Simple Summary
Cancer treatments have made remarkable advances with the introduction of immunotherapy, which recruits the body’s immune system to fight cancer. Despite these advancements, cancer can sometimes develop resistance to such treatments, diminishing their effectiveness. Our research is focused on the early detection of signs that indicate a cancer’s resistance to immunotherapy, enabling physicians to swiftly alter treatment approaches and improve the chances of patient recovery. We are particularly keen on identifying distinct markers in tumors that indicate this resistance. To achieve a deeper understanding, we utilized scaled-down models of patient tumors, including organoids and xenografts, in laboratory studies. Our goal was to discover innovative methods to combat treatment resistance, potentially enhancing patient care and providing valuable insights for ongoing cancer research.
Abstract
Cancer immunotherapy has ushered in a transformative era in oncology, offering unprecedented promise and opportunities. Despite its remarkable breakthroughs, the field continues to grapple with the persistent challenge of treatment resistance. This resistance not only undermines the widespread efficacy of these pioneering treatments, but also underscores the pressing need for further research. Our exploration into the intricate realm of cancer immunotherapy resistance reveals various mechanisms at play, from primary and secondary resistance to the significant impact of genetic and epigenetic factors, as well as the crucial role of the tumor microenvironment (TME). Furthermore, we stress the importance of devising innovative strategies to counteract this resistance, such as employing combination therapies, tailoring immune checkpoints, and implementing real-time monitoring. By championing these state-of-the-art methods, we anticipate a paradigm that blends personalized healthcare with improved treatment options and is firmly committed to patient welfare. Through a comprehensive and multifaceted approach, we strive to tackle the challenges of resistance, aspiring to elevate cancer immunotherapy as a beacon of hope for patients around the world.
1. Introduction
Cancer immunotherapy heralds a promising revolution in the realm of oncological treatments. This groundbreaking approach, rooted in historical milestones like “Coley’s toxins” [1] and, later, the identification of cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4), has consistently showcased the potential to redefine cancer treatment paradigms [2,3,4]. As we deepened our understanding of tumor antigens and immune–tumor interactions in the latter half of the 20th century, the emergence of agents targeting CTLA-4, programmed cell death protein 1 (PD-1), and programmed death-ligand 1 (PD-L1) pathways marked significant successes in treating a range of malignancies [5,6,7]. Additionally, personalized strategies, such as chimeric antigen receptor (CAR) T-cell therapies, offer compelling efficacy, particularly in hematological malignancies [8,9,10]. The scope of cancer immunotherapy has since broadened, delving into influencing factors like the tumor microenvironment (TME) and even the gut microbiome to amplify therapeutic impact [11,12].
Despite these advances, resistance to immunotherapy presents a formidable barrier, emerging from innate tumor characteristics and adaptive changes in the genetic and proteomic landscape [13]. At the heart of this challenge lies the TME, which harbors elements like regulatory T cells (Tregs) and certain cytokines that shield tumor cells, allowing them to cleverly sidestep immune detection [14,15,16].
Our objectives are to dissect the complexity of immunotherapy resistance, evaluate both primary and secondary mechanisms, and consider the profound influence of genetic, epigenetic, and environmental factors [17]. We spotlight emerging strategies to overcome resistance and highlight the necessity of an integrated approach involving real-time monitoring, precision analytics, and patient-centered care [18]. By addressing these challenges head-on, we aim to advance the efficacy of cancer immunotherapy, reinforcing its position as a cornerstone of modern cancer care.
Through navigating the intricate landscape of resistance, we present insights into both established and novel strategies to outmaneuver the adaptive nature of tumors [19]. This review encapsulates the critical need for adaptability in treatment approaches, the ongoing quest for data-driven precision in patient-focused care, and the overarching potential of immunotherapy to redefine the future of cancer treatment [20,21,22].
2. The Immune Maze: Understanding the Complex Landscape
At the heart of the challenges presented by immunotherapy lies a deep-rooted, intricate interplay between the immune system and cancerous tumors. Grasping this landscape is pivotal to addressing the ever-evolving complexities of immunotherapy resistance [23,24]. To embark on this journey, it is crucial to recognize the distinctions between primary and secondary resistance and the multifarious mechanisms that underlie them [25].
Primary resistance: Innate to certain tumors, primary resistance emerges due to various factors that hinder the immune system’s capability to detect and counteract tumor cells. Some tumors are devoid of the critical antigens essential for immune recognition, rendering them less amenable to immunotherapeutic strategies [26,27]. Another dominant culprit is the immunosuppressive TME, characterized by a plethora of inhibitory factors and cells that dampen immune responses [28,29].
Consequently, secondary resistance develops as a backlash to therapeutic interventions. This form of resistance revitalizes tumor growth even after an initial successful response to immunotherapy including nivolumab (a PD-1 inhibitor) and ipilimumab (a CTLA-4 inhibitor) [30]. The driving forces behind this resistance span a spectrum from the genetic evolution of the tumor, which can lead to the modification or loss of previously identifiable antigens, to dynamic modifications to the TME, such as the amplification of immunosuppressive molecules or the influx of inhibitory cells [26,31,32].
Building on this, recent discoveries in the field have shed light on crucial aspects of immunotherapy resistance. Cutting-edge research has delved into the genetic and epigenetic blueprints of tumors. It has been shown that genetic modifications can recalibrate a tumor’s antigenic composition, impeding its visibility to immune cells [27,33,34,35]. Moreover, epigenetic shifts can mute genes vital for immune detection without altering the DNA structure or can modify how the tumor communicates with the surrounding immune framework [36,37,38].
Simultaneously, within the TME are distinct cellular entities that have gained prominence. These include Tregs, myeloid-derived suppressor cells (MDSCs), and tumor-associated macrophages (TAMs), which play cardinal roles in dampening immune activity and forming a protective bulwark around tumors [39,40,41]. Current research endeavors are evaluating their potential as resistance biomarkers, offering a glimpse into therapeutic trajectories [42,43].
Another pivotal aspect is the TME hypoxia [44,45]. Rapid tumor growth often surpasses its vascular supply, instigating hypoxia, which in turn sparks resistance pathways [44,46,47]. This oxygen deficiency is correlated with elevated PD-L1 expression, which mutes T-cell responses, facilitating tumor evasion [48,49].
Furthermore, the interplay between tumors and major histocompatibility complex (MHC) molecules is gaining traction [9,50]. MHCs are paramount in displaying tumor-specific peptides on the tumor surface for the T-cell detection [27,51,52]. Tumors have been found to employ evasion techniques, such as downregulating MHC expression or tweaking antigen-processing systems [27,53].
On a related note, immune checkpoints continue to be a focal point in the resistance discourse [54,55]. Often regulators in the immune system, these checkpoints are manipulated by tumors to serve as barriers against immune onslaughts [56,57]. Contemporary treatments, especially checkpoint disruptors, aspire to dismantle these barriers, amplifying immune responses against malignancies [7,58,59]. The latest clinical trials are unraveling the effectiveness of and obstacles to bypassing checkpoint-triggered resistance [60,61,62,63].
In summary, a profound understanding of the intricacies of immunotherapy resistance, its genesis, current revelations, and the TME’s role is fundamental in forging ahead with innovative strategies to subvert these hurdles. Subsequent sections provide a deeper exploration of these tactics.
3. Frontline Foes: Decoding the Architects of Immunotherapy Resistance
The TME serves as a dynamic milieu, evolving continuously and influencing the efficacy of cancer immunotherapies [64]. Key cytokines, notably transforming growth factor beta (TGF-β) and IL-10, are pivotal in modulating the TME, orchestrating immunosuppressive signals that underpin tumor resilience against therapeutic strategies.
Tregs are essential players within the TME, possessing the capability to subdue robust immune responses, particularly from formidable cells like cytotoxic T cells (CTLs) [65,66,67]. This suppression presents formidable challenges for immunotherapies, with Tregs secreting TGF-β and IL-10 to augment their inhibitory functions [68,69].
MDSCs further complicate the TME dynamics. These immune cells exacerbate the suppressive atmosphere, inhibiting CTLs and natural killer (NK) cells, thus limiting their tumor-fighting abilities [43,70]. They excel in restraining CTLs and NK cells, thus curtailing the NK cells’ tumor-eradicating capabilities [43,71,72]. Additionally, the MDSCs foster Treg proliferation, intensifying the suppressive milieu [73,74].
TAMs, with their versatile roles, are noteworthy contributors to the TME. Their ability to transition between M1-like (TAM1) and M2-like (TAM2) states plays a significant role in the balance between tumor defense and progression [75,76]. While TAM1 cells act aggressively against cancer cells, TAM2 cells encourage a suppressive environment, promoting tissue repair and angiogenesis, as well as safeguarding tumors from immune attacks [77,78,79].
Tumor-associated neutrophils (TAN) also differentiate into two major phenotypes within the TME. While TAN1 cells inhibit cancer progression, TAN2 cells support tumor growth, underscoring the multifaceted interactions within the TME [80,81].
Other factors, like rapid tumor growth leading to hypoxic conditions, activate various resistance mechanisms [82,83]. This includes the upregulation of immune checkpoint molecules such as PD-L1 on tumor surfaces, hindering T-cell functionality [84,85]. Hypoxia-triggered signaling pathways further deepen the TME’s suppressive nature [45,86].
Cancer cells also deploy evasion strategies, manipulating MHC molecules to reduce their visibility to the immune system [87,88]. Despite the promise of immune checkpoint inhibitors (ICIs), challenges remain in terms of assuring sustained outcomes and managing emergent resistance [7,89,90].
In closing, a profound grasp of these pivotal agents within the TME is paramount for charting successful strategies against the immunotherapy resistance [91]. As the research community continues its quest, the hope is to modulate these elements, enhancing the potency of the cancer immunotherapy [91,92,93]. By appreciating the TME’s intricacies, we inch closer to reshaping therapeutic outcomes and offering renewed hope to countless patients.
Figure 1 below provides a schematic representation of the intricate cellular interactions within the hypoxic TME, highlighting the key players involved in immunotherapy resistance.
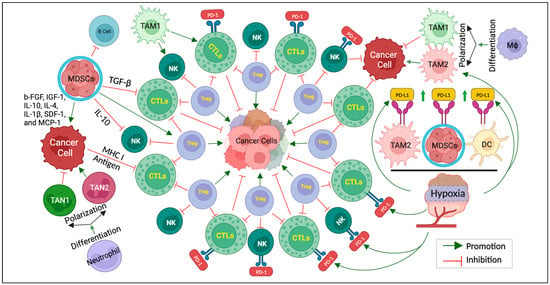
Figure 1.
The keys to overcoming immunotherapy resistance. Schematic representation of cellular interactions within the hypoxic TME. Cancer cells are surrounded by various cells, including Treg, CTLs, NK cells, TAM, TAN, MDSCs, etc. CTLs and NK cells exhibit PD-1 receptors that interact with PD-L1 expressed by TAM2, MDSCs, and DCs in the hypoxic TME. TAMs can undergo polarization and differentiation influenced via the hypoxic TME. TAM1 exhibits antitumor, while TAM2 promotes tumors. MDSCs release a series of cytokines (b-FGF, IGF-1, IL-10, IL-4, IL-1β, SDF-1, and MCP-1) affecting cancer cell behavior. TGF-β and IL-10 act as regulatory molecules inhibiting CTLs and NK cells, respectively. While the MHC I molecule and tumor antigen facilitate the interaction between cancer cells and CTLs, TAN1, and TAN2, differentiated from TAN, play the roles of inhibiting and promoting cancer cells, respectively. This figure illustrates the complex network of cellular interactions within the hypoxic TME.
4. Pioneering Strategies to Overcome Resistance
Cancer immunotherapy, while promising, is often hindered by the development of resistance. Several innovative strategies have been developed to address this, each designed to improve patient outcomes and enhance treatment efficacy.
4.1. Combination Therapies
Combination therapies represent a multi-pronged attack against cancer, targeting different aspects of tumor biology. These therapies may combine agents that halt tumor growth with those that boost the immune response. Despite the potential for increased toxicity, the benefits often outweigh the risks, necessitating careful patient management [94,95,96].
4.2. Tumor Microenvironment (TME)
Strategies that modify the TME aim to disrupt the supportive network of the tumor, including alterations in blood flow and stromal cell inhibition. Such interventions highlight the TME’s critical role in cancer therapy [97,98,99,100,101,102].
4.3. Emerging Immune Checkpoints
New research is focused on uncovering and targeting novel immune checkpoints that tumors exploit to evade immune detection. Agents targeting the ITIM domain (TIGIT), T cell immunoglobulin and mucin-domain-containing-3 (TIM-3), and lymphocyte activation gene-3 (LAG-3) are under investigation for their therapeutic potential [103,104].
4.4. Enhancing Immunotherapy with Oncolytic Viruses
Oncolytic viruses are emerging as a novel countermeasure to immunotherapy resistance. These viruses are engineered to selectively infect and destroy cancer cells while also modulating the immune environment to reverse resistance mechanisms. For example, the oncolytic virus VSV-GP, when combined with PD-1 inhibitors, has been found to effectively kill tumor cells. It also encourages the maturation of DCs and the influx of T-cells into the tumor milieu, which are crucial steps in reigniting the immune system’s attack on the cancer [105].
Furthermore, clinical trials, such as one led by Chesney et al., have revealed that T-VEC, an oncolytic virus derived from the herpes simplex virus, can significantly enhance treatment outcomes for melanoma patients, especially when administered in conjunction with ICIs [106]. This dual approach not only targets the tumor directly, but also reactivates the patient’s immune response against the tumor, providing a two-pronged attack against cancer resistance.
These developments signify a stride forward in integrating oncolytic virotherapy into the arsenal of immunotherapeutic strategies. By continuing to leverage these biological agents, researchers aim to unlock new pathways to overcome resistance and maximize the therapeutic potential of cancer immunotherapy.
4.5. Cell Therapy (ACT)
ACT personalizes treatment by using the patient’s immune cells, like TILs or chimeric antigen receptor (CAR)-T cells, to combat cancer. While effective in blood cancers, its application in solid tumors is an active area of research [107,108,109,110].
4.6. Cancer Vaccines
Cancer vaccines aim to prime the immune system to recognize and attack tumors, with DC and viral vector vaccines leading the way. This strategy is part of a broader effort to induce durable immune responses against cancer [111,112,113,114].
4.7. Navigating Medication-Induced Resistance in Immunotherapy
The interplay between certain medications and cancer immunotherapy is complex and can inadvertently contribute to treatment resistance. Corticosteroids, which are commonly prescribed to alleviate the side effects of immunotherapy, may inadvertently suppress the immune response, reducing the efficacy of treatments like ICIs [115,116]. Additionally, chemotherapeutic agents, while targeting cancer cells, may also inadvertently modify the immune environment in a way that fosters resistance [117,118]. This alteration in the immune landscape can hinder the immune system’s ability to effectively recognize and attack tumor cells.
Moreover, the use of antibiotics has been linked to disruptions in the gut microbiome, an emerging factor in the modulation of immunotherapy responses [119]. The gut microbiome plays a crucial role in maintaining a balanced immune system, and its disturbance may impact the success of immunotherapeutic strategies.
Furthermore, kinase inhibitors, used in targeted therapies, might alter critical signaling pathways that are essential for the activation and function of immune cells, contributing to a resistance scenario [120,121]. Such unintended effects underscore the necessity for clinicians to carefully consider the full spectrum of a patient’s medication regimen when administering immunotherapy.
By comprehensively understanding these drug interactions and their implications, medical professionals can devise strategies to avoid or counteract the resistance-inducing effects of these drugs. This may involve adjusting dosages, sequencing treatments, or selecting alternative therapeutic agents to maintain the robustness of the immune response [122].
Integrating advanced strategies that account for drug-induced resistance with conventional cancer therapies represents a significant step toward a new era in cancer treatment. This multifaceted approach emphasizes the need for continuous research and adaptation to refine immunotherapy regimens, ensuring they remain potent against cancer while respecting the patient’s overall well-being and minimizing unintended resistance [17,123].
Figure 2 below provides a visual representation of the different immunotherapeutic agents and their specific targets within the tumor microenvironment, illustrating the mechanisms by which they exert their effects.
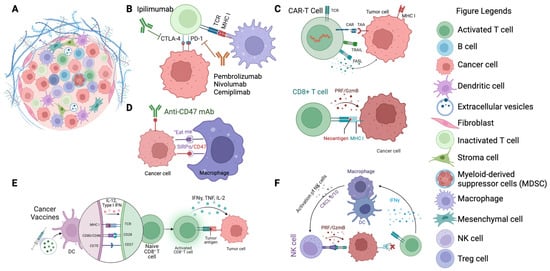
Figure 2.
Targets of immunotherapeutic agents in cancer therapy. (A) Illustration of the TME featuring cancer cells surrounded by various immune cells and extracellular matrix components. (B) Depiction of immune checkpoint inhibitors (ICIs) such as CTLA-4 and PD-1 (e.g., ipilimumab, pembrolizumab, nivolumab, cemiplimab) binding to their respective receptors on T cells, preventing immune evasion by cancer cells. (C) Representation of CAR T-cells targeting tumor-associated antigens (TAAs) on cancer cells, triggering cytotoxic responses. (D) Macrophage checkpoint inhibition: anti-CD47 mAb blocks the “don’t eat me” signal on cancer cells, promoting their phagocytosis by macrophages. (E) Depiction of dendritic cells (DCs) presenting tumor antigens to naïve T cells, leading to their activation and the initiation of an adaptive immune response against cancer cells. (F) Illustration of activated NK cells targeting cancer cells, mediated by cytokine signaling (e.g., IFNγ production), which enhances the innate immune response against tumors.
4.8. Integrated Strategies for Overcoming Resistance
To surmount the challenges presented by resistance to immunotherapy, an integrated approach is necessary. This involves not only the combination of therapeutic modalities but also the development of new agents that can tackle the evolved defense mechanisms of tumors. Precision medicine plays a crucial role in this, with targeted therapies designed to counteract specific pathways of resistance identified in a patient’s tumor profile [17]. Adopting personalized treatment regimens based on molecular diagnostics and patient-derived models, such as organoids and xenografts, is showing promise in enhancing treatment efficacy and reducing toxicity [123]. Furthermore, the implementation of real-time monitoring systems and predictive biomarkers facilitates a more responsive approach to immunotherapy adjustments [124,125]. The future of overcoming immunotherapy resistance lies in the synergy of these innovative strategies, each contributing a piece to the complex puzzle of cancer treatment [126].
In the following section, we provide an overview of pioneering strategies in cancer immunotherapy. Table 1 summarizes these strategies, including their approaches, key components, benefits, drug examples, and supporting references.

Table 1.
Overview of pioneering strategies in cancer immunotherapy.
To wrap up this exploration, the integration of these advanced strategies with traditional therapies offers a multifaceted approach to overcoming immunotherapy resistance, signaling a new era of hope for cancer treatment [129,130].
5. Recent Insights and Developments in Overcoming Immunotherapy Resistance
The endeavor to unravel and overcome resistance in cancer immunotherapy has uncovered significant genetic and epigenetic influences that affect patient outcomes [91,131,132,133].
5.1. Genetic Alterations and Immunotherapy Resistance
The emergence of resistance to immunotherapy due to genetic alterations within cancer cells is a major concern that complicates treatment outcomes. These mutations can significantly alter the immune system’s ability to recognize and destroy cancer cells. One of the key genetic changes involves mutations in the beta-2-microglobulin (B2M) gene, a critical component of the major histocompatibility complex (MHC) class I molecules. The MHC class I molecule presents tumor antigens to T cells, and any disruption in this pathway, as caused by B2M mutations, can lead to ineffective T cell-mediated tumor cell lysis [134,135].
Moreover, the Janus kinase (JAK) pathway, which includes the genes JAK1 and JAK2, plays a pivotal role in immune response signaling [136]. Mutations in these genes can have profound effects on the efficacy of immunotherapies. Shen et al.’s investigation into JAK1/JAK2 alterations revealed that such mutations can result in resistance to PD-1 blockade therapies by impairing the interferon signaling pathway, which is vital for the activation of the immune response against tumor cells [137].
Additionally, research indicates that alterations in the neoantigen landscape of cancer cells, due to genetic mutations, can influence the responsiveness to immunotherapy. The mutational burden and the quality of the neoantigens presented can either enhance or diminish the therapeutic efficacy, as the immune system may or may not recognize these neoantigens as targets [138,139].
These genetic alterations underscore the need for comprehensive genomic profiling of tumors to anticipate and overcome resistance mechanisms. By understanding and mapping these genetic changes, clinicians can personalize immunotherapy approaches, potentially restoring the sensitivity of cancer cells to treatment and improving patient prognosis.
5.2. Epigenetic Dynamics and Their Role in Resistance
The regulatory landscape of epigenetic modifications is significant in immunotherapy resistance, profoundly affecting gene expression and the immune detection of tumors. DNA methylation, which adds a methyl group to DNA and often leads to gene silencing, has been implicated in immune evasion. Mehdi et al. [140] have identified that hypermethylation of the promoter regions of Th1-type cytokine genes can result in the suppression of crucial immune signaling pathways. This hypermethylation effectively reduces the expression of cytokines necessary for a robust anti-tumor immune response, thus facilitating tumor cells’ escape from immune surveillance [141].
Histone modifications, another crucial aspect of epigenetics, involve changes to the proteins around which DNA is wound. Histone acetylation and deacetylation, controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs), can alter the accessibility of DNA to transcription machinery. Aberrations in HDAC activity have been linked to the repression of tumor suppressor genes. For example, overactivity of HDACs can lead to the tight winding of DNA around histones, effectively “hiding” tumor antigens from immune cells and contributing to resistance to immunotherapies such as checkpoint inhibitors [141,142].
Specific treatments, like the DNA methyltransferase inhibitors azacitidine and decitabine, have been shown to induce these epigenetic changes. They can enhance the effectiveness of immunotherapy by altering the expression of cancer/testis antigens and MHC molecules, heightening tumor immunogenicity [34,143]. However, they can also trigger immune evasion, necessitating a nuanced approach to their use in conjunction with immunotherapies [144].
Histone deacetylase inhibitors, such as vorinostat and romidepsin, have dual roles. While they can increase antigen presentation, they have also been implicated in promoting regulatory T-cell functions, which could dampen the immune response [145,146]. This highlights the delicate balance required when integrating epigenetic therapies with immunotherapy and underscores the need for further research to optimize these combinations.
5.3. The Microbiome’s Influence on Immunotherapy Efficacy
The interplay between the gut microbiome and the efficacy of cancer immunotherapy is a an intensively researched topic. The diverse community of microbes residing in the gastrointestinal tract exerts a substantial influence on the body’s immune responses, with significant implications for the effectiveness of immunotherapeutic agents.
In a landmark study by Derosa et al., researchers identified that the presence of specific gut bacteria, such as Akkermansia muciniphila, significantly improved the efficacy of PD-1 inhibitors. This microbe appeared to bolster the host immune system’s capacity for tumor surveillance, potentially by maintaining mucosal integrity or enhancing immune cell activation, thus increasing the effectiveness of immunotherapies [147]. Such findings have led to the proposal that the gut microbiome could serve as a predictive biomarker for immunotherapy responses, and through interventions such as diet or probiotics, could be adjusted to improve clinical outcomes.
Conversely, antibiotic use can disrupt the delicate balance of the gut microbiome, with studies like those conducted by Patel et al. demonstrating negative impacts on the efficacy of immunotherapies. Antibiotics may diminish beneficial bacteria, impair immune function, and lessen the host’s response to PD-1 inhibitors, highlighting the need for careful consideration of antibiotic use during immunotherapy [148].
This emerging research area has spurred interest in probiotics and fecal microbiota transplantation (FMT) as methods to modulate the gut microbiome favorably. Ongoing clinical trials are exploring the potential of these interventions to modulate the gut microbiome in order to improve the patient response rate to cancer immunotherapy [149,150].
Overall, a growing body of evidence supports the notion that therapeutic modulation of the microbiome could serve as an adjunct to enhance the efficacy of immunotherapy and reduce resistance. Ongoing research into microbiome-based adjuvants holds promise for refining the management of cancer through these novel interventions.
6. Clinical Implications and Translational Approaches
The recognition and early identification of biomarkers indicative of resistance is pivotal in optimizing cancer treatment protocols. Biomarkers, such as high PD-L1 expression or a significant tumor mutational burden (TMB), as well as genetic alterations like JAK1/2 mutations, are at the forefront of predicting and countering immunotherapy resistance [151]. These biomarkers not only facilitate diagnosis, but are also vital for the creation of targeted strategies that preemptively confront specific resistance pathways [152].
Translational research tools like patient-derived organoids (PDOs) and xenograft models (PDX) are instrumental in applying preclinical findings to clinical treatment design. For instance, PDOs derived from colorectal cancer patients have been utilized to evaluate the efficacy of novel drugs, replicating the complex cellular environment of the originating tumor [153,154]. These studies have led directly to clinical trials and adjustments to treatment regimens, exemplifying how PDOs can significantly influence therapeutic planning and patient management.
In the vanguard of translational research, PDX models stand out for their direct impact on clinical decision-making. By engrafting human tumor tissues into immunodeficient mice, PDX models maintain the tumor’s intrinsic heterogeneity, providing insights into the tumor’s response to new treatments. These models have significantly advanced our understanding of resistance mechanisms, guiding the design of clinical trials aimed at targeted resistance pathways.
For instance, PDX research has led to the discovery of alternative immune checkpoints and changes in antigen presentation, shaping the development of combination therapies and influencing clinical treatment modifications. Such studies have also identified biomarkers predictive of treatment response, allowing for the adaptation of clinical protocols [155].
A key example of the impact of PDX models is their use in pinpointing specific genetic mutations that confer resistance to standard therapies. Insights gained from PDX studies have informed the enrollment of patients in trials for new targeted agents, leading to improved outcomes. These translational models are thus integral to the evolution of personalized medicine, enhancing the specificity and adaptability of cancer therapies [155].
PDX models, together with PDOs, enhance therapeutic planning by replicating the complex tumor environment, thereby offering a dynamic platform for drug evaluation and the development of personalized treatment regimens [153,154].
The synergy between clinical acumen and advanced translational models is reshaping cancer therapy, increasing the precision of the current treatments, and paving the way for innovative strategies to navigate the complexities of immunotherapy resistance. This integrated approach is set to refine patient care, promising a future where cancer treatment is as personalized as it is effective.
7. Future Perspectives in Immunotherapy
The future of immunotherapy is illuminated by advancements across varied disciplines, seamlessly integrating cutting-edge technologies poised to redefine oncological breakthroughs.
At the vanguard of these advancements, the integration of artificial intelligence (AI) and machine learning offers the capability to decipher vast genetic and proteomic datasets [156,157,158]. While this technological leap revolutionizes personalized immunotherapy by predicting tumor behavior and resistance mechanisms, as well as enabling real-time patient monitoring, it also brings forth challenges. For instance, ensuring the privacy and security of patient data processed by AI becomes paramount. Moreover, the algorithms’ decision-making processes require transparency, especially when used to make clinical recommendations. Ethical considerations arise, questioning the extent of reliance on AI for treatment decisions and potential biases embedded within the algorithms.
Nanotechnology, emphasizing nanoparticles, holds significant potential to enhance the immunotherapy [8,50,52,159,160,161]. Its ability to deliver drugs precisely to tumor sites and fine-tune immune responses charts the path for groundbreaking strategies. These include modifying the TME to impede tumor growth, optimizing nutrient dynamics within the TME, and propelling the development of neoantigen vaccines. However, the use of nanoparticles raises concerns regarding long-term safety, potential off-target effects, and their interactions with the body’s natural systems. Ethical discussions also surround the equitable distribution of such advanced treatments and the potential high costs associated with them.
Tumor epigenetics is a rising domain, with research directed toward harnessing epigenetic modulators to manipulate gene expression patterns. This tactic could potentially combat immunotherapeutic resistance, thus diversifying treatment avenues.
Simultaneously, telemedicine platforms are bridging geographical chasms, ensuring that specialized care becomes universally accessible [162]. Such platforms empower individuals in regions with constrained specialty resources to receive optimal treatment recommendations. The prevailing transformative phase in immunotherapy flourishes with interdisciplinary collaboration. Disciplines like genetics, immunology, bioengineering, and sociology coalesce, exemplified by the amalgamation of genomic sequencing, microfluidic technologies, and 3D tumor modeling to sharpen therapeutic strategies.
In summation, the dynamic realm of immunotherapy intertwines an array of disciplines, pioneering technologies, and global partnerships. The forthcoming epoch promises unmatched precision and flexibility, as well as a rejuvenated wave of oncological innovations, albeit not without its challenges and ethical dilemmas.
8. Conclusions
Throughout our journey into the complex landscape of immunotherapy, we confronted a myriad of challenges and opportunities. The foremost among these was the issue of immunotherapy resistance. While such challenges might seem daunting, they also serve as gateways to novel innovations. Our increasingly profound comprehension, bolstered by advancements in AI, nanotechnology, and epigenetics, is propelling us toward solutions that were once considered beyond reach.
Immunotherapy heralds a paradigm shift in oncological treatments, emphasizing the body’s intrinsic defenses against malignancies. Yet, the ever-present shadow of resistance reminds us of the continuous need for exploration, adaptation, and innovation. It is the collective endeavors of researchers, clinicians, and pioneers across disciplines that underpin the remarkable breakthroughs we witness today. These efforts inch us closer to the overarching goal: to overcome cancer resistance and elevate patient outcomes.
However, like all scientific pursuits, our research has its confines. Future studies might focus on deeper dives into molecular mechanisms, patient-specific factors, or even socio-economic considerations that could influence resistance. Expanding on these areas would undeniably enrich our understanding.
In summary, our journey through the complexities of immunotherapy resistance is continuous, but the advancements made signal a hopeful future. Here, cancer treatments are envisioned to be not only more personalized and powerful, but also characterized by fewer adverse effects. The crux of this progress lies in persistent research, international cooperation, and a steadfast commitment to revolutionizing the story of cancer treatment.
Author Contributions
L.Y. and W.M. conceptualized the article and prepared figures. L.Y., Q.W., and W.M. wrote and edited the manuscript. W.M. finalized the figures and manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing does not apply to this article.
Conflicts of Interest
The authors confirm that the research was undertaken without any commercial or financial affiliations that might be perceived as potential conflicts of interest.
Abbreviations
| ACT | adoptive cell therapy |
| AI | artificial intelligence |
| CAR | chimeric antigen receptor |
| CTLs | cytotoxic T cells |
| CTLA-4 | cytotoxic T-lymphocyte-associated antigen 4 |
| cfDNA | cell-free DNA |
| CTCs | circulating tumor cells |
| EGFR | epidermal growth factor receptor |
| LAG-3 | lymphocyte activation gene-3 |
| MDSCs | myeloid-derived suppressor cells |
| MHC | major histocompatibility complex |
| NK | natural killer |
| NSCLC | non-small-cell lung cancer |
| PBMC | peripheral blood mononuclear cells |
| PD-1 | programmed cell death protein 1 |
| PD-L1 | programmed death-ligand 1 |
| PDO | patient-derived organoids |
| PDX | patient-derived xenograft |
| TAMs | tumor-associated macrophages |
| TAM1 | type-1 TAM |
| TAM2 | type-2 TAM |
| TAN1 | type-1 TAN |
| TAN2 | type-2 TAN |
| TANs | tumor-associated neutrophils |
| TIGIT | T cell immunoreceptor with immunoglobulin and ITIM domain |
| TIM-3 | T cell immunoglobulin and mucin-domain-containing-3 |
| TGF | transforming growth factor |
| Tregs | regulatory T cells |
| TMB | tumor mutational burden |
| TME | tumor microenvironment |
References
- Shin, Y.H.; Bang, S.; Park, S.M.; Ma, X.; Cassilly, C.; Graham, D.; Xavier, R.; Clardy, J. Revisiting Coley’s Toxins: Immunogenic Cardiolipins from Streptococcus pyogenes. J. Am. Chem. Soc. 2023, 145, 21183–21188. [Google Scholar] [CrossRef]
- McCarthy, E.F. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop. J. 2006, 26, 154–158. [Google Scholar]
- Coley, W.B. The treatment of malignant tumors by repeated inoculations of erysipelas; with a report of ten original cases. Am. J. Med. Sci. 1893, 105, 487–511. [Google Scholar] [CrossRef]
- Brunet, J.F.; Denizot, F.; Luciani, M.F.; Roux-Dosseto, M.; Suzan, M.; Mattei, M.G.; Golstein, P. A new member of the immunoglobulin superfamily—CTLA-4. Nature 1987, 328, 267–270. [Google Scholar] [CrossRef]
- Zahavi, D.; Weiner, L. Monoclonal Antibodies in Cancer Therapy. Antibodies 2020, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.T.; Lee, S.H.; Heo, Y.S. Molecular Interactions of Antibody Drugs Targeting PD-1, PD-L1, and CTLA-4 in Immuno-Oncology. Molecules 2019, 24, 1190. [Google Scholar] [CrossRef]
- Yao, L.; Jia, G.; Lu, L.; Bao, Y.; Ma, W. Factors affecting tumor responders and predictive biomarkers of toxicities in cancer patients treated with immune checkpoint inhibitors. Int. Immunopharmacol. 2020, 85, 106628. [Google Scholar] [CrossRef]
- Bae, J.; Parayath, N.; Ma, W.; Amiji, M.; Munshi, N.; Anderson, K.C. BCMA peptide-engineered nanoparticles enhance induction and function of antigen-specific CD8(+) cytotoxic T lymphocytes against multiple myeloma: Clinical applications. Leukemia 2020, 34, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lu, L.; Ma, W. Efficacy, Safety, and Challenges of CAR T-Cells in the Treatment of Solid Tumors. Cancers 2022, 14, 5983. [Google Scholar] [CrossRef] [PubMed]
- Jogalekar, M.P.; Rajendran, R.L.; Khan, F.; Dmello, C.; Gangadaran, P.; Ahn, B.C. CAR T-Cell-Based gene therapy for cancers: New perspectives, challenges, and clinical developments. Front. Immunol. 2022, 13, 925985. [Google Scholar] [CrossRef] [PubMed]
- Murciano-Goroff, Y.R.; Warner, A.B.; Wolchok, J.D. The future of cancer immunotherapy: Microenvironment-targeting combinations. Cell Res. 2020, 30, 507–519. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Guo, G.; Han, J.; Yu, J. Gut microbiome in modulating immune checkpoint inhibitors. EBioMedicine 2022, 82, 104163. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.M.; Choi, J.; Lim, M. Mechanisms of immunotherapy resistance: Lessons from glioblastoma. Nat. Immunol. 2019, 20, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, K.; Hana, D.; Chou, J.T.; Singh, C.; Mackiewicz, A.; Kaczmarek, M. Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance. Front. Immunol. 2021, 12, 656364. [Google Scholar] [CrossRef] [PubMed]
- Haist, M.; Stege, H.; Grabbe, S.; Bros, M. The Functional Crosstalk between Myeloid-Derived Suppressor Cells and Regulatory T Cells within the Immunosuppressive Tumor Microenvironment. Cancers 2021, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- Iliadi, C.; Verset, L.; Bouchart, C.; Martinive, P.; Van Gestel, D.; Krayem, M. The current understanding of the immune landscape relative to radiotherapy across tumor types. Front. Immunol. 2023, 14, 1148692. [Google Scholar] [CrossRef]
- Piper, M.; Kluger, H.; Ruppin, E.; Hu-Lieskovan, S. Immune Resistance Mechanisms and the Road to Personalized Immunotherapy. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e390290. [Google Scholar] [CrossRef]
- Fountzilas, E.; Tsimberidou, A.M.; Vo, H.H.; Kurzrock, R. Clinical trial design in the era of precision medicine. Genome Med. 2022, 14, 101. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, X.; Jin, S.; Chen, Y.; Guo, R. Ferroptosis in cancer therapy: A novel approach to reversing drug resistance. Mol. Cancer 2022, 21, 47. [Google Scholar] [CrossRef]
- Liao, J.; Li, X.; Gan, Y.; Han, S.; Rong, P.; Wang, W.; Li, W.; Zhou, L. Artificial intelligence assists precision medicine in cancer treatment. Front. Oncol. 2022, 12, 998222. [Google Scholar] [CrossRef]
- Brown, C.E.; Bucktrout, S.; Butterfield, L.H.; Futer, O.; Galanis, E.; Hormigo, A.; Lim, M.; Okada, H.; Prins, R.; Marr, S.S.; et al. The future of cancer immunotherapy for brain tumors: A collaborative workshop. J. Transl. Med. 2022, 20, 236. [Google Scholar] [CrossRef]
- Luo, J.; Li, X.; Wei, K.L.; Chen, G.; Xiong, D.D. Advances in the application of computational pathology in diagnosis, immunomicroenvironment recognition, and immunotherapy evaluation of breast cancer: A narrative review. J. Cancer Res. Clin. Oncol. 2023, 149, 12535–12542. [Google Scholar] [CrossRef] [PubMed]
- Abaza, A.; Sid Idris, F.; Anis Shaikh, H.; Vahora, I.; Moparthi, K.P.; Al Rushaidi, M.T.; Muddam, M.R.; Obajeun, O.A.; Jaramillo, A.P.; Khan, S. Programmed Cell Death Protein 1 (PD-1) and Programmed Cell Death Ligand 1 (PD-L1) Immunotherapy: A Promising Breakthrough in Cancer Therapeutics. Cureus 2023, 15, e44582. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Guo, Z.; Wang, L. Progress and Challenges of Immunotherapy Predictive Biomarkers for Triple Negative Breast Cancer in the Era of Single-Cell Multi-Omics. Life 2023, 13, 1189. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.N.; Tian, Q.; Teng, Q.X.; Wurpel, J.N.D.; Zeng, L.; Pan, Y.; Chen, Z.S. Understanding and targeting resistance mechanisms in cancer. MedComm (2020) 2023, 4, e265. [Google Scholar] [CrossRef]
- Kim, S.K.; Cho, S.W. The Evasion Mechanisms of Cancer Immunity and Drug Intervention in the Tumor Microenvironment. Front. Pharmacol. 2022, 13, 868695. [Google Scholar] [CrossRef] [PubMed]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, Z.; Dang, Q.; Xu, H.; Lv, J.; Li, H.; Han, X. Immunosuppression in tumor immune microenvironment and its optimization from CAR-T cell therapy. Theranostics 2022, 12, 6273–6290. [Google Scholar] [CrossRef]
- Tie, Y.; Tang, F.; Wei, Y.Q.; Wei, X.W. Immunosuppressive cells in cancer: Mechanisms and potential therapeutic targets. J. Hematol. Oncol. 2022, 15, 61. [Google Scholar] [CrossRef]
- Trujillo, J.A.; Luke, J.J.; Zha, Y.; Segal, J.P.; Ritterhouse, L.L.; Spranger, S.; Matijevich, K.; Gajewski, T.F. Secondary resistance to immunotherapy associated with beta-catenin pathway activation or PTEN loss in metastatic melanoma. J. Immunother. Cancer 2019, 7, 295. [Google Scholar] [CrossRef]
- Cendrowicz, E.; Sas, Z.; Bremer, E.; Rygiel, T.P. The Role of Macrophages in Cancer Development and Therapy. Cancers 2021, 13, 1946. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, B.V.; Nepal, S.; Varambally, S. Genomic and Epigenomic Alterations in Cancer. Am. J. Pathol. 2016, 186, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Dai, E.; Zhu, Z.; Wahed, S.; Qu, Z.; Storkus, W.J.; Guo, Z.S. Epigenetic modulation of antitumor immunity for improved cancer immunotherapy. Mol. Cancer 2021, 20, 171. [Google Scholar] [CrossRef]
- Martinez-Jimenez, F.; Priestley, P.; Shale, C.; Baber, J.; Rozemuller, E.; Cuppen, E. Genetic immune escape landscape in primary and metastatic cancer. Nat. Genet. 2023, 55, 820–831. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Cancer Epigenetics, Tumor Immunity, and Immunotherapy. Trends Cancer 2020, 6, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Turcan, S. Epigenetic Drugs and Their Immune Modulating Potential in Cancers. Biomedicines 2022, 10, 211. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Wang, W.; Zhang, B.; Yu, X.; Shi, S. Epigenetic regulation in the tumor microenvironment: Molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 210. [Google Scholar] [CrossRef]
- Lindau, D.; Gielen, P.; Kroesen, M.; Wesseling, P.; Adema, G.J. The immunosuppressive tumour network: Myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology 2013, 138, 105–115. [Google Scholar] [CrossRef]
- Ma, T.; Renz, B.W.; Ilmer, M.; Koch, D.; Yang, Y.; Werner, J.; Bazhin, A.V. Myeloid-Derived Suppressor Cells in Solid Tumors. Cells 2022, 11, 310. [Google Scholar] [CrossRef]
- Sun, R.; Zhao, H.; Gao, D.S.; Ni, A.; Li, H.; Chen, L.; Lu, X.; Chen, K.; Lu, B. Amphiregulin couples IL1RL1(+) regulatory T cells and cancer-associated fibroblasts to impede antitumor immunity. Sci. Adv. 2023, 9, eadd7399. [Google Scholar] [CrossRef]
- Shi, H.; Li, K.; Ni, Y.; Liang, X.; Zhao, X. Myeloid-Derived Suppressor Cells: Implications in the Resistance of Malignant Tumors to T Cell-Based Immunotherapy. Front. Cell Dev. Biol. 2021, 9, 707198. [Google Scholar] [CrossRef]
- Li, K.; Shi, H.; Zhang, B.; Ou, X.; Ma, Q.; Chen, Y.; Shu, P.; Li, D.; Wang, Y. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct. Target. Ther. 2021, 6, 362. [Google Scholar] [CrossRef]
- Kopecka, J.; Salaroglio, I.C.; Perez-Ruiz, E.; Sarmento-Ribeiro, A.B.; Saponara, S.; De Las Rivas, J.; Riganti, C. Hypoxia as a driver of resistance to immunotherapy. Drug Resist. Updat. 2021, 59, 100787. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, Q.; Zhang, Y.; Liu, Z.; Zheng, Z.; Liu, S.; Meng, L.; Xin, Y.; Jiang, X. Targeting hypoxia in the tumor microenvironment: A potential strategy to improve cancer immunotherapy. J. Exp. Clin. Cancer Res. 2021, 40, 24. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Zhu, J.; Zhang, X.; Mao, X. The Role of Hypoxia and Cancer Stem Cells in Development of Glioblastoma. Cancers 2023, 15, 2613. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Calar, K.; de la Puente, P. Mimicking tumor hypoxia and tumor-immune interactions employing three-dimensional in vitro models. J. Exp. Clin. Cancer Res. 2020, 39, 75. [Google Scholar] [CrossRef] [PubMed]
- Shurin, M.R.; Umansky, V. Cross-talk between HIF and PD-1/PD-L1 pathways in carcinogenesis and therapy. J. Clin. Investig. 2022, 132, e159473. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; You, L.; Nepovimova, E.; Heger, Z.; Wu, W.; Kuca, K.; Adam, V. Hypoxia-inducible factors: Master regulators of hypoxic tumor immune escape. J. Hematol. Oncol. 2022, 15, 77. [Google Scholar] [CrossRef]
- Li, H.; Shao, S.; Cai, J.; Burner, D.; Lu, L.; Chen, Q.; Minev, B.; Ma, W. Artificial human antigen-presenting cells are superior to dendritic cells at inducing cytotoxic T-cell responses. Immunology 2017, 152, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Ma, W.; Johnson, C.H.; Khan, S.A.; Irwin, M.L.; Pusztai, L. In silico designed mRNA vaccines targeting CA-125 neoantigen in breast and ovarian cancer. Vaccine 2023, 41, 2073–2083. [Google Scholar] [CrossRef]
- Ma, W.; Smith, T.; Bogin, V.; Zhang, Y.; Ozkan, C.; Ozkan, M.; Hayden, M.; Schroter, S.; Carrier, E.; Messmer, D.; et al. Enhanced presentation of MHC class Ia, Ib and class II-restricted peptides encapsulated in biodegradable nanoparticles: A promising strategy for tumor immunotherapy. J. Transl. Med. 2011, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef] [PubMed]
- Haddad, A.F.; Young, J.S.; Gill, S.; Aghi, M.K. Resistance to immune checkpoint blockade: Mechanisms, counter-acting approaches, and future directions. Semin. Cancer Biol. 2022, 86, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Han, Y.; Zhang, Y.; Zhao, Q.; Wang, H.; Wei, J.; Meng, L.; Xin, Y.; Jiang, X. Overcoming acquired resistance to cancer immune checkpoint therapy: Potential strategies based on molecular mechanisms. Cell Biosci. 2023, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Marin-Acevedo, J.A.; Dholaria, B.; Soyano, A.E.; Knutson, K.L.; Chumsri, S.; Lou, Y. Next generation of immune checkpoint therapy in cancer: New developments and challenges. J. Hematol. Oncol. 2018, 11, 39. [Google Scholar] [CrossRef]
- Naimi, A.; Mohammed, R.N.; Raji, A.; Chupradit, S.; Yumashev, A.V.; Suksatan, W.; Shalaby, M.N.; Thangavelu, L.; Kamrava, S.; Shomali, N.; et al. Tumor immunotherapies by immune checkpoint inhibitors (ICIs); the pros and cons. Cell Commun. Signal. 2022, 20, 44. [Google Scholar] [CrossRef]
- Webb, E.S.; Liu, P.; Baleeiro, R.; Lemoine, N.R.; Yuan, M.; Wang, Y.H. Immune checkpoint inhibitors in cancer therapy. J. Biomed. Res. 2018, 32, 317–326. [Google Scholar] [CrossRef]
- Xie, Q.; Zhang, P.; Wang, Y.; Mei, W.; Zeng, C. Overcoming resistance to immune checkpoint inhibitors in hepatocellular carcinoma: Challenges and opportunities. Front. Oncol. 2022, 12, 958720. [Google Scholar] [CrossRef]
- Fares, C.M.; Van Allen, E.M.; Drake, C.G.; Allison, J.P.; Hu-Lieskovan, S. Mechanisms of Resistance to Immune Checkpoint Blockade: Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 147–164. [Google Scholar] [CrossRef]
- Passaro, A.; Brahmer, J.; Antonia, S.; Mok, T.; Peters, S. Managing Resistance to Immune Checkpoint Inhibitors in Lung Cancer: Treatment and Novel Strategies. J. Clin. Oncol. 2022, 40, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Metropulos, A.E.; Munshi, H.G.; Principe, D.R. The difficulty in translating the preclinical success of combined TGFbeta and immune checkpoint inhibition to clinical trial. EBioMedicine 2022, 86, 104380. [Google Scholar] [CrossRef]
- Tiwari, A.; Trivedi, R.; Lin, S.Y. Tumor microenvironment: Barrier or opportunity towards effective cancer therapy. J. Biomed. Sci. 2022, 29, 83. [Google Scholar] [CrossRef]
- Chen, M.L.; Pittet, M.J.; Gorelik, L.; Flavell, R.A.; Weissleder, R.; von Boehmer, H.; Khazaie, K. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, Y.; Liu, S.; Wang, H.; Zhu, J.; Ou, L.; Xu, X. Targeting regulatory T cells for immunotherapy in melanoma. Mol. Biomed. 2021, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Itahashi, K.; Irie, T.; Nishikawa, H. Regulatory T-cell development in the tumor microenvironment. Eur. J. Immunol. 2022, 52, 1216–1227. [Google Scholar] [CrossRef]
- Nishikawa, H.; Koyama, S. Mechanisms of regulatory T cell infiltration in tumors: Implications for innovative immune precision therapies. J. Immunother. Cancer 2021, 9, e002591. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, X.; Wu, S.; Cui, D.; Xu, Z. Myeloid-derived suppressor cells: Key immunosuppressive regulators and therapeutic targets in hematological malignancies. Biomark. Res. 2023, 11, 34. [Google Scholar] [CrossRef]
- Zalfa, C.; Paust, S. Natural Killer Cell Interactions with Myeloid Derived Suppressor Cells in the Tumor Microenvironment and Implications for Cancer Immunotherapy. Front. Immunol. 2021, 12, 633205. [Google Scholar] [CrossRef]
- Jakos, T.; Pislar, A.; Jewett, A.; Kos, J. Myeloid-Derived Suppressor Cells Hamper Natural Killer Cell Activity in Cancer: Role of Peptidases. Crit. Rev. Immunol. 2021, 41, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, H.; Jihu, R.; Zhou, J.; Zeng, R.; Yan, H. Novel Characterization of Myeloid-Derived Suppressor Cells in Tumor Microenvironment. Front. Cell Dev. Biol. 2021, 9, 698532. [Google Scholar] [CrossRef] [PubMed]
- Mehdizadeh, R.; Shariatpanahi, S.P.; Goliaei, B.; Ruegg, C. Targeting myeloid-derived suppressor cells in combination with tumor cell vaccination predicts anti-tumor immunity and breast cancer dormancy: An in silico experiment. Sci. Rep. 2023, 13, 5875. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, S. Tumor-Associated Macrophages and Their Functional Transformation in the Hypoxic Tumor Microenvironment. Front. Immunol. 2021, 12, 741305. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Saeed, A.; Liu, Q.; Jiang, Q.; Xu, H.; Xiao, G.G.; Rao, L.; Duo, Y. Macrophages in immunoregulation and therapeutics. Signal Transduct. Target. Ther. 2023, 8, 207. [Google Scholar] [CrossRef]
- Li, M.; He, L.; Zhu, J.; Zhang, P.; Liang, S. Targeting tumor-associated macrophages for cancer treatment. Cell Biosci. 2022, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.K.; Kadel, S.; Townsend, M.G.; Oliwa, M.; Guerriero, J.L. Macrophage Biology and Mechanisms of Immune Suppression in Breast Cancer. Front. Immunol. 2021, 12, 643771. [Google Scholar] [CrossRef]
- Feng, Y.; Ye, Z.; Song, F.; He, Y.; Liu, J. The Role of TAMs in Tumor Microenvironment and New Research Progress. Stem Cells Int. 2022, 2022, 5775696. [Google Scholar] [CrossRef]
- Yan, M.; Zheng, M.; Niu, R.; Yang, X.; Tian, S.; Fan, L.; Li, Y.; Zhang, S. Roles of tumor-associated neutrophils in tumor metastasis and its clinical applications. Front. Cell Dev. Biol. 2022, 10, 938289. [Google Scholar] [CrossRef]
- Que, H.; Fu, Q.; Lan, T.; Tian, X.; Wei, X. Tumor-associated neutrophils and neutrophil-targeted cancer therapies. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188762. [Google Scholar] [CrossRef] [PubMed]
- Wicks, E.E.; Semenza, G.L. Hypoxia-inducible factors: Cancer progression and clinical translation. J. Clin. Investig. 2022, 132, e159839. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, L.; Li, X.F. Hypoxia and the Tumor Microenvironment. Technol. Cancer Res. Treat. 2021, 20, 15330338211036304. [Google Scholar] [CrossRef]
- Patsoukis, N.; Wang, Q.; Strauss, L.; Boussiotis, V.A. Revisiting the PD-1 pathway. Sci. Adv. 2020, 6, eabd2712. [Google Scholar] [CrossRef]
- Yi, M.; Jiao, D.; Xu, H.; Liu, Q.; Zhao, W.; Han, X.; Wu, K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer 2018, 17, 129. [Google Scholar] [CrossRef]
- Zhang, Y.; Coleman, M.; Brekken, R.A. Perspectives on Hypoxia Signaling in Tumor Stroma. Cancers 2021, 13, 3070. [Google Scholar] [CrossRef] [PubMed]
- Shklovskaya, E.; Rizos, H. MHC Class I Deficiency in Solid Tumors and Therapeutic Strategies to Overcome It. Int. J. Mol. Sci. 2021, 22, 6741. [Google Scholar] [CrossRef]
- Wen, M.; Li, Y.; Qin, X.; Qin, B.; Wang, Q. Insight into Cancer Immunity: MHCs, Immune Cells and Commensal Microbiota. Cells 2023, 12, 1882. [Google Scholar] [CrossRef]
- Zhou, J.; Bashey, A.; Zhong, R.; Corringham, S.; Messer, K.; Pu, M.; Ma, W.; Chut, T.; Soiffer, R.; Mitrovich, R.C.; et al. CTLA-4 blockade following relapse of malignancy after allogeneic stem cell transplantation is associated with T cell activation but not with increased levels of T regulatory cells. Biol. Blood Marrow Transplant. 2011, 17, 682–692. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, W.; Yang, W.; Zhou, M.; Liu, F. Acquired resistance for immune checkpoint inhibitors in cancer immunotherapy: Challenges and prospects. Aging 2022, 14, 1048–1064. [Google Scholar] [CrossRef]
- Said, S.S.; Ibrahim, W.N. Cancer Resistance to Immunotherapy: Comprehensive Insights with Future Perspectives. Pharmaceutics 2023, 15, 1143. [Google Scholar] [CrossRef]
- Lao, Y.; Shen, D.; Zhang, W.; He, R.; Jiang, M. Immune Checkpoint Inhibitors in Cancer Therapy—How to Overcome Drug Resistance? Cancers 2022, 14, 3575. [Google Scholar] [CrossRef]
- Baxter, M.A.; Middleton, F.; Cagney, H.P.; Petty, R.D. Resistance to immune checkpoint inhibitors in advanced gastro-oesophageal cancers. Br. J. Cancer 2021, 125, 1068–1079. [Google Scholar] [CrossRef]
- Dutta, S.; Ganguly, A.; Chatterjee, K.; Spada, S.; Mukherjee, S. Targets of Immune Escape Mechanisms in Cancer: Basis for Development and Evolution of Cancer Immune Checkpoint Inhibitors. Biology 2023, 12, 218. [Google Scholar] [CrossRef] [PubMed]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef] [PubMed]
- Flies, D.B.; Langermann, S.; Jensen, C.; Karsdal, M.A.; Willumsen, N. Regulation of tumor immunity and immunotherapy by the tumor collagen extracellular matrix. Front. Immunol. 2023, 14, 1199513. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, X.; Chang, C.Y.; Wang, H.Y.; Wang, R.F. The Interplay between T Cells and Cancer: The Basis of Immunotherapy. Genes 2023, 14, 1008. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022, 21, 799–820. [Google Scholar] [CrossRef]
- Park, K.; Veena, M.S.; Shin, D.S. Key Players of the Immunosuppressive Tumor Microenvironment and Emerging Therapeutic Strategies. Front. Cell Dev. Biol. 2022, 10, 830208. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, T.; Gu, J.; Lu, L. Targeting the metabolism of tumor-infiltrating regulatory T cells. Trends Immunol. 2023, 44, 598–612. [Google Scholar] [CrossRef]
- Bejarano, L.; Jordao, M.J.C.; Joyce, J.A. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov. 2021, 11, 933–959. [Google Scholar] [CrossRef]
- Lee, J.B.; Ha, S.J.; Kim, H.R. Clinical Insights into Novel Immune Checkpoint Inhibitors. Front. Pharmacol. 2021, 12, 681320. [Google Scholar] [CrossRef]
- Dulal, D.; Boring, A.; Terrero, D.; Johnson, T.; Tiwari, A.K.; Raman, D. Tackling of Immunorefractory Tumors by Targeting Alternative Immune Checkpoints. Cancers 2023, 15, 2774. [Google Scholar] [CrossRef]
- Zhu, X.; Fan, C.; Xiong, Z.; Chen, M.; Li, Z.; Tao, T.; Liu, X. Development and application of oncolytic viruses as the nemesis of tumor cells. Front. Microbiol. 2023, 14, 1188526. [Google Scholar] [CrossRef]
- Chesney, J.A.; Ribas, A.; Long, G.V.; Kirkwood, J.M.; Dummer, R.; Puzanov, I.; Hoeller, C.; Gajewski, T.F.; Gutzmer, R.; Rutkowski, P.; et al. Randomized, Double-Blind, Placebo-Controlled, Global Phase III Trial of Talimogene Laherparepvec Combined with Pembrolizumab for Advanced Melanoma. J. Clin. Oncol. 2023, 41, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Feldman, S.A.; Assadipour, Y.; Kriley, I.; Goff, S.L.; Rosenberg, S.A. Adoptive Cell Therapy—Tumor-Infiltrating Lymphocytes, T-Cell Receptors, and Chimeric Antigen Receptors. Semin. Oncol. 2015, 42, 626–639. [Google Scholar] [CrossRef]
- Li, J.; Xiao, Z.; Wang, D.; Jia, L.; Nie, S.; Zeng, X.; Hu, W. The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells. Mol. Cancer 2023, 22, 141. [Google Scholar] [CrossRef] [PubMed]
- Ingram, Z.; Madan, S.; Merchant, J.; Carter, Z.; Gordon, Z.; Carey, G.; Webb, T.J. Targeting Natural Killer T Cells in Solid Malignancies. Cells 2021, 10, 1329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, T.; Zheng, L.; Liu, H.; Song, W.; Liu, D.; Li, Z.; Pan, C.X. Combination strategies to maximize the benefits of cancer immunotherapy. J. Hematol. Oncol. 2021, 14, 156. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, M.; Wang, M.; Wan, D.; Wei, Y.; Wei, X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, M.; Poznanska, J.; Fechner, F.; Michalska, N.; Paszkowska, S.; Napierala, A.; Mackiewicz, A. Cancer Vaccine Therapeutics: Limitations and Effectiveness—A Literature Review. Cells 2023, 12, 2159. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Yam, J.W.P.; Mao, X. Dendritic Cell Vaccines: A Shift from Conventional Approach to New Generations. Cells 2023, 12, 2147. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Wahi, A.; Sharma, P.; Nagpal, R.; Raina, N.; Kaurav, M.; Bhattacharya, J.; Rodrigues Oliveira, S.M.; Dolma, K.G.; Paul, A.K.; et al. Recent Advances in Cancer Vaccines: Challenges, Achievements, and Futuristic Prospects. Vaccines 2022, 10, 2011. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.S.; Johnson, D.B.; Balko, J.M. Corticosteroids and Cancer Immunotherapy. Clin. Cancer Res. 2023, 29, 2580–2587. [Google Scholar] [CrossRef] [PubMed]
- Kalfeist, L.; Galland, L.; Ledys, F.; Ghiringhelli, F.; Limagne, E.; Ladoire, S. Impact of Glucocorticoid Use in Oncology in the Immunotherapy Era. Cells 2022, 11, 770. [Google Scholar] [CrossRef]
- Meng, L.; Wei, Y.; Xiao, Y. Chemo-immunoablation of solid tumors: A new concept in tumor ablation. Front. Immunol. 2022, 13, 1057535. [Google Scholar] [CrossRef]
- Li, J.Y.; Chen, Y.P.; Li, Y.Q.; Liu, N.; Ma, J. Chemotherapeutic and targeted agents can modulate the tumor microenvironment and increase the efficacy of immune checkpoint blockades. Mol. Cancer 2021, 20, 27. [Google Scholar] [CrossRef]
- Eng, L.; Sutradhar, R.; Niu, Y.; Liu, N.; Liu, Y.; Kaliwal, Y.; Powis, M.L.; Liu, G.; Peppercorn, J.M.; Bedard, P.L.; et al. Impact of Antibiotic Exposure before Immune Checkpoint Inhibitor Treatment on Overall Survival in Older Adults with Cancer: A Population-Based Study. J. Clin. Oncol. 2023, 41, 3122–3134. [Google Scholar] [CrossRef]
- Peng, C.; Rabold, K.; Mulder, W.J.M.; Jaeger, M.; Netea-Maier, R.T. Kinase Inhibitors’ Effects on Innate Immunity in Solid Cancers. Cancers 2021, 13, 5695. [Google Scholar] [CrossRef]
- Castelo-Soccio, L.; Kim, H.; Gadina, M.; Schwartzberg, P.L.; Laurence, A.; O’Shea, J.J. Protein kinases: Drug targets for immunological disorders. Nat. Rev. Immunol. 2023, 23, 787–806. [Google Scholar] [CrossRef]
- Fogli, L.K.; Aurigemma, R.; Sommers, C.L.; Singh, A.; Bourcier, K.; Ernstoff, M.S.; NCI Cell Therapy Workshop Committee. Challenges and next steps in the advancement of immunotherapy: Summary of the 2018 and 2020 National Cancer Institute workshops on cell-based immunotherapy for solid tumors. J. Immuno Ther. Cancer 2021, 9, e003048. [Google Scholar] [CrossRef]
- Xu, S.; Tan, S.; Guo, L. Patient-Derived Organoids as a Promising Tool for Multimodal Management of Sarcomas. Cancers 2023, 15, 4339. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A.; Carini, C. Insights and Strategies of Melanoma Immunotherapy: Predictive Biomarkers of Response and Resistance and Strategies to Improve Response Rates. Int. J. Mol. Sci. 2022, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Lv, Z.; Xu, D.; Cui, J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 2020, 8, 34. [Google Scholar] [CrossRef]
- Shao, J.; Jin, Y.; Jin, C. A new approach to overcoming resistance to immunotherapy: Nanotechnology. Front. Oncol. 2023, 13, 1210245. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhang, T.; Xia, R.; Wei, Y.; Wei, X. Targeting the tumor stroma for cancer therapy. Mol. Cancer 2022, 21, 208. [Google Scholar] [CrossRef]
- Curigliano, G.; Gelderblom, H.; Mach, N.; Doi, T.; Tai, D.; Forde, P.M.; Sarantopoulos, J.; Bedard, P.L.; Lin, C.C.; Hodi, F.S.; et al. Phase I/Ib Clinical Trial of Sabatolimab, an Anti-TIM-3 Antibody, Alone and in Combination with Spartalizumab, an Anti-PD-1 Antibody, in Advanced Solid Tumors. Clin. Cancer Res. 2021, 27, 3620–3629. [Google Scholar] [CrossRef]
- Finck, A.V.; Blanchard, T.; Roselle, C.P.; Golinelli, G.; June, C.H. Engineered cellular immunotherapies in cancer and beyond. Nat. Med. 2022, 28, 678–689. [Google Scholar] [CrossRef]
- Kiaie, S.H.; Salehi-Shadkami, H.; Sanaei, M.J.; Azizi, M.; Shokrollahi Barough, M.; Nasr, M.S.; Sheibani, M. Nano-immunotherapy: Overcoming delivery challenge of immune checkpoint therapy. J. Nanobiotechnol. 2023, 21, 339. [Google Scholar] [CrossRef]
- Bai, R.; Chen, N.; Li, L.; Du, N.; Bai, L.; Lv, Z.; Tian, H.; Cui, J. Mechanisms of Cancer Resistance to Immunotherapy. Front. Oncol. 2020, 10, 1290. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.M.; Yang, M.F.; Liang, Y.J.; Peng, Q.Z.; Zhang, Y.; Tian, C.M.; Wang, L.S.; Yao, J.; Nie, Y.Q.; et al. New Insights Into the Epigenetic Regulation of Inflammatory Bowel Disease. Front. Pharmacol. 2022, 13, 813659. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Z.; Yao, T.; Zhou, J.; Wang, Z. The immune-related role of beta-2-microglobulin in melanoma. Front. Oncol. 2022, 12, 944722. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhong, F.; Wu, H.; Che, K.; Shi, J.; Wu, N.; Fu, Y.; Wang, Y.; Hu, J.; Qian, X.; et al. Prevalence and Associations of Beta2-Microglobulin Mutations in MSI-H/dMMR Cancers. Oncologist 2023, 28, e136–e144. [Google Scholar] [CrossRef]
- Hu, Q.; Bian, Q.; Rong, D.; Wang, L.; Song, J.; Huang, H.S.; Zeng, J.; Mei, J.; Wang, P.Y. JAK/STAT pathway: Extracellular signals, diseases, immunity, and therapeutic regimens. Front. Bioeng. Biotechnol. 2023, 11, 1110765. [Google Scholar] [CrossRef]
- Shen, H.; Huang, F.; Zhang, X.; Ojo, O.A.; Li, Y.; Trummell, H.Q.; Anderson, J.C.; Fiveash, J.; Bredel, M.; Yang, E.S.; et al. Selective suppression of melanoma lacking IFN-gamma pathway by JAK inhibition depends on T cells and host TNF signaling. Nat. Commun. 2022, 13, 5013. [Google Scholar] [CrossRef]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Blass, E.; Ott, P.A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021, 18, 215–229. [Google Scholar] [CrossRef]
- Mehdi, A.; Rabbani, S.A. Role of Methylation in Pro- and Anti-Cancer Immunity. Cancers 2021, 13, 545. [Google Scholar] [CrossRef]
- Desaulniers, D.; Vasseur, P.; Jacobs, A.; Aguila, M.C.; Ertych, N.; Jacobs, M.N. Integration of Epigenetic Mechanisms into Non-Genotoxic Carcinogenicity Hazard Assessment: Focus on DNA Methylation and Histone Modifications. Int. J. Mol. Sci. 2021, 22, 10969. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Basdra, E.K.; Papavassiliou, A.G.; Piperi, C. Prominent Role of Histone Modifications in the Regulation of Tumor Metastasis. Int. J. Mol. Sci. 2021, 22, 2778. [Google Scholar] [CrossRef]
- Xiong, D.; Zhang, L.; Sun, Z.J. Targeting the epigenome to reinvigorate T cells for cancer immunotherapy. Mil. Med. Res. 2023, 10, 59. [Google Scholar] [CrossRef]
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: Mechanism and clinical application. Clin. Epigenet. 2021, 13, 166. [Google Scholar] [CrossRef]
- Shen, C.; Li, M.; Duan, Y.; Jiang, X.; Hou, X.; Xue, F.; Zhang, Y.; Luo, Y. HDAC inhibitors enhance the anti-tumor effect of immunotherapies in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1170207. [Google Scholar] [CrossRef]
- Lu, G.; Jin, S.; Lin, S.; Gong, Y.; Zhang, L.; Yang, J.; Mou, W.; Du, J. Update on histone deacetylase inhibitors in peripheral T-cell lymphoma (PTCL). Clin. Epigenet. 2023, 15, 124. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Thomas, A.M.; Iebba, V.; Zalcman, G.; Friard, S.; Mazieres, J.; Audigier-Valette, C.; Moro-Sibilot, D.; Goldwasser, F.; et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 2022, 28, 315–324. [Google Scholar] [CrossRef]
- Patel, P.; Poudel, A.; Kafle, S.; Thapa Magar, M.; Cancarevic, I. Influence of Microbiome and Antibiotics on the Efficacy of Immune Checkpoint Inhibitors. Cureus 2021, 13, e16829. [Google Scholar] [CrossRef]
- Najmi, M.; Tran, T.; Witt, R.G.; Nelson, K.C. Modulation of the Gut Microbiome to Enhance Immunotherapy Response in Metastatic Melanoma Patients: A Clinical Review. Dermatol. Ther. 2022, 12, 2489–2497. [Google Scholar] [CrossRef]
- Villemin, C.; Six, A.; Neville, B.A.; Lawley, T.D.; Robinson, M.J.; Bakdash, G. The heightened importance of the microbiome in cancer immunotherapy. Trends Immunol. 2023, 44, 44–59. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Seyhan, A.A.; Carini, C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? J. Transl. Med. 2019, 17, 114. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Lin, K.; Wang, X.; Tu, Y.; Zhuo, Z. Progress in building clinically relevant patient-derived tumor xenograft models for cancer research. Anim. Model. Exp. Med. 2023, 6, 381–398. [Google Scholar] [CrossRef]
- Chen, K.; Li, Y.; Wang, B.; Yan, X.; Tao, Y.; Song, W.; Xi, Z.; He, K.; Xia, Q. Patient-derived models facilitate precision medicine in liver cancer by remodeling cell-matrix interaction. Front. Immunol. 2023, 14, 1101324. [Google Scholar] [CrossRef]
- Chitrangi, S.; Vaity, P.; Jamdar, A.; Bhatt, S. Patient-derived organoids for precision oncology: A platform to facilitate clinical decision making. BMC Cancer 2023, 23, 689. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, R.; Payra, S.; Singh, S.K. Artificial Intelligence and Machine Learning in Pharmacological Research: Bridging the Gap Between Data and Drug Discovery. Cureus 2023, 15, e44359. [Google Scholar] [CrossRef]
- Dlamini, Z.; Francies, F.Z.; Hull, R.; Marima, R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct. Biotechnol. J. 2020, 18, 2300–2311. [Google Scholar] [CrossRef]
- Subbiah, V. The next generation of evidence-based medicine. Nat. Med. 2023, 29, 49–58. [Google Scholar] [CrossRef]
- Chen, Q.; Jia, G.; Zhao, X.; Bao, Y.; Zhang, Y.; Ozkan, C.; Minev, B.; Ma, W. Novel Survivin Peptides Screened with Computer Algorithm Induce Cytotoxic T Lymphocytes with Higher Cytotoxic Efficiency to Cancer Cells. Front. Mol. Biosci. 2020, 7, 570003. [Google Scholar] [CrossRef]
- Chen, Q.; Bao, Y.; Burner, D.; Kaushal, S.; Zhang, Y.; Mendoza, T.; Bouvet, M.; Ozkan, C.; Minev, B.; Ma, W. Tumor growth inhibition by mSTEAP peptide nanovaccine inducing augmented CD8(+) T cell immune responses. Drug Deliv. Transl. Res. 2019, 9, 1095–1105. [Google Scholar] [CrossRef]
- Ma, W.; Chen, M.; Kaushal, S.; McElroy, M.; Zhang, Y.; Ozkan, C.; Bouvet, M.; Kruse, C.; Grotjahn, D.; Ichim, T.; et al. PLGA nanoparticle-mediated delivery of tumor antigenic peptides elicits effective immune responses. Int. J. Nanomed. 2012, 7, 1475–1487. [Google Scholar] [CrossRef] [PubMed]
- Haimi, M. The tragic paradoxical effect of telemedicine on healthcare disparities—A time for redemption: A narrative review. BMC Med. Inform. Decis. Mak. 2023, 23, 95. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).