Feasibility Study Utilizing NanoString’s Digital Spatial Profiling (DSP) Technology for Characterizing the Immune Microenvironment in Barrett’s Esophagus Formalin-Fixed Paraffin-Embedded Tissues
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Selection
2.2. Tissue Processing and Hematoxylin and Eosin (H and E) Staining
2.3. Sample Preparation and GeoMx Digital Spatial Profiling
2.3.1. Spatial Transcriptomics Analysis
2.3.2. Spatial Proteomics Analysis
2.4. Strategies for Region of Interest Selection and Segmentation in Corresponding Areas of Interest
2.5. Data Quality Control of Regions of Interest/Areas of Interest and Data Normalization
2.5.1. Protein Quality Control and Normalization
2.5.2. Cancer Transcriptome Analysis Quality Control and Normalization
2.6. Statistical Analysis
3. Results
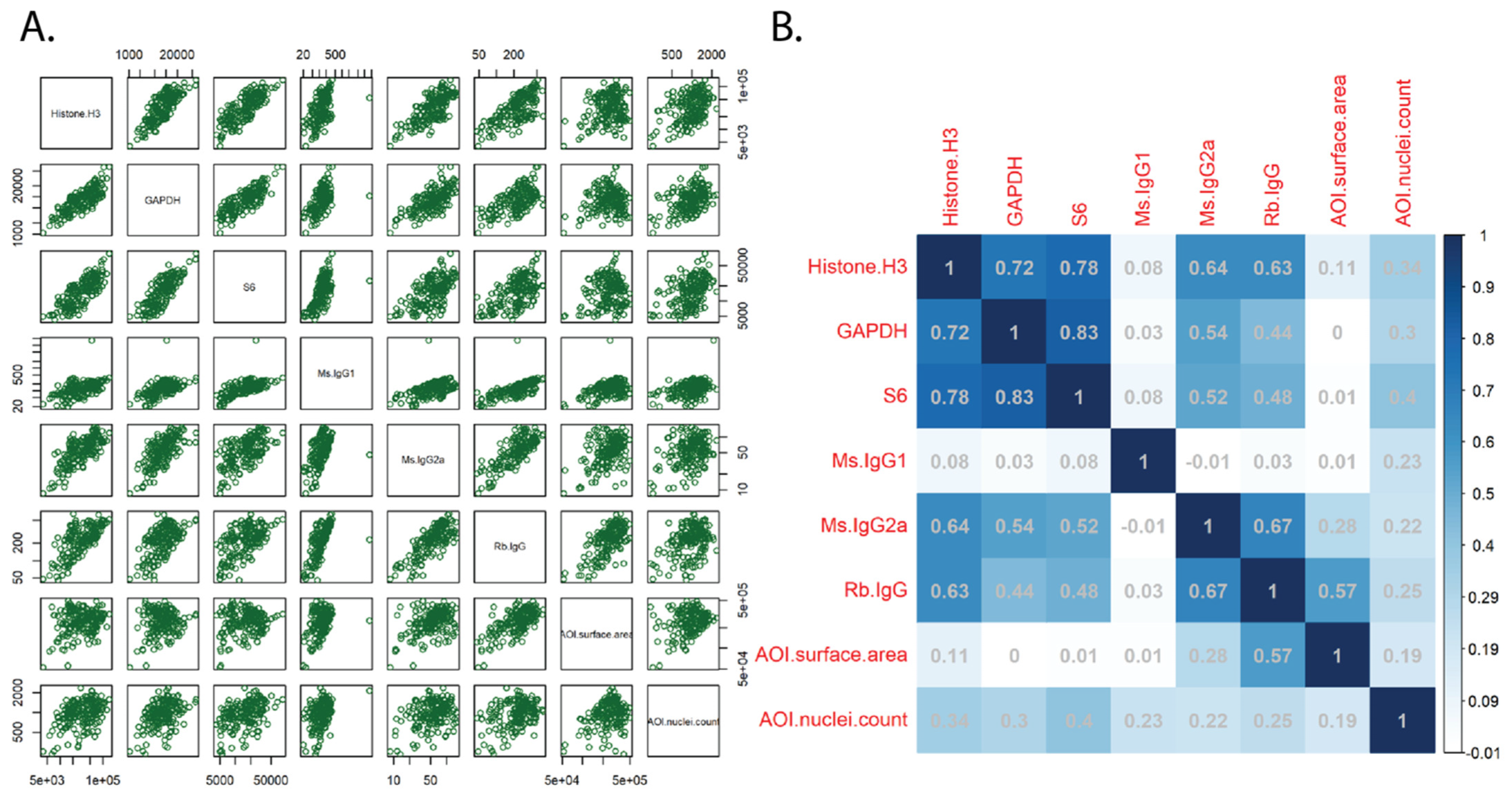
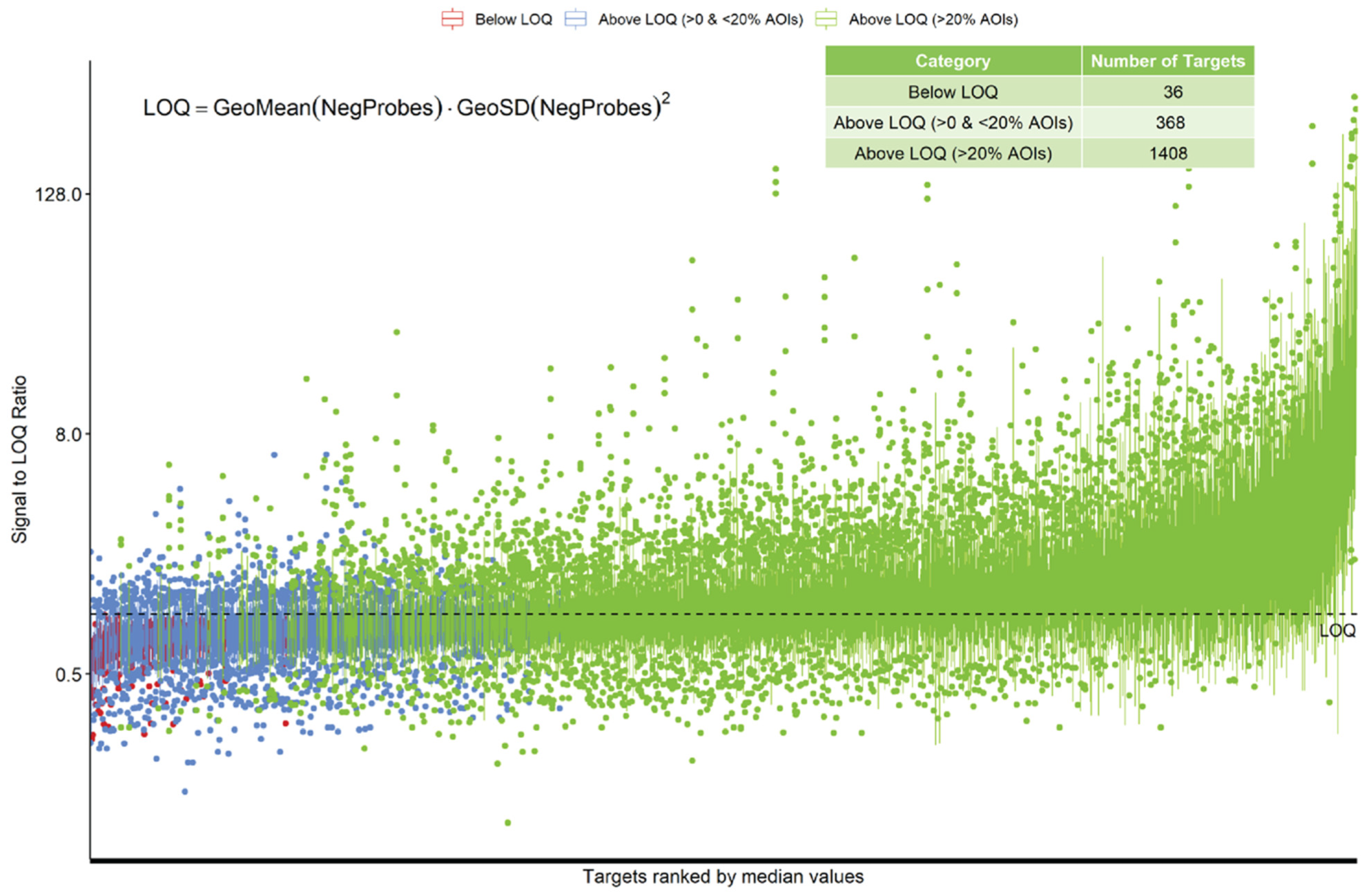
3.1. Feasibility, Normalization, and Quality Control
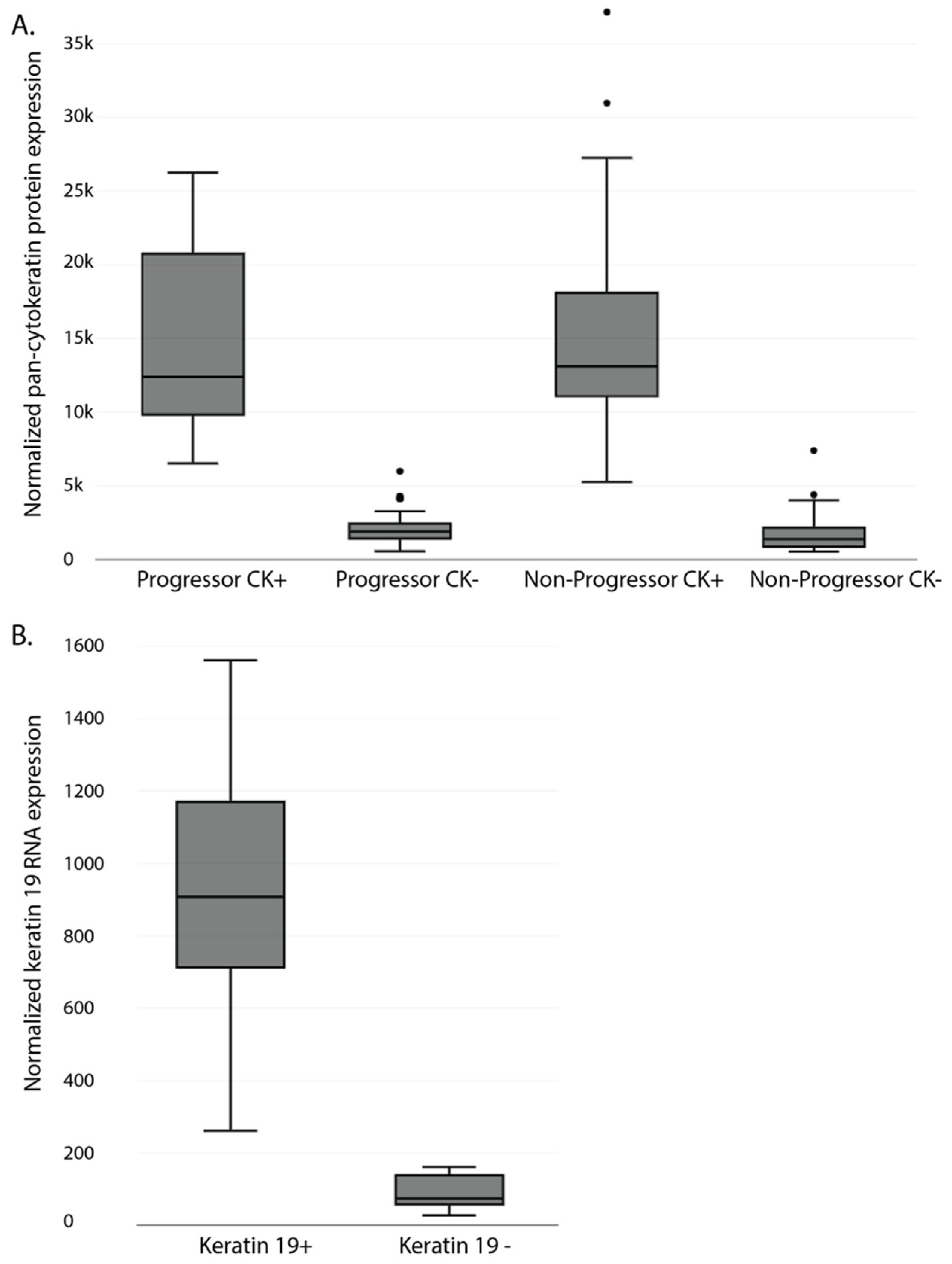
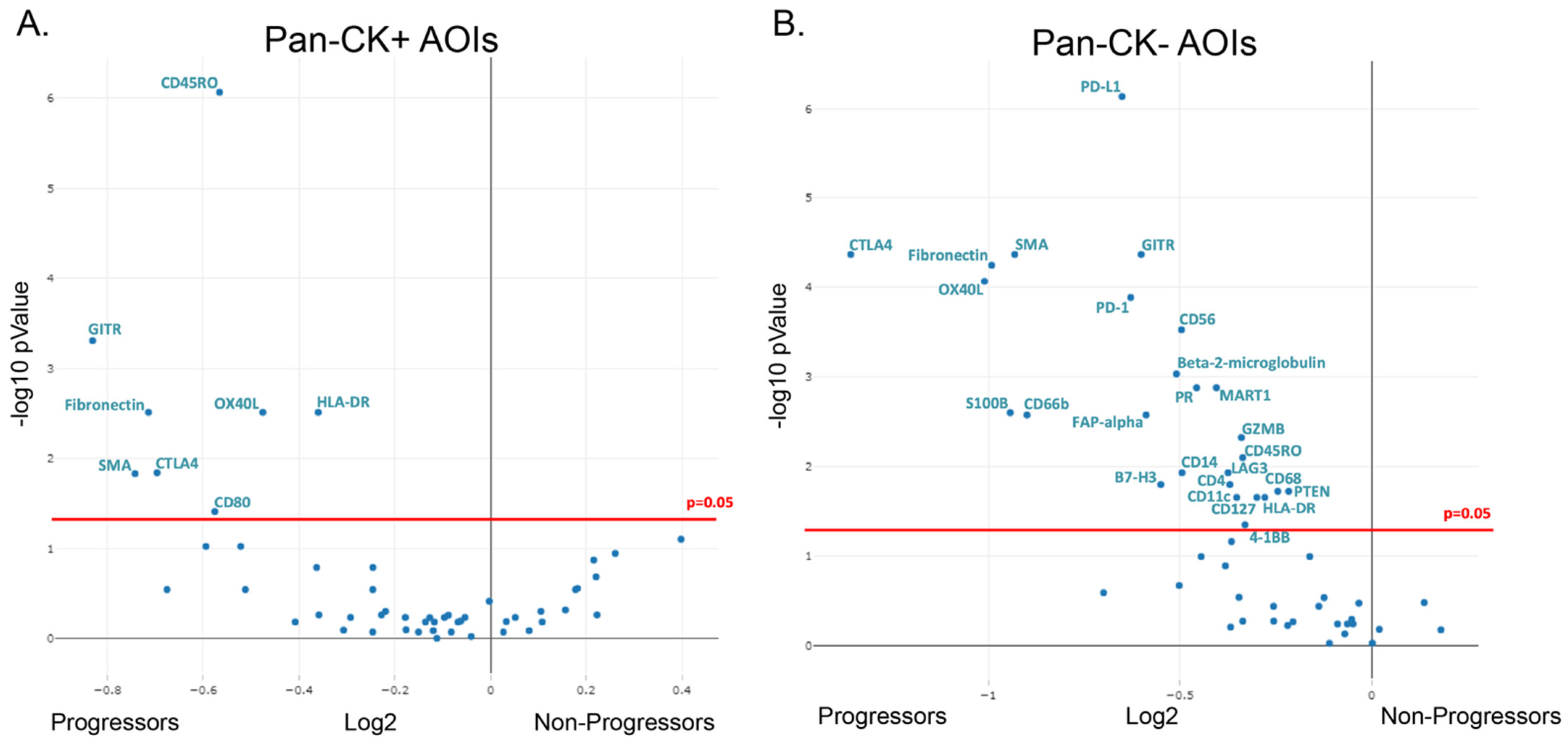
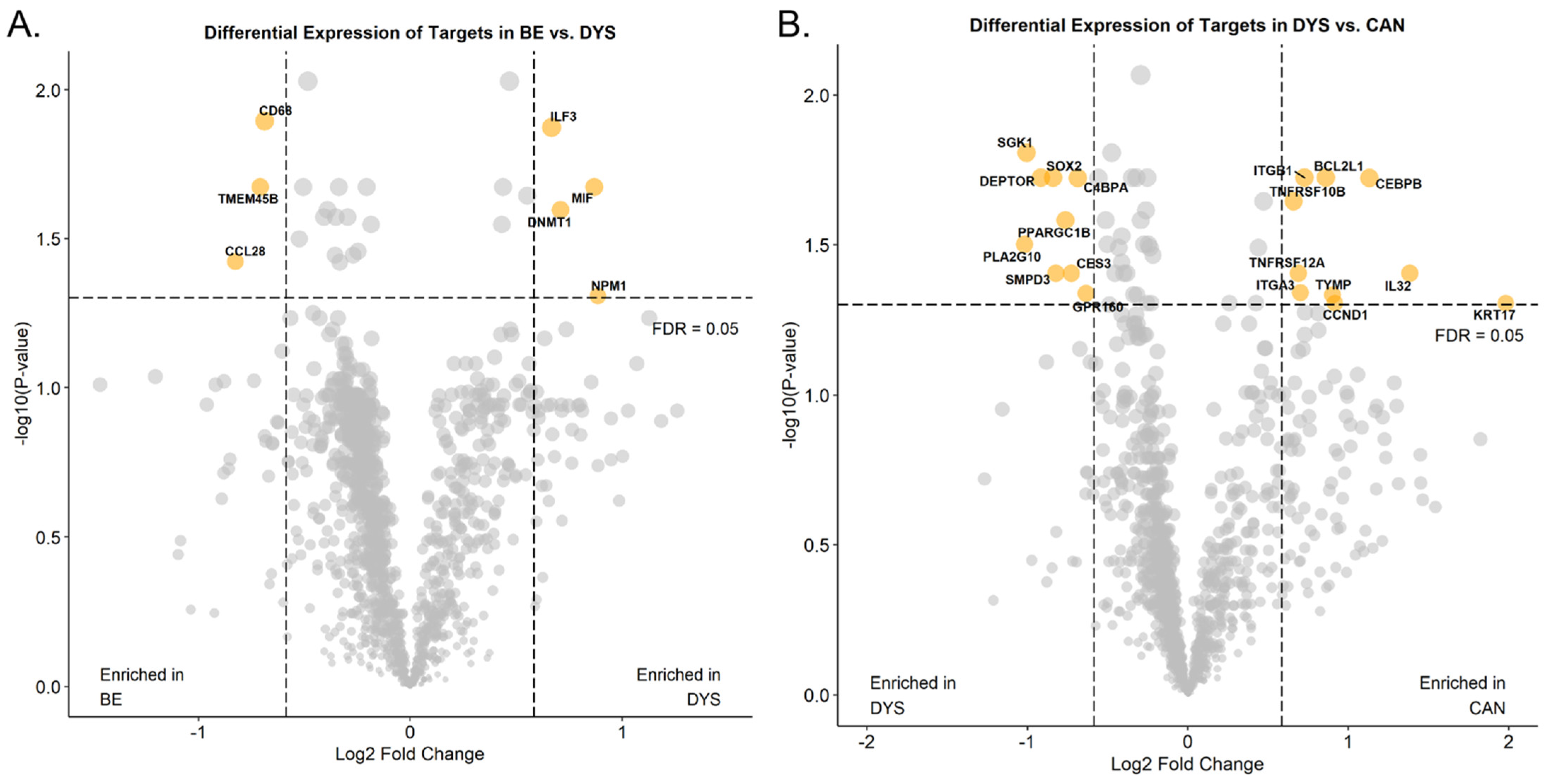
3.2. Differential Protein Expression in Formalin-Fixed Paraffin-Embedded Samples of Non-Dysplastic Barrett’s Esophagus from Progressing and Non-Progression Patients
3.3. Feasibility of Spatial RNA Detection in Formalin-Fixed Paraffin-Embedded Barrett’s Esophagus Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atherfold, P.A.; Janusz, A.J. Molecular biology of Barrett’s Cancer. Best Pract. Res. Clin. Gastroenterol. 2006, 20, 813–827. [Google Scholar] [CrossRef]
- Loos, M.; Langer, R.; Schuster, T.; Gertler, R.; Walch, A.; Rauser, S.; Friess, H.; Feith, M. Clinical Significance of the Costimulatory Molecule B7-H1 in Barrett Carcinoma. Ann Thorac Surg. 2011, 91, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Rai, V.; Abdo, J.; Agrawal, D.K. Biomarkers for Early Detection, Prognosis, and Therapeutics of Esophageal Cancers. Int. J. Mol. Sci. 2023, 24, 3316. [Google Scholar] [CrossRef] [PubMed]
- Uhlenhopp, D.J.; Then, E.O.; Sunkara, T.; Gaduputi, V. Epidemiology of Esophageal Cancer: Update in Global Trends, Etiology and Risk Factors. Clin. J. Gastroenterol. 2020, 13, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.K.; Fitzgerald, R.C. The Horizon of Screening for Barrett’s Esophagus and Esophageal Cancer. Tech. Gastrointest. Endosc. 2023, 25, 146–156. [Google Scholar] [CrossRef]
- Stawinski, P.M.; Dziadkowiec, K.N.; Kuo, L.A.; Echavarria, J.; Saligram, S. Barrett’s Esophagus: An Updated Review. Diagnostics 2023, 13, 321. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, E.W.; Luna, R.A.; Harrison, V.L.; Hunter, J.G. Barrett’s Esophagus: A Review of the Literature. World J. Gastrointest. Surg. 2011, 15, 708–718. [Google Scholar] [CrossRef]
- Kaz, A.M.; Grady, W.M.; Stachler, M.D.; Bass, A.J. Genetic and Epigenetic Alterations in Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterol. Clin. N. Am. 2015, 44, 473–489. [Google Scholar] [CrossRef]
- Luebeck, E.G.; Curtius, K.; Hazelton, W.D.; Maden, S.; Yu, M.; Thota, P.N.; Patil, D.T.; Chak, A.; Willis, J.E.; Grady, W.M. Identification of a Key Role of Widespread Epigenetic Drift In Barrett’s Esophagus and Esophageal Adenocarcinoma. Clin. Epigenetics 2017, 9, 113. [Google Scholar] [CrossRef]
- Stachler, M.D.; Camarda, N.D.; Deitrick, C.; Kim, A.; Agoston, A.T.; Odze, R.D.; Hornick, J.L.; Nag, A.; Thorner, A.R.; Ducar, M.; et al. Detection of Mutations in Barrett’s Esophagus Before Progression to High-Grade Dysplasia or Adenocarcinoma. Gastroenterology 2018, 155, 156–167. [Google Scholar] [CrossRef]
- Maslyonkina, K.S.; Konyukova, A.K.; Alexeeva, D.Y.; Sinelnikov, M.Y.; Mikhaleva, L.M. Barrett’s esophagus: The Pathomorphological and Molecular Genetic Keystones of Neoplastic Progression. Cancer Med. 2022, 11, 447–478. [Google Scholar] [CrossRef] [PubMed]
- Stachler, M.D.; Taylor-Weiner, A.; Peng, S.; McKenna, A.; Agoston, A.T.; Odze, R.D.; Davison, J.M.; Nason, K.S.; Loda, M.; Leshchiner, I.; et al. Paired Exome Analysis of Barrett’s Esophagus and Adenocarcinoma. Nat. Genet. 2015, 47, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.C.; Abdalla, S.; Onwuegbusi, B.A.; Sirieix, P.; Saeed, I.T.; Burnham, W.R.; Farthing, M.J.G. Inflammatory Gradient in Barrett’s Oesophagus: Implications for Disease Complications. Gut 2002, 51, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Shi, J.; Zheng, J.; Li, Y.; Wang, H.; Mi, Y.; Dong, Z. Research Progress in Relationship Between Chronic Inflammation and Development of Esophageal Cancer. Front. Med. 2022, 4, 34–40. [Google Scholar] [CrossRef]
- Kavanagh, M.E.; Conroy, M.J.; Clarke, N.E.; Gilmartin, N.T.; O’Sullivan, K.E.; Feighery, R.; MacCarthy, F.; O'Toole, D.; Ravi, N.; Reynolds, J.V.; et al. Impact of the Inflammatory Microenvironment on T-cell Phenotype in the Progression from Reflux Oesophagitis to Barrett Oesophagus and Oesophageal Adenocarcinoma. Cancer Lett. 2016, 370, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Lagisetty, K.H.; McEwen, D.P.; Nancarrow, D.J.; Schiebel, J.G.; Ferrer-Torres, D.; Ray, D.; Frankel, T.L.; Lin, J.; Chang, A.C.; Kresty, L.A.; et al. Immune Determinants of Barrett’s Progression to Esophageal Adenocarcinoma. JCI Insight 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex Digital Spatial Profiling of Proteins and RNA in Fixed Tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef]
- Loussouarn, D.; Oliver, L.; Salaud, C.; Samarut, E.; Bourgade, R.; Béroud, C.; Morenton, E.; Heymann, D.; Vallette, F.M. Spatial Distribution of Immune Cells in Primary and Recurrent Glioblastoma: A Small Case Study. Cancers 2023, 15, 3256. [Google Scholar] [CrossRef]
- Van, T.M.; Blank, C.U. A User’s Perspective on GeoMxTM Digital Spatial Profiling. Immunooncol. Technol. 2019, 1, 11–18. [Google Scholar] [CrossRef]
- Frei, N.F.; Konté, K.; Duits, L.C.; Klaver, E.; Ten Kate, F.J.; Offerhaus, G.J.; Meijer, S.L.; Visser, M.; Seldenrijk, C.A.; Schoon, E.J.; et al. The SpaTemp Cohort: 168 Nondysplastic Barrett’s Esophagus Surveillance Patients with and without Progression to Early Neoplasia to Evaluate the Distribution of Biomarkers Over Space and time. Dis. Esophagus 2021, 34, doaa095. [Google Scholar] [CrossRef]
- Larson, K.; Ho, H.H.; Anumolu, P.L.; Chen, T.M. Hematoxylin and Eosin Tissue Stain in Mohs Micrographic Surgery: A Review. Dermatol. Surg. 2011, 37, 1089–1099. [Google Scholar] [CrossRef]
- Bergholtz, H.; Carter, J.M.; Cesano, A.; Cheang, M.C.U.; Church, S.E.; Divakar, P.; Fuhrman, C.A.; Goel, S.; Gong, J.; Guerriero, J.L.; et al. On Behalf of the GeoMx Breast Cancer Consortium. Best Practices for Spatial Profiling for Breast Cancer Research with the GeoMx® Digital Spatial Profiler. Cancers 2021, 13, 4456. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chen, T.; Destenaves, B. Quantitative RNA Assessment and Long-Term Stability in the FFPE Tumor Samples using Digital Spatial Profiler. Immuno-Oncol. Technol. 2022, 13, 100069. [Google Scholar] [CrossRef] [PubMed]
- Beasley, G.M.; Therien, A.D.; Holl, E.K.; Al-Rohil, R.; Selim, M.A.; Farrow, N.E.; Pan, L.; Haynes, P.; Liang, Y.; Tyler, D.S.; et al. Dissecting the Immune Landscape of Tumor Draining Lymph Nodes in Melanoma with High-Plex Spatially Resolved Protein Detection. Cancer Immunol. Immunother. 2021, 70, 475–483. [Google Scholar] [CrossRef] [PubMed]
- van Hijfte, L.; Geurts, M.; Vallentgoed, W.R.; Eilers, P.H.C.; Sillevis Smitt, P.A.E.; Debets, R.; French, P.J. Alternative Normalization and Analysis Pipeline to Address Systematic Bias in Nano String GeoMx Digital Spatial Profiling Data. iScience 2022, 26, 105760. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Farooqi, M.S.; Habeebu, S.S.; Gonzalez, E.; Flatt, T.G.; Wilson, A.L.; Barr, F.G. NanoString Digital Molecular Profiling of Protein and microRNA in Rhabdomyosarcoma. Cancers 2022, 14, 522. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Monkman, J.; Shah, E.T.; Matigian, N.; Adams, M.N.; O’byrne, K. Spatial Profiling Identifies Prognostic Features of Response to Adjuvant Therapy in Triple Negative Breast Cancer (TNBC). Front. Oncol. 2022, 11, 798296. [Google Scholar] [CrossRef]
- Monkman, J.; Taheri, T.; Warkiani, M.E.; O’leary, C.; Ladwa, R.; Richard, D.; O’byrne, K.; Kulasinghe, A. High-Plex and High-Throughput Digital Spatial Profiling of Non-Small-Cell Lung Cancer (NSCLC). Cancers 2020, 12, 3551. [Google Scholar] [CrossRef]
- Brady, L.; Kriner, M.; Coleman, I.; Morrissey, C.; Roudier, M.; True, L.D.; Gulati, R.; Plymate, S.R.; Zhou, Z.; Birditt, B.; et al. Inter- and Intra-Tumor Heterogeneity of Metastatic Prostate Cancer Determined by Digital Spatial Gene Expression Profiling. Nat. Commun. 2021, 12, 1426. [Google Scholar] [CrossRef]
- Toki, M.I.; Merritt, C.R.; Wong, P.F.; Smithy, J.W.; Kluger, H.M.; Syrigos, K.N.; Ong, G.T.; Warren, S.E.; Beechem, J.M.; Rimm, D.L. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy-Treated Patients Using Digital Spatial Profiling. Clin. Cancer Res. 2019, 25, 5503–5512. [Google Scholar] [CrossRef]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B Cells and Tertiary Lymphoid Structures Promote Immunotherapy Response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Zimmerman, S.M.; Fropf, R.; Kulasekara, B.R.; Griswold, M.; Appelbe, O.; Bahrami, A.; Boykin, R.; Buhr, D.L.; Fuhrman, K.; Hoang, M.L.; et al. Spatially Resolved Whole Transcriptome Profiling in Human and Mouse Tissue Using Digital Spatial Profiling. Genome Res. 2022, 32, 1892–1905. [Google Scholar] [CrossRef]
- Peleg, N.; Schmilovitz-Weiss, H.; Shamah, S.; Schwartz, A.; Dotan, I.; Sapoznikov, B. Neutrophil to lymphocyte ratio and risk of neoplastic progression in patients with Barrett’s esophagus. Endoscopy 2021, 53, 774–781. [Google Scholar] [CrossRef]
- Zhang, H.; Dai, Z.; Wu, W.; Wang, Z.; Zhang, N.; Zhang, L.; Zeng, W.J.; Liu, Z.; Cheng, Q. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 1–22. [Google Scholar] [CrossRef]
- Derks, S.; Nason, K.S.; Liao, X.; Stachler, M.D.; Liu, K.X.; Bin Liu, J.; Sicinska, E.; Goldberg, M.S.; Freeman, G.J.; Rodig, S.J.; et al. Epithelial PD-L2 Expression Marks Barrett’s Esophagus and Esophageal Adenocarcinoma. Caner Immunol. Res. 2015, 3, 1123–1129. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qurat-ul-Ain; Frei, N.F.; Khoshiwal, A.M.; Stougie, P.; Odze, R.; Camilleri-Broet, S.; Ferri, L.; Duits, L.C.; Bergman, J.; Stachler, M.D. Feasibility Study Utilizing NanoString’s Digital Spatial Profiling (DSP) Technology for Characterizing the Immune Microenvironment in Barrett’s Esophagus Formalin-Fixed Paraffin-Embedded Tissues. Cancers 2023, 15, 5895. https://doi.org/10.3390/cancers15245895
Qurat-ul-Ain, Frei NF, Khoshiwal AM, Stougie P, Odze R, Camilleri-Broet S, Ferri L, Duits LC, Bergman J, Stachler MD. Feasibility Study Utilizing NanoString’s Digital Spatial Profiling (DSP) Technology for Characterizing the Immune Microenvironment in Barrett’s Esophagus Formalin-Fixed Paraffin-Embedded Tissues. Cancers. 2023; 15(24):5895. https://doi.org/10.3390/cancers15245895
Chicago/Turabian StyleQurat-ul-Ain, Nicola F. Frei, Amir M. Khoshiwal, Pim Stougie, Robert Odze, Sophie Camilleri-Broet, Lorenzo Ferri, Lucas C. Duits, Jacques Bergman, and Matthew D. Stachler. 2023. "Feasibility Study Utilizing NanoString’s Digital Spatial Profiling (DSP) Technology for Characterizing the Immune Microenvironment in Barrett’s Esophagus Formalin-Fixed Paraffin-Embedded Tissues" Cancers 15, no. 24: 5895. https://doi.org/10.3390/cancers15245895
APA StyleQurat-ul-Ain, Frei, N. F., Khoshiwal, A. M., Stougie, P., Odze, R., Camilleri-Broet, S., Ferri, L., Duits, L. C., Bergman, J., & Stachler, M. D. (2023). Feasibility Study Utilizing NanoString’s Digital Spatial Profiling (DSP) Technology for Characterizing the Immune Microenvironment in Barrett’s Esophagus Formalin-Fixed Paraffin-Embedded Tissues. Cancers, 15(24), 5895. https://doi.org/10.3390/cancers15245895






