J.; McKiernan, J.M. Epidemiology, clinical staging, and presentation of renal cell carcinoma. Urol. Clin. N. Am. 2008, 35, 581–592. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Carrasco, J.C.; Cheng, L.; Scarpelli, M.; Kirkali, Z.; Montironi, R. 2009 update on the classification of renal epithelial tumors in adults. Int. J. Urol. 2009, 16, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Dabestani, S.; Thorstenson, A.; Lindblad, P.; Harmenberg, U.; Ljungberg, B.; Lundstam, S. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: A population-based study. World J. Urol. 2016, 34, 1081–1086. [Google Scholar] [CrossRef]
- Klatte, T.; Rossi, S.H.; Stewart, G.D. Prognostic factors and prognostic models for renal cell carcinoma: A literature review. World J. Urol. 2018, 36, 1943–1952. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Chang, Y.-H.; Hajek, J.; Symeonides, S.N.; Lee, J.L.; Sarwar, N.; et al. Adjuvant Pembrolizumab after Nephrectomy in Renal-Cell Carcinoma. N. Engl. J. Med. 2021, 385, 683–694. [Google Scholar] [CrossRef]
- Powles, T.; Albiges, L.; Bex, A.; Grünwald, V.; Porta, C.; Procopio, G.; Schmidinger, M.; Suárez, C.; de Velasco, G. ESMO Clinical Practice Guideline update on the use of immunotherapy in early stage and advanced renal cell carcinoma. Ann. Oncol. 2021, 32, 1511–1519. [Google Scholar] [CrossRef]
- Powles, T.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Symeonides, S.N.; Hajek, J.; Gurney, H.; Chang, Y.-H.; Lee, J.L.; et al. Pembrolizumab versus placebo as post-nephrectomy adjuvant therapy for clear cell renal cell carcinoma (KEYNOTE-564): 30-month follow-up analysis of a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 1133–1144. [Google Scholar] [CrossRef]
- Pal, S.K.; Uzzo, R.; Karam, J.A.; A Master, V.; Donskov, F.; Suarez, C.; Albiges, L.; Rini, B.; Tomita, Y.; Kann, A.G.; et al. Adjuvant atezolizumab versus placebo for patients with renal cell carcinoma at increased risk of recurrence following resection (IMmotion010): A multicentre, randomised, double-blind, phase 3 trial. Lancet 2022, 400, 1103–1116. [Google Scholar] [CrossRef]
- Nishida, K.; Kawashima, A.; Kanazawa, T.; Kidani, Y.; Yoshida, T.; Hirata, M.; Yamamoto, K.; Yamamoto, Y.; Sawada, M.; Kato, R.; et al. Clinical importance of the expression of CD4+CD8+ T cells in renal cell carcinoma. Int. Immunol. 2020, 32, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Xia, Y.; Lin, Z.; Qu, Y.; Qi, Y.; Chen, Y.; Zhou, Q.; Zeng, H.; Wang, J.; Chang, Y.; et al. Tumor-infiltrating CD39+CD8+ T cells determine poor prognosis and immune evasion in clear cell renal cell carcinoma patients. Cancer Immunol. Immunother. 2020, 69, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Becht, E.; Pagès, F.; Skliris, G.P.; Verkarre, V.; Vano, Y.; Mejean, A.; Saint-Aubert, N.; Lacroix, L.; Natario, I.; et al. Orchestration and Prognostic Significance of Immune Checkpoints in the Microenvironment of Primary and Metastatic Renal Cell Cancer. Clin. Cancer Res. 2015, 21, 3031–3040. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Montero, C.M.; Rini, B.I.; Finke, J.H. The immunology of renal cell carcinoma. Nat. Rev. Nephrol. 2020, 16, 721–735. [Google Scholar] [CrossRef]
- Kurachi, M.; Kurachi, J.; Suenaga, F.; Tsukui, T.; Abe, J.; Ueha, S.; Tomura, M.; Sugihara, K.; Takamura, S.; Kakimi, K.; et al. Chemokine receptor CXCR3 facilitates CD8(+) T cell differentiation into short-lived effector cells leading to memory degeneration. J. Exp. Med. 2011, 208, 1605–1620. [Google Scholar] [CrossRef] [PubMed]
- Bangs, D.J.; Tsitsiklis, A.; Steier, Z.; Chan, S.W.; Kaminski, J.; Streets, A.; Yosef, N.; Robey, E.A. CXCR3 regulates stem and proliferative CD8+ T cells during chronic infection by promoting interactions with DCs in splenic bridging channels. Cell Rep. 2022, 38, 110266. [Google Scholar] [CrossRef]
- Hickman, H.D.; Reynoso, G.V.; Ngudiankama, B.F.; Cush, S.S.; Gibbs, J.; Bennink, J.R.; Yewdell, J.W. CXCR3 chemokine receptor enables local CD8(+) T cell migration for the destruction of virus-infected cells. Immunity 2015, 42, 524–537. [Google Scholar] [CrossRef]
- Liu, L.; Callahan, M.K.; Huang, D.; Ransohoff, R.M. Chemokine receptor CXCR3: An unexpected enigma. Curr. Top. Dev. Biol. 2005, 68, 149–181. [Google Scholar]
- Baggiolini, M. Chemokines and leukocyte traffic. Nature 1998, 392, 565–568. [Google Scholar] [CrossRef]
- Lasagni, L.; Francalanci, M.; Annunziato, F.; Lazzeri, E.; Giannini, S.; Cosmi, L.; Sagrinati, C.; Mazzinghi, B.; Orlando, C.; Maggi, E.; et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J. Exp. Med. 2003, 197, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Sgadari, C.; Farber, J.M.; Angiolillo, A.L.; Liao, F.; Teruya-Feldstein, J.; Burd, P.R.; Yao, L.; Gupta, G.; Kanegane, C.; Tosato, G. Mig, the monokine induced by interferon-gamma, promotes tumor necrosis in vivo. Blood 1997, 89, 2635–2643. [Google Scholar] [CrossRef] [PubMed]
- Luster, A.D.; Leder, P. IP-10, a -C-X-C- chemokine, elicits a potent thymus-dependent antitumor response in vivo. J. Exp. Med. 1993, 178, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Billottet, C.; Quemener, C.; Bikfalvi, A. CXCR3, a double-edged sword in tumor progression and angiogenesis. Biochim. Biophys. Acta 2013, 1836, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Gudowska-Sawczuk, M.; Kudelski, J.; Mroczko, B. The Role of Chemokine Receptor CXCR3 and Its Ligands in Renal Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 8582. [Google Scholar] [CrossRef]
- Suyama, T.; Furuya, M.; Nishiyama, M.; Kasuya, Y.; Kimura, S.; Ichikawa, T.; Ueda, T.; Nikaido, T.; Ito, H.; Ishikura, H. Up-regulation of the interferon gamma (IFN-gamma)-inducible chemokines IFN-inducible T-cell alpha chemoattractant and monokine induced by IFN-gamma and of their receptor CXC receptor 3 in human renal cell carcinoma. Cancer 2005, 103, 258–267. [Google Scholar] [CrossRef]
- Jöhrer, K.; Zelle-Rieser, C.; Perathoner, A.; Moser, P.; Hager, M.; Ramoner, R.; Gander, H.; Höltl, L.; Bartsch, G.; Greil, R.; et al. Up-regulation of functional chemokine receptor CCR3 in human renal cell carcinoma. Clin. Cancer Res. 2005, 11, 2459–2465. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Y.; Fu, Q.; Zhou, L.; Chang, Y.; Xu, L.; Zhang, W.; Xu, J. Elevated expression of IFN-inducible CXCR3 ligands predicts poor prognosis in patients with non-metastatic clear-cell renal cell carcinoma. Oncotarget 2016, 7, 13976–13983. [Google Scholar] [CrossRef]
- Utsumi, T.; Suyama, T.; Imamura, Y.; Fuse, M.; Sakamoto, S.; Nihei, N.; Ueda, T.; Suzuki, H.; Seki, N.; Ichikawa, T. The association of CXCR3 and renal cell carcinoma metastasis. J. Urol. 2014, 192, 567–574. [Google Scholar] [CrossRef]
- Wightman, S.C.; Uppal, A.; Pitroda, S.P.; Ganai, S.; Burnette, B.; Stack, M.; Oshima, G.; Khan, S.; Huang, X.; Posner, M.C.; et al. Oncogenic CXCL10 signalling drives metastasis development and poor clinical outcome. Br. J. Cancer 2015, 113, 327–335. [Google Scholar] [CrossRef]
- Kondo, T.; Ito, F.; Nakazawa, H.; Horita, S.; Osaka, Y.; Toma, H. High expression of chemokine gene as a favorable prognostic factor in renal cell carcinoma. J. Urol. 2004, 171, 2171–2175. [Google Scholar] [CrossRef]
- Koh, H.M.; Hyun, C.L.; Jang, B.G.; Lee, H.J. CXCR3 expression as a prognostic factor in gastric cancer: A meta-analysis. Transl. Cancer Res. 2021, 10, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Leibovich, B.C.; Lohse, C.M.; Cheville, J.C.; Zaid, H.B.; Boorjian, S.A.; Frank, I.; Thompson, R.H.; Parker, W.P. Predicting Oncologic Outcomes in Renal Cell Carcinoma After Surgery. Eur. Urol. 2018, 73, 772–780. [Google Scholar] [CrossRef]
- Leibovich, B.C.; Blute, M.L.; Cheville, J.C.; Lohse, C.M.; Frank, I.; Kwon, E.D.; Weaver, A.L.; Parker, A.S.; Zincke, H. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: A stratification tool for prospective clinical trials. Cancer 2003, 97, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Paner, G.P.; Stadler, W.M.; Hansel, D.E.; Montironi, R.; Lin, D.W.; Amin, M.B. Updates in the Eighth Edition of the Tumor-Node-Metastasis Staging Classification for Urologic Cancers. Eur. Urol. 2018, 73, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, H.; Miller, J.S.; Fajardo, D.A.; Lee, T.K.; Netto, G.J.; Epstein, J.I. Non-invasive papillary urothelial neoplasms: The 2004 WHO/ISUP classification system. Pathol. Int. 2010, 60, 1–8. [Google Scholar] [CrossRef]
- Li, R.; Ferdinand, J.R.; Loudon, K.W.; Bowyer, G.S.; Laidlaw, S.; Muyas, F.; Mamanova, L.; Neves, J.B.; Bolt, L.; Fasouli, E.S.; et al. Mapping single-cell transcriptomes in the intra-tumoral and associated territories of kidney cancer. Cancer Cell 2022, 40, 1583–1599.e10. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.; Li, R. Mapping single cell transcriptomes in kidney cancer. Mendeley Data 2022, V1. [Google Scholar] [CrossRef]
- Xu, C.; Lopez, R.; Mehlman, E.; Regier, J.; Jordan, M.I.; Yosef, N. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst Biol. 2021, 17, e9620. [Google Scholar] [CrossRef]
- Gayoso, A.; Lopez, R.; Xing, G.; Boyeau, P.; Pour Amiri, V.V.; Hong, J.; Wu, K.; Jayasuriya, M.; Mehlman, E.; Langevin, M.; et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 2022, 40, 163–166. [Google Scholar] [CrossRef]
- Fridman, W.H.; Zitvogel, L.; Sautès-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Tanaka, N.; Takamatsu, K.; Hakozaki, K.; Fukumoto, K.; Masuda, T.; Mikami, S.; Shinojima, T.; Kakimi, K.; Tsunoda, T.; et al. Multiplexed single-cell pathology reveals the association of CD8 T-cell heterogeneity with prognostic outcomes in renal cell carcinoma. Cancer Immunol. Immunother. 2021, 70, 3001–3013. [Google Scholar] [CrossRef]
- Woods, A.N.; Wilson, A.L.; Srivinisan, N.; Zeng, J.; Dutta, A.B.; Peske, J.D.; Tewalt, E.F.; Gregg, R.K.; Ferguson, A.R.; Engelhard, V.H. Differential Expression of Homing Receptor Ligands on Tumor-Associated Vasculature that Control CD8 Effector T-cell Entry. Cancer Immunol. Res. 2017, 5, 1062–1073. [Google Scholar] [CrossRef]
- Gunderson, A.J.; Yamazaki, T.; McCarty, K.; Fox, N.; Phillips, M.; Alice, A.; Blair, T.; Whiteford, M.; O’Brien, D.; Ahmad, R.; et al. TGFβ suppresses CD8+ T cell expression of CXCR3 and tumor trafficking. Nat. Commun. 2020, 11, 1749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Liu, F.; Yang, K. Role of CD68 in tumor immunity and prognosis prediction in pan-cancer. Sci. Rep. 2022, 12, 7844. [Google Scholar] [CrossRef] [PubMed]
- Saleh, R.; Elkord, E. FoxP3+ T regulatory cells in cancer: Prognostic biomarkers and therapeutic targets. Cancer Lett. 2020, 490, 174–185. [Google Scholar] [CrossRef]
- Huang, A.C.; Postow, M.A.; Orlowski, R.J.; Mick, R.; Bengsch, B.; Manne, S.; Xu, W.; Harmon, S.; Giles, J.R.; Wenz, B.; et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017, 545, 60–65. [Google Scholar] [CrossRef]
- Zisman, A.; Pantuck, A.J.; Dorey, F.; Said, J.W.; Shvarts, O.; Quintana, D.; Gitlitz, B.J.; Dekernion, J.B.; Figlin, R.A.; Belldegrun, A.S. Improved prognostication of renal cell carcinoma using an integrated staging system. J. Clin. Oncol. 2001, 19, 1649–1657. [Google Scholar] [CrossRef]
- Sternberg, C.N.; Hawkins, R.E.; Wagstaff, J.; Salman, P.; Mardiak, J.; Barrios, C.H.; Zarba, J.J.; Gladkov, O.A.; Lee, E.; Szczylik, C.; et al. A randomised, double-blind phase III study of pazopanib in patients with advanced and/or metastatic renal cell carcinoma: Final overall survival results and safety update. Eur. J. Cancer 2013, 49, 1287–1296. [Google Scholar] [CrossRef]
- Chow, M.T.; Ozga, A.J.; Servis, R.L.; Frederick, D.T.; Lo, J.A.; Fisher, D.E.; Freeman, G.J.; Boland, G.M.; Luster, A.D. Intratumoral Activity of the CXCR3 Chemokine System Is Required for the Efficacy of Anti-PD-1 Therapy. Immunity 2019, 50, 1498–1512.e5. [Google Scholar] [CrossRef]
- Chuah, S.; Lee, J.; Song, Y.; Kim, H.-D.; Wasser, M.; Kaya, N.A.; Bang, K.; Lee, Y.J.; Jeon, S.H.; Suthen, S.; et al. Uncoupling immune trajectories of response and adverse events from anti-PD-1 immunotherapy in hepatocellular carcinoma. J. Hepatol. 2022, 77, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Tsoi, J.; Onyshchenko, M.; Abril-Rodriguez, G.; Ross-Macdonald, P.; Wind-Rotolo, M.; Champhekar, A.; Medina, M.; Torrejon, D.Y.; Shin, D.S.; et al. Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell 2020, 38, 500–515.e3. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Lin, A.; Le Sun, L.; Wei, T.; Ying, H.; Zhang, J.; Luo, P.; Zhu, W. Activation of the chemokine receptor 3 pathway leads to a better response to immune checkpoint inhibitors in patients with metastatic urothelial carcinoma. Cancer Cell Int. 2022, 22, 186. [Google Scholar] [CrossRef] [PubMed]

| Antibodies | ||||
|---|---|---|---|---|
| Name | Citation | Supplier | Cat No. | Clone No. |
| CXCR3 | IF | Abcam | ab288437 | EPR25373-32 |
| CD4 | IF | Cell Marque | 104R-26 | EP204 |
| CD8 | IF | Dako | M710301-2 | C8\144B |
| FoxP3 | IF | Abcam | ab20034 | 236A/E7 |
| CD68 | IF | Dako | M087601-2 | PG-M1 |
| Cytokeratin | IF | Thermo Fisher | MA5-13156 | AE1/AE3 |
| Software | ||||
| Name | Manufacturer | Version | ||
| Mantra Snap | Akoya Biosciences | 1.0.4 | ||
| inForm Analysis | Akoya Biosciences | 2.4.10 | ||
| Reagents & Materials | ||||
| Name | Supplier | Cat no. | ||
| Opal 6-plex Detection Kit | Akoya Biosciences | NEL821001KT | ||
| BOND Polymer Refine Detection | Leica Biosystems | DS9800 | ||
| BOND Epitope Retrival 1 | Leica Biosystems | AR9961 | ||
| BOND Epitope Retrival 2 | Leica Biosystems | AR9640 | ||
| BOND Dewax Solution | Leica Biosystems | AR9222 | ||
| BOND Wash Solution 10× | Leica Biosystems | AR9590 | ||
| Spectral DAPI | Akoya Biosciences | FP1490 | ||
| Prolong Diamond Antifade | Thermo Fisher | P36961 | ||
| BOND Research Detection System | Leica Biosystems | DS9455 | ||
| BOND Titration Kit | Leica Biosystems | OPT9049 | ||
| Opal 6-plex Detection Kit | Akoya Biosciences | NEL821001KT | ||
| BOND Polymer Refine Detection | Leica Biosystems | DS9800 | ||
| BOND Epitope Retrival 1 | Leica Biosystems | AR9961 | ||
| BOND Epitope Retrival 2 | Leica Biosystems | AR9640 | ||
| Factor | Total (n = 42) | Recurrence (n = 20) | No Recurrence (n = 22) | p Value # |
|---|---|---|---|---|
| Age (years), mean (SD) | 62.1 (12.3) | 61 (10.2) | 63.1 (14.1) | 0.588 |
| Male sex, n (%) | 31 (73.8%) | 17 (85%) | 15 (68.2%) | 0.284 |
| BMI, mean (SD) | 26.0 (4.6) | 25.7 (4.7) | 26.4 (4.5) | 0.472 |
| Cigarette Smoking, n (%) | 8 (19%) | 3 (15%) | 5 (22.8%) | 0.700 |
| CKD > grade 3a, n (%) | 6 (14.2%) | 1 (5%) | 5 (22.8%) | 0.187 |
| IDDM type II, n (%) | 8 (19%) | 5 (25%) | 3 (13.6%) | 0.445 |
| Alcohol intake > 3×/month | 23 (54.7%) | 9 (45%) | 12 (54.5%) | 0.232 |
| pT, n (%) | 0.0003 | |||
| pT1/T2 | 27 (64.3%) | 7 (35%) | 20 (90.9%) | |
| pT3/T4 | 15 (35.7%) | 13 (65%) | 2 (9.1%) | |
| Tumor grade, n (%) | 0.002 ** | |||
| G1/G2 | 29 (69%) | 9 (45%) | 20 (90.9%) | |
| G3/G4 | 13 (31%) | 11 (55%) | 2 (9.1%) | |
| Histology, n (%) | 1.000 | |||
| ccRCC | 25 (59.5%) | 12 (60%) | 13 (59.1%) | |
| nccRCC | 17 (40.5%) | 8 (40%) | 9 (40.9%) | |
| Sarcomatoid differentiation, n (%) | 2 (4.8%) | 2 (10%) | 0 (0%) | 0.527 |
| CXCR3, mean (SD) | 74.8 (112.7) | 113.3 (141.1) | 36.4 (55) | 0.0251 * |
| CD4, mean (SD) | 46.9 (82.4) | 38.8 (99) | 54.3 (65.2) | 0.548 |
| CD8, mean (SD) | 99.8 (142.3) | 120.3 (172.6) | 81.3 (108.7) | 0.381 |
| CD68, mean (SD) | 72.8 (70.9) | 72.1 (80.3) | 73.4 (63.1) | 0.957 |
| FoXP3, mean (SD) | 2.2 (2.6) | 1.7 (2.8) | 2.6 (2.3) | 0.263 |
| Follow-up (months), mean (SD) | 96.4 (57) | 76.2 (44.8) | 114.5 (22) | 0.001 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindner, A.K.; Martowicz, A.; Untergasser, G.; Haybaeck, J.; Compérat, E.; Kocher, F.; Seeber, A.; Thurnher, M.; Pichler, R. CXCR3 Expression Is Associated with Advanced Tumor Stage and Grade Influencing Survival after Surgery of Localised Renal Cell Carcinoma. Cancers 2023, 15, 1001. https://doi.org/10.3390/cancers15041001
Lindner AK, Martowicz A, Untergasser G, Haybaeck J, Compérat E, Kocher F, Seeber A, Thurnher M, Pichler R. CXCR3 Expression Is Associated with Advanced Tumor Stage and Grade Influencing Survival after Surgery of Localised Renal Cell Carcinoma. Cancers. 2023; 15(4):1001. https://doi.org/10.3390/cancers15041001
Chicago/Turabian StyleLindner, Andrea Katharina, Agnieszka Martowicz, Gerold Untergasser, Johannes Haybaeck, Eva Compérat, Florian Kocher, Andreas Seeber, Martin Thurnher, and Renate Pichler. 2023. "CXCR3 Expression Is Associated with Advanced Tumor Stage and Grade Influencing Survival after Surgery of Localised Renal Cell Carcinoma" Cancers 15, no. 4: 1001. https://doi.org/10.3390/cancers15041001
APA StyleLindner, A. K., Martowicz, A., Untergasser, G., Haybaeck, J., Compérat, E., Kocher, F., Seeber, A., Thurnher, M., & Pichler, R. (2023). CXCR3 Expression Is Associated with Advanced Tumor Stage and Grade Influencing Survival after Surgery of Localised Renal Cell Carcinoma. Cancers, 15(4), 1001. https://doi.org/10.3390/cancers15041001









