Simple Summary
Medulloblastoma is the most prevalent intracerebellar pediatric brain tumor, accounting for approximately 20% of all childhood brain tumors and over 60% of embryonal brain tumors. MYC-driven medulloblastoma has extreme metastatic potential and is often resistant to multipronged treatment. PRMT5 plays a key role in cell functions and processes in MYC-driven medulloblastoma by stabilizing the MYC protein. RMT5 inhibitors can potentially disrupt MYC’s function, impeding tumor progression and offering a target therapeutic approach to treat MYC-amplified medulloblastoma. Here, we highlight the challenges that must be addressed in future drug development.
Abstract
MYC amplification or overexpression is most common in Group 3 medulloblastomas and is positively associated with poor clinical outcomes. Recently, protein arginine methyltransferase 5 (PRMT5) overexpression has been shown to be associated with tumorigenic MYC functions in cancers, particularly in brain cancers such as glioblastoma and medulloblastoma. PRMT5 regulates oncogenes, including MYC, that are often deregulated in medulloblastomas. However, the role of PRMT5-mediated post-translational modification in the stabilization of these oncoproteins remains poorly understood. The potential impact of PRMT5 inhibition on MYC makes it an attractive target in various cancers. PRMT5 inhibitors are a promising class of anti-cancer drugs demonstrating preclinical and preliminary clinical efficacies. Here, we review the publicly available preclinical and clinical studies on PRMT5 targeting using small molecule inhibitors and discuss the prospects of using them in medulloblastoma therapy.
1. Introduction
Medulloblastoma is the most prevalent intracerebellar pediatric brain tumor, accounting for approximately 20% of all childhood brain tumors and over 60% of embryonal brain tumors [1]. Medulloblastoma is classified into four major molecularly diverse subgroups including wingless (WNT), Sonic hedgehog (SHH, p53 mutant and p53 wild type), Group 3, and Group 4 medulloblastomas [2,3,4]. The WNT subgroup comprises approximately 10% of the medulloblastoma cases and has the most favorable clinical outcomes, with a 5-year overall survival surpassing 95% [5,6,7]. The SHH subgroup typically displays deregulation of the SHH signaling pathway and represents approximately one-third of childhood patients with medulloblastomas [2,8]. Group 3 medulloblastomas often exhibit MYC overexpression and have the most dismal clinical diagnosis of the four medulloblastoma subgroups, with a survival rate of less than 60%. MYC-driven medulloblastomas have extreme metastatic potential and are often resistant to multipronged treatment [9,10,11]. Group 4 is the most prevalent subgroup, accounting for nearly 40% of all medulloblastoma tumors, and is normally seen in children aged 5–10 years and rarely in infants [2]. Although progress has been made in understanding medulloblastoma at the molecular and genetic level, comparatively few targeted therapies have achieved clinical success. Current therapies for medulloblastoma have progressed in favor of patient survival to about 70% [8]. However, this comes with consequences, as standard treatment or medications like chemotherapy, brain and spinal cord radiation, and surgical removal leave patients at risk for permanent mental disabilities [1,12].
Post-translational modification (PTM) is one targetable regulatory mechanism of MYC and other proteins, with the potential to be developed therapeutically. While the roles of PTMs like phosphorylation [13], ubiquitinoylation [14], and acetylation [15] in controlling these proteins responsible for medulloblastoma have received significant attention, arginine methylation has only recently been investigated. Arginine methylation is one of the common PTM processes that are catalyzed by a member of the protein arginine methyltransferase (PRMT) family; this group of nine enzymes is responsible for the methylation of arginine, using S-adenosylmethionine (SAM) as a methyl group donor. The physiological control of many cellular processes, including splicing transcription and mitosis, depends on the activity of PRMT family enzymes [16]. PRMTs have also been revealed to be involved in the progression of various types of cancers [17,18]. In humans, PRMT members can be divided into various classes based on their enzymatic role, i.e., type I (PRMT1-4, PRMT6, and PRMT8) that catalyze the formation of monomethyl arginine (MMA) and asymmetric dimethyl arginine (ADMA); type II (PRMT5 and PRMT9) that catalyze the formation of MMA and symmetric dimethyl arginine (SDMA); and type III (PRMT7) which is responsible for the formation of MMA [19]. As the most prevalent type II SDMA methyltransferase, PRMT5 forms a heterotetrametric complex with a protein called methylosome protein 50 (MEP50) that can catalyze symmetric demethylation of various histone and non-histone proteins [20]. Remarkably, PRMT5 was proven to regulate the function of glioma-associated oncogene homolog 1(GLI1) protein in an SHH-responsive cell line [21]. PRMT5 also represents a requisite driver of tumor progression in SHH-medulloblastoma and MYC-amplified medulloblastoma [22,23]. During conversion to malignancy or metastasis, PRMT5 acts as an oncogene. PRMT5 enzyme inhibition or its catalytic depletion frequently reduces or halts cellular proliferation, while its hyperexpression leads to hyper-proliferation [24,25,26]. Consequently, PRMT5 is emerging as a novel target for the treatment of various cancers, including medulloblastoma.
Recently, PRMT5 inhibitors have been credited with inhibiting the growth of cancerous cells in vitro and in vivo. Various PRMT5 inhibitors with different functions have undergone clinical trials for the treatment of advanced cancer or recurrent solid tumors [27,28]. The effects of PRMT5 inhibition on cancerous cells’ proliferation, invasion, and migration can contribute to anti-cancer efficacy [29,30]. This review explores the current knowledge of the effectiveness of PRMT5 inhibitors in preclinical and preliminary clinical settings, which may aid in understanding how to treat MYC-amplified medulloblastoma more effectively and safely.
2. PRMT5 Structure, Function, and Localization
2.1. Structure
PRMT5 is a primary type II arginine methyl transferase that forms a prominent methylosome complex with distinctive binding oligopeptides, such as the WD (Trp-Asp) repeat-containing 50-kilodalton methylosome protein (MEP50). PRMT5 requires the existence of diverse substrate adapters such as rio-domain-containing protein 1 (RioK1), chloride channel nucleotide-sensitive 1A protein (pIC1n) and cooperator of PRTM5 (COPR5) to detect and catalyze the SDMA on histone and non-histone proteins via PTMs [31,32,33]. PRMT5′s structure consists of a triphosphate isomerase (TIM) barrel, an intermediate Rossmann-fold, and a C-terminal β-barrel [34]. Four PRMT5 units generate a hetero octameric complex by binding with four MEP50s (Figure 1). Studies have demonstrated that PRMT5 alone has minimal methyltransferase activity; it must be complexed with MEP50 to achieve normal catalysis of SDMA on proteins [35]. This could be because MEP50 enhances the stability of PRMT5 for a long time by binding with proteins and acting as a metastable cofactor.
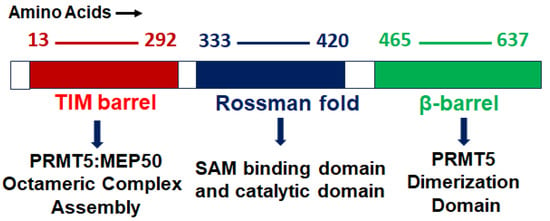
Figure 1.
PRMT5 protein structure: structural and functional domains.
2.2. Function
PRMT5 plays a key role in cell functions and processes by regulating the methylation of cellular proteins, which affects oncogenic cellular processes such as cell proliferation and differentiation [29,30,36]. PRMT5 regulates these processes by modifying gene expression to stabilize histones H4R3, H3R2, H3R8, and H2AR3 and non-histone proteins via the SDMA process [37,38]. An extensive range of nonhistone proteins have also been revealed as PRMT5 substrates, including androgen receptor (AR), EGFR, GATA4, C-MYC, N-MYC, IL-2, E2F1, GM130, HOXA9, KLF4, KLF5, NOTCH, NFkB(p65), PDCD4, POLR2A, P53, RAF proteins, SPT5, SREBP1a, Sm proteins, nucleolin, and others [12,36,39,40,41,42,43,44,45,46,47,48,49,50,51,52]. In addition, some substrates such as certain nonhistone oncogenic transcription factors are symmetrically dimethylated by PRMT5. PRMT5-regulated cellular processes are shown in Figure 2. The importance of arginine methylation by PRMT5 in cancer progression has only recently become apparent [17]. PRMT5 knockout mice exhibited embryonic lethality, demonstrating the role of PRTM5 in embryonic development and crucial biological functions. In mouse embryonic stem cells (ESCs), PRMT5 maintains pluripotency, whereas in human ESCs, it influences only proliferation [53,54]. PRMT5 is needed for neural stem cell persistence and its deletion causes premature death of the mouse by disrupting the development of the central nervous system [55]. PRMT5 promotes SWI/SNF-mediated chromatin remodeling and controls the process of myogenesis. Deletion of PRMT5 causes an obstacle in developmental processes, uncontrolled proliferation, and impairment of adult tissue differentiation [53,54,55,56,57,58]. Notably, PRMT5 is overexpressed in a number of cancers, including melanoma, multiple myeloma, lymphoma, glioblastoma, breast, lung, pancreas, prostate, ovarian, and colorectal cancers, and high expression of PRMT5 often correlates with poor patient clinical outcomes [38,59]. Organ-specific functions of PRMT5 are shown in Table 1. The higher expression of PRMT5 in cancer is thought to epigenetically suppress tumor suppressor and cell cycle genes [17,60]. Recently, the association of PRMT5 with MYC was found in numerous cancers, including brain tumors such as glioblastoma; this association creates abnormalities in MYC function [61,62,63]. Consequently, PRMT5 has been recognized as an oncogenic function and has received extensive interest as a potential target for better clinical outcomes. To this end, numerous potent therapeutic agents have been developed to inhibit PRMT5 and their antitumor effects are now being assessed in preclinical models and clinical trials [27,64].
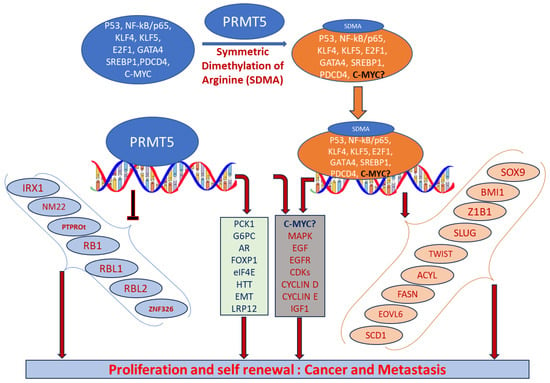
Figure 2.
Biological functions of PRMT5 that regulate cellular processes. Elevated expression of PRMT5 can cause post-translational modification of several transcription factors by symmetrically dimethylating arginine residues of proteins and regulate the expression of their corresponding targeted genes. When recruited to the promoter regions of precise target genes in the nucleus, they can promote cell proliferation and tumorigenesis.
2.3. Localization
Cytosolic and nuclear localization of PRMT5 helps to determine its role in the cell. PRMT5 is predominantly localized in the cytoplasm in lung [65], prostate [66], and melanoma cancer [67]. Diffused cellular localization of PRMT5 was confirmed in both the cytoplasm and the nucleus of brain tumor glioblastoma cells [61]. Cytoplasmic and nuclear localization of PRMT5 has also been confirmed in various preclinical mouse models and primary human cancer tissues [68]. In adult mice, PRMT5 is expressed predominantly in the nucleus of the neurons in the cerebrum and spinal cord [55]. Han et al. demonstrated the high expression of PRMT5 as a marker of malignant progression in glioblastoma and its crucial role in tumor growth [63]. Our lab recently analyzed the localization of the PRMT5 in tumor tissues of medulloblastoma patients as well as in MYC-amplified cell lines. PRMT5 demonstrated predominantly nuclear localization in both HD-MB03 and primary tumor cells [22].


Table 1.
Organ-specific roles of PRMT5.
Table 1.
Organ-specific roles of PRMT5.
| Organ | Cellular Function | Mechanism | References |
|---|---|---|---|
| Brain | Cell cycle progression, apoptosis | Altered expression and stability of MYC | [22] |
| Phase separation | Methylation of FUS | [69] | |
| GSK3β-NF-kβ signaling | Altered expression of E2F1 | [70] | |
| HTT toxicity | Altered expression of HTT | [71] | |
| AKT-ERK signaling, cell cycle progression | Altered expression of PTEN | [72] | |
| Cell cycle progression, stemness | Altered RNA Splicing | [73,74] | |
| mTOR signaling | Methylation (hnRNPA1) | [75] | |
| DNA instability response | Altered expression of RNF168 | [76] | |
| Cell migration, cell cycle progression, and apoptosis | Altered expression of LRP12 | [62] | |
| AKT signaling and metastasis | Methylation of PKB | [77] | |
| Lungs | Metastasis | Altered expression of EMT genes | [78] |
| Metastasis | Altered expression of SHARPIN | [79] | |
| Metastasis | Altered expression of FGFR3/miR-99 family | [80] | |
| Metastasis | Methylation of KLF5 | [81] | |
| Liver | Lipid metabolism | Methylation of SREBP | [47] |
| ERK signaling | Altered expression of BTG2 | [82] | |
| PRMY5 deprivation | PRMT5 activity of LINC01138 | [83] | |
| WNT-β-Catenin signaling | Altered cofactor binding of LYRIC | [84] | |
| Spleen | NA | Altered stability of MYC | [85] |
| Pancreas | Glucose metabolism, cell cycle progression | Altered stability of MYC | [86,87] |
| Bone | Type I interferon signaling | Altered expression (interferon gene) | [88,89] |
| Prostate | AR, ERG signaling | Altered methylation (AR) | [90,91] |
| Ovary | NA | Altered methylation (E2F1) | [92] |
| Heart | Transcriptional activity | Methylation (GATA4) | [45] |
| Breast | Stemness | Altered expression of C-MYC, KLF4, and OCT4 | [93] |
| Stemness | Altered expression of FOXP1 | [94] | |
| Metastasis and invasion | Altered expression of AKT genes | [78] | |
| Metastasis and AKT signaling | Methylation of AKT | [95] | |
| Cell cycle progression | Methylation of KLF4 | [96] | |
| Cell migration | Methylation of ZNF326 | [97] | |
| NA | Methylation of PDCD4 | [98] |
Abbreviations: AKT-ERK, alpha serine/threonine-protein-extracellular-regulated kinase; AR, androgen receptor; E2F1, E2 promoter binding factor 1; EMT, epithelial–mesenchymal transition; ERG, ETS-related gene; FGFR3, fibroblast growth factor 3; FOXP1, forkhead box protein P1; FUS, fused in sarcoma; GSK3β-NF-kβ, glycogen synthase kinase; hnRNPA1, heterogeneous nuclear ribonucleoprotein A1; HTT, huntingtin protein; KLF4, Kruppel-like factor 4; LRP12, low-density lipoprotein receptor-related protein 12; LINC01138, long non-coding RNA; miR-99, microRNA99; mTOR, mammalian target of rapamycin; OCT4, octamer binding protein 4; PKB, protein kinase B; PDCD4, program cell death protein 4; SREBP, sterol regulatory element-binding protein; ZNF326, zinc finger protein 326.
3. PRMT5 Association with MYC-Driven Medulloblastoma
Epigenetic deregulation plays a key role in medulloblastoma tumorigenesis, especially in aggressive Group 3 and Group 4 medulloblastomas [99,100,101,102], where germline mutations in known cancer predisposition genes are rare. Indeed, epigenetic deregulator or chromatin modifiers, including histone acetylase or methylation/methyltransferase activities, are very common in Group 3 and 4 medulloblastomas compared to other subgroups. This emphasizes the need to discover and understand the pertinent mechanisms of epigenetic regulation or PTMs and the corresponding therapeutic targets. We recently reported that PRTM5 is a critical regulator MYC oncoprotein in an MYC-amplified (Group 3) medulloblastoma [22]. We found that high levels of PRMT5 not only mirror MYC expression but also correlate with poor outcomes in Group 3 medulloblastoma patients. Mechanistically, we showed that PRMT5 stabilizes the MYC protein by physically interacting with it, raising the intriguing possibility that PRMT5 can regulate MYC function at both the transcriptional and translational/post-translational levels. The exact MYC oncogenic programs regulated by PRMT5 in medulloblastoma are largely unknown. Therefore, exploring the regulation of MYC-driven oncogenic progresses by PRMT5 is crucial to identify effective therapeutics for these high-risk patients.
The involvement of PRMT5 has been verified in the epigenetic regulation of chromatin complexes following interaction with numerous proteins, including transcription factors [42], and their activities are dysregulated in various cancers [59]. In recent studies, high levels of PRMT5 and MYC corelate with glioma malignancy [61,62,63]. Furthermore, PRMT5 is physically associated with N-MYC (an MYC homologue) and enhances the stability of N-MYC in neuroblastoma cells [51]. Nonetheless, the function of PRMT5 and its interaction with MYC in MYC-driven medulloblastoma have not been fully investigated. Favia et al. reported that the association of PRMT1 and PRMT5 with MYC in glioblastoma stem cells resulted in MYC being dimethylated symmetrically and asymmetrically by both enzymes, respectively [103]. MYC-driven cellular processes resulting from symmetric dimethylation by PRMT5 are shown in Figure 3. The colocalization of PRMT5 and MYC suggests that PRMT5 forms a complex with MYC and supports its stabilization in MYC-amplified medulloblastoma cells. This physical interaction of PRMT5 and MYC implies a potential role of PRMT5 in medulloblastoma tumorigenesis.
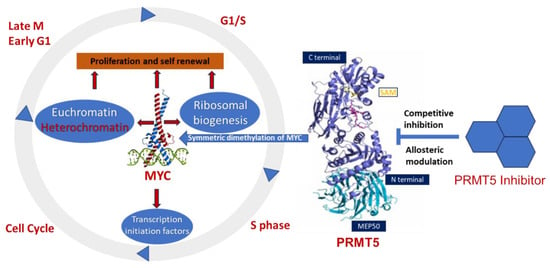
Figure 3.
Overexpression of PRMT5 causes symmetric demethylation and stabilization of MYC, leading to reduced apoptosis and enhanced cell proliferation. As indicated, various steps in this process can be modulated by PRMT5 inhibitors.
Highly expressed PRMT5 stabilizes MYC and promotes its expression in medulloblastomas. Studies support the predictive value of PRMT5 overexpression as a biomarker for aggressive tumorigenesis in cancer patients. Knockdown of PRMT5 in medulloblastoma cells suppresses cell growth by diminishing MYC stability, supporting the functional role of the PRMT5–MYC interaction complex in medulloblastoma [22]. Since MYC and PRMT5 co-expression and colocalization were observed in the nucleus, PRMT5 could also regulate MYC function at the transcriptional level. Further studies are needed to investigate PRMT5’s roles in the regulation of the transcription and translation of MYC.
PRMT5 is a stemness factor crucial in maintaining the balance between quiescence, proliferation, and generation for cancer stem cells and non-cancer cells. The role of PRMT5 in stemness has been demonstrated in embryonic (ESCs) and neural stem cells (NSCs) [53,72,104]. Provided that NCCs or cancer stem cells have a great influence on medulloblastoma recurrence and tumorigenesis, there might be a role for PRMT5 in regulating the self-renewal of tumor initiation in medulloblastoma. Recently, the methylation of stemness factor KLF-4 (Kruppel-like factor-4) by PRTM5 was shown in breast cancer [96]. This methylation leads to KLF4 protein stabilization, promoting tumorigenesis. In another study, researchers synthesized a novel compound that has the potency to inhibit PRMT5, disrupt the interaction of PRMT5 and KLF4, and suppress breast cancer development [105]. KLFs are evolutionarily conserved zinc-finger-associated transcription factors with distinct regulatory functions in cell growth, proliferation, and differentiation. Moreover, PRMT5 interacts with KLF5 (another member of KLF family proteins) and accelerates its dimethylation, a mechanism that depends on methyltransferase activity [81]. Further investigation to understand the mechanism of PRTM5–KLF4/KL5 interactions could uncover another new strategy to elucidate therapeutic targets for MYC-amplified medulloblastoma.
4. Potential Inhibitors of PRMT5
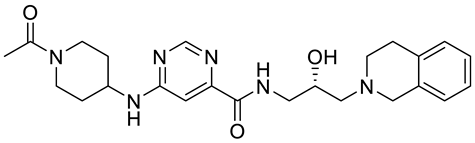
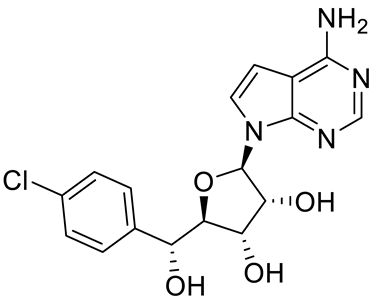
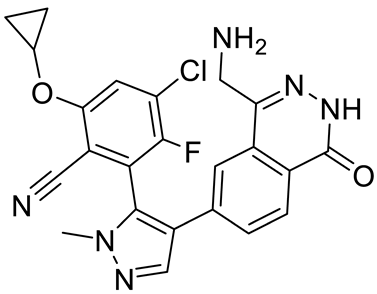
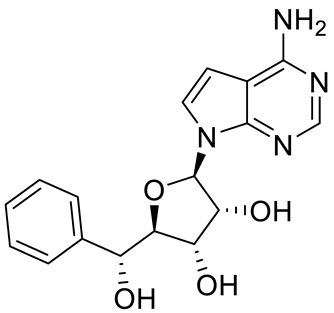
PRMT5 inhibitors have been proven to prevent the growth of cancerous cells in vitro and in vivo. Many PRMT5 inhibitors have entered clinical trials for the treatment of multiple types of cancer [34,106,107]. The pharmacological effects of these inhibitors with their targets in various cancers are summarized in Table 2, and details about corresponding clinical trials are given in Table 3.

Table 2.
Pharmacologically active PRMT5 inhibitors.

Table 3.
PRMT5 inhibitors in clinical trials.
4.1. JNJ-64619178
JNJ-64619178 (International Patent Number: WO/2017/032840 A1) is a potent PRMT5 inhibitor that irreversibly binds to the SAM pocket of the PRMT5/MEP50 and establishes a short kinetic constant of target unbinding, resulting in prolonged trapping of PRMT5/MEP50 in an inactive transition that impedes arginine methylation of histone proteins to reduce cellular proliferation [130,131]. The pharmacokinetic (PK) profile of JNJ-64619178 on a single post-oral dose (PO; 10 mg/kg) and intravenous (IV; 2.5 mg/kg) administration led to a low clearance (CL = 6.6 mL/min/kg) in mice and reasonable oral bioavailability (F = 36%). It is presently in clinical trials (NCT03573310) for patients with advanced solid tumors, non-Hodgkin lymphoma, and lower-risk myelodysplastic syndrome [28,132]. Initial clinical results revealed evidence that JNJ-64619178 has manageable toxicity and antitumor activity at a dose of 1.5 mg QD [132]. A phase 1 dose escalation study involving 90 patients was conducted to identify recommended phase 2 dose (RP2D) levels for JNJ-64619178. Based on safety, clinical activity, and PK and pharmacodynamic (PD) outcomes, two RP2Ds (1.5 mg intermittently and 1 mg once daily) were selected to inhibit PRMT5 activity in patients with cancerous tumors [133].
4.2. PF06939999
PF06939999 is another potent, selective SAM-competitive inhibitor whose complete mechanism is still unknown. PF06939999 displayed superior in vitro and in vivo antitumor activity with concomitant loss of SDMA [109]. The drug sensitivity to PF06939999 in non-small cell lung cancer (NSCLC) is associated with signaling pathways involving MYC, cell cycles, and spliceosomes and with mutations in splicing factors. The PK profile of PF06939999 in a single dose (PO, 10 mg/kg; IV, 2 mg/kg) revealed a reasonable plasma clearance (CL = 40 mL/min/kg) and steady-state volume of distribution (Vss 3.8 L/kg) in rodents with moderate oral bioavailability (F = 40%). A phase I dose escalation clinical trial (NCT03854227) showed promising results in patients with various cancers, including NSCLC, head and neck squamous cell carcinoma (HNSCC), and others [132,134]. The results of the NCT03854227 were described in the ASCO annual meeting in 2021 [135].
4.3. EPZ015666
EPZ015666 is a selective substrate-competitive inhibitor of PRMT5 with potential antiproliferative and antineoplastic activity [110,136]. Previously, it was known as GSK53235025 [106]. This inhibitor was first employed in mantle cell lymphoma. It was also used in multiple myeloma and medulloblastoma [137]. The efficacy of EPZ015666 was determined on the three MYC-amplified medulloblastoma cell lines (HD-MB03, D-283, and D-341). Medulloblastoma cells were treated with EPZ015666 in a dose-dependent manner for 72 hr and the results of cell growth assays confirmed that EPZ015666 induced growth inhibition directly proportionate to the dose in all MYC-driven medulloblastoma cell lines at a low micromolar potency, with an IC50 of 1.5–2.5 μM [22]. The PK profile of EPZ015666 in a single dose (oral, 100 mg/kg) revealed a low plasma clearance and a satisfactory brain distribution in mice. EPZ015666 significantly downregulates the higher expression of PRMT5 and MYC in medulloblastoma cells [22], suggesting it has therapeutic potential for MYC-driven medulloblastoma.
4.4. GSK3326595
GSK3326595 is a selective substrate-competitive PRMT5 inhibitor with potential antitumor and antiproliferative activity, which has shown efficacy in various tumor models [77]. Two clinical trials [138,139,140] are assessing this compound in patients with solid tumor cancers, primarily adenoid cystic carcinoma and colorectal and breast cancer. A phase I (NCT02783300) clinical trial is underway to assess the safety, PK, and PD in adults with solid tumors. General adverse events in this study were common but mild. Another clinical trial (NCT03614728) on reverted or refractory myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML) from MDS is active [141]. A third trial, designed to evaluate the drug in patients with early breast cancer, has been completed but no results have been posted yet [142].
4.5. AMG 193
AMG 193 is a methylthioadenosine (MTA)-cooperative PRMT5 inhibitor that specifically targets the MTS-bound state of PRMT5 [119]. This state is enhanced in methylthioadenosine phosphorylase (MTAP)-null tumors. AMG 193 has shown potential inhibition in a patient-derived xenograft model as well as MTAP-null cancer cell lines. NCT05094336, a first-in-human (FIH), open-label, multicenter phase I/II trial, is enrolling patients with MTAP-null NSCLC to evaluate the safety, tolerability, PK, PD, and efficacy of AMG 193. Docetaxel is used as a combination drug for this clinical trial [143]. However, MTAP-null mutations are still not identified in medulloblastoma, so AMG 193 might be less relevant for treatment.
4.6. PRT543
PRT543 is a potent PRMT5 inhibitor that inhibits the methyltransferase activity of PRMT5 by selectively binding to it, causing potent inhibition of cellular proliferation and SDMA formation in various cancerous cell lines [107,144]. PRT543 is currently under assessment in a phase I (NCT03886831) clinical trial that has been completed in patients with advanced solid tumors and hematologic malignancies. The purpose of the study was to define a safe dose and timetable for consumption in successive developments of PRT543. This dose-escalation, open-label study initially provided favorable results. Target engagement was confirmed by measuring serum SDMA. Phase I dose escalation and expansion studies are continuing to enroll patients.
4.7. PRT811
PRT811 is a selective and orally bioavailable PRMT5 inhibitor that passes the blood–brain barrier and shows effectiveness in high-grade glioma. PRT811 is currently under evaluation in a multicenter, open-level, phase I clinical trial (NCT04089449) in patients with central nervous system (CNS) lymphomas, recurrent high-grade gliomas, and advanced solid tumors. PRT811 has excellent PK properties in multiple preclinical species with a >two-fold higher brain vs. plasma exposure in rodents. PRT811 quickly penetrates the blood–brain barrier in rodents with higher exposure. PRT811 inhibits SDMA and cell proliferation of brain tumor cells [145]. PRT811 is broadly active against brain cancer cells and cancers with high rates of brain metastases [145]. Initial data were presented at the AACR-NCI-EORTC conference held in 2021 [121].
4.8. TNG908
TNG908 is also an MTA-cooperative PRMT5 inhibitor. The MTA-cooperative binding process has demonstrated the synthetic lethal relationship between MTAP losses and PRMT5 inhibition. TNG908 demonstrated a 15-fold higher potency in MTAP-null cancer cell lines. Pharmacokinetically, it is not a substrate of efflux transporters like Pgp and BCRP, which is a favorable predictor of the ability to cross the blood–brain barrier [122]. However, we still need to measure its exposure in the brain and determine the Kp. The PD properties of NTG908 allow for PRMT5 inhibition, which was confirmed by decreased levels of SDMA-modified proteins in a dose-dependent manner in a glioblastoma xenograft model. TNG908 demonstrated antineoplastic activity against MTAP-null selective tumors in various xenograft models, including tumor regression in a model representing NSCLC and cholangial and urothelial carcinomas [14,146]. One clinical trial (NCT05275478) is going to recruit patients with locally advanced solid tumors.
4.9. MRTX1719
MRTX1719 is another MTA-cooperative inhibitor of PRMT5. MRTX1719 catalytically binds the PRMT5–MTA complex and stabilizes it in an inactive form. In vitro, MRTX1719 demonstrates a long-lasting therapeutic effect in MTAP cells. Additionally, in vivo studies verified that MRTX1719 demonstrates potent and enduring inhibition of PRMT5 in a MTAP-deleted tumor xenograft model, reducing its SDMA activity [123]. An ongoing phase I/II clinical trial (NCT05245500) is evaluating the safety, tolerability, PK/PD, and antineoplastic activity against advanced and metastatic solid tumors. Preliminary data have been presented, including objective responses in patients with mesothelioma, MTAP-deleted melanoma, gallbladder adenocarcinoma, NSCLC, malignant peripheral nerve sheath tumors, solid tumors, and pancreatic adenocarcinoma [124].
4.10. LLY 283
LLY 283 has the potential to inhibit PRMT5 by binding competitively to the SAM binding site of PRMT5 as a cofactor competitive inhibitor [125]. LLY 283 can efficiently permeate the blood–brain barrier, as the compound is eliminated more quickly from the plasma than from the brain [74]. LLY 283 significantly decreases SDMA levels in cancerous cells. Pharmacologically, LLY 283 suppressed the growth of glioblastoma cell cultures derived from a cohort of 46 patients. Importantly, LLY 283 has shown significant survival benefits in mice implanted with a patient-derived xenograft (PDX), a preclinical orthotopic model of glioblastoma, even though more preclinical and clinical studies are warranted. LLY 283 showed a satisfactory PK profile, including a high metabolic stability and moderate permeability with oral bioavailability (F = 50%), which makes it an effective probe molecule for in vivo assessment.
4.11. Compound1a
Compound1a has also been recognized as a potential PRMT5 inhibitor. It binds allosterically to PRMT5 and competes with SAM at the binding site. Compound1a has previously been described as a human β-secretase (BACE1) and BACE2 inhibitor [147]. It demonstrated targeted effectiveness and cell-based inhibition of MCF7 cells, based on quantitation of symmetrically dimethylated nuclear protein levels. Compound1a makes an enzyme–substrate complex to bind with the co-crystal system of PRMT5–MEPP50. This complex reveals that a distinctive binding mode and considerable structural changes in the backbone of PRMT5 result from SAM-competitive inhibition [106,127,137].
4.12. CMP5
CMP5 is another molecule identified as a PRMT5 inhibitor [72]. By reducing the recruitment of PRMT5 in the glioblastoma cell line, it reduces the methylation of histone. However, it does not demethylate histones that have already been methylated by PRMT5. CMP5 has shown the capability to control differentiated and undifferentiated cancerous cell populations [148] and induce senescence and apoptosis of cancerous cells [136]. CMP5 has been shown to have anti-cancer efficacy against a glioblastoma xenograft model. In preclinical PK studies, CMP5 was revealed to accumulate in brain tissue without causing toxicity [149]. Chromatin histone methylation in the promoter region of DKK1 and DKK3 was hindered by CMP5-based inhibition of PRMT5, which decreased the expression of cyclin D1 and SUBRVIVIN [137]. Overexpression of cyclin D1 is directly linked to cancer progression.
4.13. GSK591
Previously known as EPZ015866, GSK591 was characterized as a potent inhibitor of PRMT5, including in vivo [27,110]. Proliferation of CRC cells is directly related to PRMT5 activity, and the inhibition of PRMT5 activity by GSK591 can stop proliferation and cell cycle progression. GSK591 significantly decreases SDMA in a dose-dependent manner and decreases the viability of neuroblastoma cell lines in a nanomolar range [77]. In one study, GSK591, in combination with LLY283, showed substantial survival benefits in an orthotopic PDX mouse model, although more preclinical and clinical studies are warranted in the future [74].
4.14. PRT382
PRT382 is a selective PRTM5 inhibitor with an adenosine backbone structurally similar to other PRMT5 inhibitors (JNJ-64619178, PF-06855800, LLY-283) [85]. PRT382 appears to have a similar SAM-competitive mechanism and optimal enzymatic kinetics in vitro that produces an IC50 of 2.8 nM with PRMT5/MEP50. It reduces SDMA with an IC50 of 27 nM and has antiproliferative activity in leukemia and lymphoma cancerous cell lines [85]. PRT382 displays low clearance and a high oral bioavailability in preclinical models. It is important to delineate the distribution of PRT382 in the brain and its efficiency in crossing the blood–brain barrier.
4.15. JBI-778
JBI-778 is a potent and strong inhibitor of PRMT5 [128] that reduces SDMA at an effective concentration of <10 nM. It exerts a strong antiproliferative activity in selected cell lines like NSCLC, neuroblastoma, glioblastoma, and medulloblastoma, with an IC50 ranging from 27 to 700 nM. JBI-778 can penetrate the blood–brain barrier with very high brain exposure in rodents, and it showed a favorable oral bioavailability in mice (F = 66%), rats (F = 52%), and dogs (F = 47%). JBI-778 showed strong tumor growth inhibition in a glioblastoma orthotopic model that mimics human GBM, with a significant extension in survival. Its differentiated mechanism makes it a potential option to treat brain metastasis cancers. Jubilant Therapeutics has received FDA clearance for an investigational new drug application (IND) to recruit patients for a phase I/II clinical trial for the assessment of safety, optimal doses, and PK properties of JBI-778 in patients.
4.16. SH3765
SH3765 is an orally bioavailable selective inhibitor of PRMT5 with antineoplastic activity that binds to PRMT5 and inhibits its methyltransferase activity at both monomethylated and dimethylated arginine residues in histone proteins. SH3765 modulates the gene expression implied in several cellular processes and decreases the growth of rapidly proliferating cells, including cancer cells. A phase I clinical trial (NCT05015309) will begin shortly to assess the safety, tolerability, and PK profile in patients with solid tumors with advanced malignancy to finalize the maximum tolerated dose (MTD) and RP2D [129].
4.17. SCR6920
SCR6920 is another orally bioavailable selective PRMT5 inhibitor with antiproliferative activity. A phase I open-label multicenter clinical trial (NCT05528055) will assess the dose escalation, safety, tolerability, and preliminary efficacy of SCR6920 in patients with advanced malignant tumors following oral administration. The dose-limiting toxicity must be the priority of this trial. The purpose of this clinical trial is to find the MTD, identify the RP2D, and accrue preliminary efficacy data in the participants [129].
5. Future Perspective and Conclusions
PRMT5-regulated oncogenes, such as C-MYC and N-MYC, are often deregulated in medulloblastomas. PRMT5 symmetrically dimethylates many proteins to regulate their stability and control activity in subcellular locations. The inactivation of PRMT5 has been shown to prevent MYC-driven lymphomagenesis [26]. PRMT5 is highly overexpressed in multiple aggressive metastatic cancers [150]. The promising role of PRMT5 in solid tumors has provoked the discovery and development of candidate drugs targeted to PRMT5 that display competitive and uncompetitive inhibition of SAM-mediated enzymatic activity. Critically, it is known that the genomic instability and catalytic activity of PRMT5 in MYC-amplified medulloblastoma cells decrease cell proliferation and induce apoptosis, which supports PRMT5 inhibition as a therapeutic option for MYC-driven medulloblastoma. More studies are needed to understand the mechanisms of PRMT5 overexpression that may cooperate with recurrent genomic lesions to contribute to medulloblastoma progression. Exploring the mechanisms of interaction between PRMT5 and MYC should give us further insights into how the two are engaged in promoting medulloblastoma aggressiveness. In addition, our in vitro and in vivo analyses of the inhibition of PRMT5, either with gene therapy or pharmacologically active small molecules as PRMT5 inhibitors, have indicated the potential of the PRMT5–MYC axis as a novel therapeutic approach in MYC-amplified medulloblastoma. As mentioned, the function of PRMT5 contributes to various physiological cellular processes to preserve cancer phenotypes and promote cancer progression in various cancer types. There is a strong rationale that the perturbation of PRMT5 can be a broadly effective means to treat cancer.
The development of PRMT5 inhibitors to achieve supportive efficacy is still in progress, and many PRMT5 inhibitors developed as SAM-competitive drugs are under clinical evaluation. However, most PRMT5 inhibitors have unwanted cytotoxicity in non-cancerous cells and in healthy tissues in clinical settings. This issue should be addressed in the context of MYC-amplified medulloblastoma. PRMT5 inhibitors could affect the signal transduction and reinstate the function of tumor suppressors via inhibition of the SDMA process. Inhibitors targeting PRMT5-mediated dimethylation may be attractive as single agents or in combination with other agents targeting MYC-amplified medulloblastoma to induce a durable response and prevent or delay acquired resistance. As PRMT5 is essential for normal cellular processes, clinical evaluation of the PRMT5 inhibitors in cancer therapy must carefully examine safety outcomes [151].
Limitations: Most investigational drugs, including some PRMT5 inhibitors, are prevented from efficiently entering into the brain. PRMT5 inhibitors are apparently transported back to the systemic circulation by the multidrug efflux pump action of proteins like P-glycoprotein (P-gp) [152,153]. Insufficient transport of drugs into the brain leads to a diminished therapeutic effect and aggravated organ toxicity side effects due to the deposition of the drug in other organs and tissues. Hence, novel PRMT5 inhibitors with satisfactory PK and PD profiles deserve additional refinement to confer more potent PRMT5 inhibition so they can be administered in minimum doses with the maximum effective concentration (MEC). It is urgent to address the issue of brain-targeted therapeutics by developing effective and safe drug delivery strategies for PRMT5 inhibitors. Several PRMT5 inhibitors are under clinical evaluation and are currently being examined in cancer patients with solid tumors, including neuroblastoma and glioblastoma. First-generation PRMT5 inhibitors cause side effects, including anemia, neutropenia, and thrombocytopenia [107,133], which can limit the capacity to reach the dose and exposure necessary to drive tumor regression in patients. The outstanding question is which PRMT5 targets should be traced in MYC-driven medulloblastoma to monitor and predict the response. GSK3326595 showed a significant inhibitory effect on the growth of MYC-driven medulloblastoma cell lines at a low micromolar potency and showed PRMT5 downregulation. Thus, GSK3326595 could potentially be further investigated at the clinical level for MYC-driven medulloblastoma. Interestingly, TNG908 has been advanced in a clinical trial as a PRMT5 inhibitor that is able to penetrate the blood–brain barrier. Two other compounds, LLY283 and CMP5, have shown favorable PK properties, along with brain distributions that suggest efficient penetration of the blood–brain barrier. JBI-778 demonstrated strong tumor growth inhibition in a glioblastoma orthotopic model and a favorable oral bioavailability. Some PRMT5 inhibitors have a very low IC50 in vitro but cannot cross the blood–brain barrier. MTA-cooperative PRMT5 inhibitors have favorable PK properties and efficiently penetrate the blood–brain barrier. However, MTAP-null mutations have still not been detected in medulloblastomas, so MTA-cooperative PRMT5 inhibitors might have less relevance compared to other PRMT5 inhibitors. JNJ-64619178 and PF06939999 have 30–40% oral bioavailability, although they are very effective in vitro. Enhancing blood–brain barrier penetration is crucial to improving the therapeutic efficacy and lowering toxicity.
Combining PRMT5 inhibitors with other drugs (e.g., chemotherapy, immune checkpoint inhibitors, or anti-EGFR drugs) in medulloblastoma treatment may hold promise by synergistically targeting cancer cells through different mechanisms. This approach may enhance the treatment efficacy, overcome drug resistance, and reduce the potential side effects associated with higher doses of individual agents. Research suggests that combining PRMT5 inhibitors with standard chemotherapy regimens could provide a more comprehensive and effective strategy for specific cancer types, including those with MYC amplification [154,155,156,157]. Clinical trials are underway to further explore the safety and efficacy of such combination treatments.
This review provides a comprehensive survey of possible PRMT5 inhibitor therapeutics to treat MYC-amplified medulloblastoma and highlights the challenges that must be addressed in future drug development.
Author Contributions
D.K.: Conceptualization, data curation, formal analysis, writing—original draft, writing—review and editing. S.J.: writing—review and editing. D.W.C.: writing—review and editing. S.S.J.: writing—review and editing. N.K.C.: conceptualization, project administration, funding acquisition, supervision, writing—original draft and review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the State of Nebraska through a grant from the Pediatric Cancer Research Group—Child Health Research Institute and by a Power 5 Grant from the Team Jack Foundation to N. K. Chaturvedi, PhD. The sponsor had no role in the writing or content of the manuscript or the decision to publish.
Data Availability Statement
This article does not report any original data.
Acknowledgments
The authors thank Matthew Sandbulte, of the Child Health Research Institute at Children’s Nebraska and the University of Nebraska Medical Center for his help in editing this manuscript.
Conflicts of Interest
The authors declare that they have no competing interests that could have appeared to influence the work reported in this article.
References
- Packer, R.J.; Macdonald, T.; Vezina, G.; Keating, R.; Santi, M. Medulloblastoma and primitive neuroectodermal tumors. Handb. Clin. Neurol. 2012, 105, 529–548. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e736. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Grobner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.; Schlanstein, M.; Northcott, P.A.; Cho, Y.J.; Koster, J.; Schouten-van Meeteren, A.; van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484. [Google Scholar] [CrossRef]
- Min, H.S.; Lee, J.Y.; Kim, S.K.; Park, S.H. Genetic grouping of medulloblastomas by representative markers in pathologic diagnosis. Transl. Oncol. 2013, 6, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Juraschka, K.; Taylor, M.D. Medulloblastoma in the age of molecular subgroups: A review. J. Neurosurg. Pediatr. 2019, 24, 353–363. [Google Scholar] [CrossRef]
- Gajjar, A.J.; Robinson, G.W. Medulloblastoma-translating discoveries from the bench to the bedside. Nat. Rev. Clin. Oncol. 2014, 11, 714–722. [Google Scholar] [CrossRef]
- Cho, Y.J.; Tsherniak, A.; Tamayo, P.; Santagata, S.; Ligon, A.; Greulich, H.; Berhoukim, R.; Amani, V.; Goumnerova, L.; Eberhart, C.G.; et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 2011, 29, 1424–1430. [Google Scholar] [CrossRef]
- Hill, R.M.; Kuijper, S.; Lindsey, J.C.; Petrie, K.; Schwalbe, E.C.; Barker, K.; Boult, J.K.; Williamson, D.; Ahmad, Z.; Hallsworth, A.; et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015, 27, 72–84. [Google Scholar] [CrossRef]
- Pei, Y.; Moore, C.E.; Wang, J.; Tewari, A.K.; Eroshkin, A.; Cho, Y.J.; Witt, H.; Korshunov, A.; Read, T.A.; Sun, J.L.; et al. An animal model of MYC-driven medulloblastoma. Cancer Cell 2012, 21, 155–167. [Google Scholar] [CrossRef]
- MacDonald, T.J.; Rood, B.R.; Santi, M.R.; Vezina, G.; Bingaman, K.; Cogen, P.H.; Packer, R.J. Advances in the diagnosis, molecular genetics, and treatment of pediatric embryonal CNS tumors. Oncologist 2003, 8, 174–186. [Google Scholar] [CrossRef]
- Zhou, M.; Jiang, J. Gli Phosphorylation Code in Hedgehog Signal Transduction. Front. Cell Dev. Biol. 2022, 10, 846927. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, J. Regulation of Hedgehog Signal Transduction by Ubiquitination and Deubiquitination. Int. J. Mol. Sci. 2021, 22, 13338. [Google Scholar] [CrossRef] [PubMed]
- Canettieri, G.; Di Marcotullio, L.; Greco, A.; Coni, S.; Antonucci, L.; Infante, P.; Pietrosanti, L.; De Smaele, E.; Ferretti, E.; Miele, E.; et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010, 12, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Zhou, W.; Shindiapina, P.; Sif, S.; Li, C.; Baiocchi, R.A. Recent advances in targeting protein arginine methyltransferase enzymes in cancer therapy. Expert. Opin. Ther. Targets 2018, 22, 527–545. [Google Scholar] [CrossRef]
- Wolf, S.S. The protein arginine methyltransferase family: An update about function, new perspectives and the physiological role in humans. Cell Mol. Life Sci. 2009, 66, 2109–2121. [Google Scholar] [CrossRef]
- Musiani, D.; Bok, J.; Massignani, E.; Wu, L.; Tabaglio, T.; Ippolito, M.R.; Cuomo, A.; Ozbek, U.; Zorgati, H.; Ghoshdastider, U.; et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 2019, 12, eaat8388. [Google Scholar] [CrossRef]
- Abe, Y.; Suzuki, Y.; Kawamura, K.; Tanaka, N. MEP50/PRMT5-mediated methylation activates GLI1 in Hedgehog signalling through inhibition of ubiquitination by the ITCH/NUMB complex. Commun. Biol. 2019, 2, 23. [Google Scholar] [CrossRef]
- Chaturvedi, N.K.; Mahapatra, S.; Kesherwani, V.; Kling, M.J.; Shukla, M.; Ray, S.; Kanchan, R.; Perumal, N.; McGuire, T.R.; Sharp, J.G.; et al. Role of protein arginine methyltransferase 5 in group 3 (MYC-driven) Medulloblastoma. BMC Cancer 2019, 19, 1056. [Google Scholar] [CrossRef] [PubMed]
- Wynn, D.T.; Rodriguez-Blanco, J.; Long, J.; Yang, F.; Shen, C.; Fei, D.; Tang, H.Y.; Hanson, D.; Robbins, D.J. Protein arginine methyltransferase 5 regulates SHH-subgroup medulloblastoma progression. Neurooncol. Adv. 2022, 4, vdac144. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Baiocchi, R.A.; Byrd, J.C.; Grever, M.R.; Jacob, S.T.; Sif, S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007, 26, 3558–3569. [Google Scholar] [CrossRef]
- Wang, L.; Pal, S.; Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell Biol. 2008, 28, 6262–6277. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Bezzi, M.; Low, D.H.; Ang, W.X.; Teo, S.X.; Gay, F.P.; Al-Haddawi, M.; Tan, S.Y.; Osato, M.; Sabo, A.; et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015, 523, 96–100. [Google Scholar] [CrossRef]
- Chan-Penebre, E.; Kuplast, K.G.; Majer, C.R.; Boriack-Sjodin, P.A.; Wigle, T.J.; Johnston, L.D.; Rioux, N.; Munchhof, M.J.; Jin, L.; Jacques, S.L.; et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015, 11, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Brehmer, D.; Beke, L.; Wu, T.; Millar, H.J.; Moy, C.; Sun, W.; Mannens, G.; Pande, V.; Boeckx, A.; van Heerde, E.; et al. Discovery and Pharmacological Characterization of JNJ-64619178, a Novel Small-Molecule Inhibitor of PRMT5 with Potent Antitumor Activity. Mol. Cancer Ther. 2021, 20, 2317–2328. [Google Scholar] [CrossRef]
- Litzler, L.C.; Zahn, A.; Meli, A.P.; Hebert, S.; Patenaude, A.M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 2019, 10, 22. [Google Scholar] [CrossRef]
- Barczak, W.; Jin, L.; Carr, S.M.; Munro, S.; Ward, S.; Kanapin, A.; Samsonova, A.; La Thangue, N.B. PRMT5 promotes cancer cell migration and invasion through the E2F pathway. Cell Death Dis. 2020, 11, 572. [Google Scholar] [CrossRef]
- Guderian, G.; Peter, C.; Wiesner, J.; Sickmann, A.; Schulze-Osthoff, K.; Fischer, U.; Grimmler, M. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J. Biol. Chem. 2011, 286, 1976–1986. [Google Scholar] [CrossRef]
- Krzyzanowski, A.; Gasper, R.; Adihou, H.; Hart, P.; Waldmann, H. Biochemical Investigation of the Interaction of pICln, RioK1 and COPR5 with the PRMT5-MEP50 Complex. Chembiochem 2021, 22, 1908–1914. [Google Scholar] [CrossRef]
- Mulvaney, K.M.; Blomquist, C.; Acharya, N.; Li, R.; Ranaghan, M.J.; O’Keefe, M.; Rodriguez, D.J.; Young, M.J.; Kesar, D.; Pal, D.; et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol. Cell 2021, 81, 3481–3495.e7. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Zheng, Q.; Zhang, D.; Lin, C.; Ouyang, L.; Zhang, J.; Chen, L. Medicinal chemistry strategies targeting PRMT5 for cancer therapy. Eur. J. Med. Chem. 2022, 244, 114842. [Google Scholar] [CrossRef] [PubMed]
- Burgos, E.S.; Wilczek, C.; Onikubo, T.; Bonanno, J.B.; Jansong, J.; Reimer, U.; Shechter, D. Histone H2A and H4 N-terminal tails are positioned by the MEP50 WD repeat protein for efficient methylation by the PRMT5 arginine methyltransferase. J. Biol. Chem. 2015, 290, 9674–9689. [Google Scholar] [CrossRef] [PubMed]
- Scoumanne, A.; Zhang, J.; Chen, X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009, 37, 4965–4976. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Mundade, R.; Lange, K.C.; Lu, T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle 2014, 13, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell Mol. Life Sci. 2015, 72, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Richard, S.; Morel, M.; Cleroux, P. Arginine methylation regulates IL-2 gene expression: A role for protein arginine methyltransferase 5 (PRMT5). Biochem. J. 2005, 388, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sun, X.; Zou, Z.; Sun, L.; Zhang, T.; Guo, S.; Wen, Y.; Liu, L.; Wang, Y.; Qin, J.; et al. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 2010, 20, 1023–1033. [Google Scholar] [CrossRef]
- Hsu, J.M.; Chen, C.T.; Chou, C.K.; Kuo, H.P.; Li, L.Y.; Lin, C.Y.; Lee, H.J.; Wang, Y.N.; Liu, M.; Liao, H.W.; et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol. 2011, 13, 174–181. [Google Scholar] [CrossRef]
- Karkhanis, V.; Hu, Y.J.; Baiocchi, R.A.; Imbalzano, A.N.; Sif, S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 2011, 36, 633–641. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Harris, D.P.; Adams, G.N.; Lause, G.E.; McHugh, A.; Tillmaand, E.G.; Money, A.; Willard, B.; Fox, P.L.; Dicorleto, P.E. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol. Cell Biol. 2012, 32, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.C.; Zheng, S.; Munro, S.; Liu, G.; Carr, S.M.; Moehlenbrink, J.; Lu, Y.C.; Stimson, L.; Khan, O.; Konietzny, R.; et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012, 31, 1785–1797. [Google Scholar] [CrossRef]
- Chen, M.; Yi, B.; Sun, J. Inhibition of cardiomyocyte hypertrophy by protein arginine methyltransferase 5. J. Biol. Chem. 2014, 289, 24325–24335. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.M.; Clegg, J.M.; Uchida, K.A.; Powers, M.A.; Ullman, K.S. Enhanced arginine methylation of programmed cell death 4 protein during nutrient deprivation promotes tumor cell viability. J. Biol. Chem. 2014, 289, 17541–17552. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, X.; Zhao, L.; Li, J.; Yang, H.; Zhu, Z.; Liu, J.; Huang, G. Arginine Methylation of SREBP1a via PRMT5 Promotes De Novo Lipogenesis and Tumor Growth. Cancer Res. 2016, 76, 1260–1272. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Gish, G.; Braunschweig, U.; Li, Y.; Ni, Z.; Schmitges, F.W.; Zhong, G.; Liu, K.; Li, W.; Moffat, J.; et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2016, 529, 48–53. [Google Scholar] [CrossRef]
- Fong, J.Y.; Pignata, L.; Goy, P.A.; Kawabata, K.C.; Lee, S.C.; Koh, C.M.; Musiani, D.; Massignani, E.; Kotini, A.G.; Penson, A.; et al. Therapeutic Targeting of RNA Splicing Catalysis through Inhibition of Protein Arginine Methylation. Cancer Cell 2019, 36, 194–209.e199. [Google Scholar] [CrossRef]
- Checa-Rodriguez, C.; Cepeda-Garcia, C.; Ramon, J.; Lopez-Saavedra, A.; Balestra, F.R.; Dominguez-Sanchez, M.S.; Gomez-Cabello, D.; Huertas, P. Methylation of the central transcriptional regulator KLF4 by PRMT5 is required for DNA end resection and recombination. DNA Repair 2020, 94, 102902. [Google Scholar] [CrossRef]
- Park, J.H.; Szemes, M.; Vieira, G.C.; Melegh, Z.; Malik, S.; Heesom, K.J.; Von Wallwitz-Freitas, L.; Greenhough, A.; Brown, K.W.; Zheng, Y.G.; et al. Protein arginine methyltransferase 5 is a key regulator of the MYCN oncoprotein in neuroblastoma cells. Mol. Oncol. 2015, 9, 617–627. [Google Scholar] [CrossRef]
- Wei, H.; Wang, B.; Miyagi, M.; She, Y.; Gopalan, B.; Huang, D.B.; Ghosh, G.; Stark, G.R.; Lu, T. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc. Natl. Acad. Sci. USA 2013, 110, 13516–13521. [Google Scholar] [CrossRef] [PubMed]
- Tee, W.W.; Pardo, M.; Theunissen, T.W.; Yu, L.; Choudhary, J.S.; Hajkova, P.; Surani, M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010, 24, 2772–2777. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Li, Z.; Chin, C.J.; Lee, S.A.; Clark, A.T. PRMT5 is required for human embryonic stem cell proliferation but not pluripotency. Stem Cell Rev. Rep. 2014, 10, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Bezzi, M.; Teo, S.X.; Muller, J.; Mok, W.C.; Sahu, S.K.; Vardy, L.A.; Bonday, Z.Q.; Guccione, E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013, 27, 1903–1916. [Google Scholar] [CrossRef]
- Dacwag, C.S.; Bedford, M.T.; Sif, S.; Imbalzano, A.N. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell Biol. 2009, 29, 1909–1921. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, J.; Hosohama, L.; Nee, K.; Gkountela, S.; Chaudhari, S.; Cass, A.A.; Xiao, X.; Clark, A.T. The Sm protein methyltransferase PRMT5 is not required for primordial germ cell specification in mice. EMBO J. 2015, 34, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gunther, S.; Looso, M.; Kunne, C.; Kruger, M.; Kim, J.; Zhou, Y.; Braun, T. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat. Commun. 2015, 6, 7140. [Google Scholar] [CrossRef]
- Shailesh, H.; Zakaria, Z.Z.; Baiocchi, R.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) dysregulation in cancer. Oncotarget 2018, 9, 36705–36718. [Google Scholar] [CrossRef]
- Pal, S.; Vishwanath, S.N.; Erdjument-Bromage, H.; Tempst, P.; Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell Biol. 2004, 24, 9630–9645. [Google Scholar] [CrossRef]
- Mongiardi, M.P.; Savino, M.; Bartoli, L.; Beji, S.; Nanni, S.; Scagnoli, F.; Falchetti, M.L.; Favia, A.; Farsetti, A.; Levi, A.; et al. Myc and Omomyc functionally associate with the Protein Arginine Methyltransferase 5 (PRMT5) in glioblastoma cells. Sci. Rep. 2015, 5, 15494. [Google Scholar] [CrossRef]
- Yan, F.; Alinari, L.; Lustberg, M.E.; Martin, L.K.; Cordero-Nieves, H.M.; Banasavadi-Siddegowda, Y.; Virk, S.; Barnholtz-Sloan, J.; Bell, E.H.; Wojton, J.; et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014, 74, 1752–1765. [Google Scholar] [CrossRef]
- Han, X.; Li, R.; Zhang, W.; Yang, X.; Wheeler, C.G.; Friedman, G.K.; Province, P.; Ding, Q.; You, Z.; Fathallah-Shaykh, H.M.; et al. Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J. Neurooncol. 2014, 118, 61–72. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, W.; Yuan, Y. Protein Arginine Methyltransferase 5 (PRMT5) as an Anticancer Target and Its Inhibitor Discovery. J. Med. Chem. 2018, 61, 9429–9441. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, R.; Matsubara, D.; Osman, W.; Morikawa, T.; Goto, A.; Morita, S.; Ishikawa, S.; Aburatani, H.; Takai, D.; Nakajima, J.; et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum. Pathol. 2014, 45, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Li, Y.; Lee, P.; Liu, T.; Wan, C.; Wang, Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS ONE 2012, 7, e44033. [Google Scholar] [CrossRef]
- Nicholas, C.; Yang, J.; Peters, S.B.; Bill, M.A.; Baiocchi, R.A.; Yan, F.; Sif, S.; Tae, S.; Gaudio, E.; Wu, X.; et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1). PLoS ONE 2013, 8, e74710. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Bezzi, M.; Guccion, E. The Where and the How of PRMT5. Curr. Mol. Bio Rep. 2015, 1, 19–28. [Google Scholar] [CrossRef]
- Chitiprolu, M.; Jagow, C.; Tremblay, V.; Bondy-Chorney, E.; Paris, G.; Savard, A.; Palidwor, G.; Barry, F.A.; Zinman, L.; Keith, J.; et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat. Commun. 2018, 9, 2794. [Google Scholar] [CrossRef]
- Quan, X.; Yue, W.; Luo, Y.; Cao, J.; Wang, H.; Wang, Y.; Lu, Z. The protein arginine methyltransferase PRMT5 regulates Abeta-induced toxicity in human cells and Caenorhabditis elegans models of Alzheimer’s disease. J. Neurochem. 2015, 134, 969–977. [Google Scholar] [CrossRef]
- Ratovitski, T.; Arbez, N.; Stewart, J.C.; Chighladze, E.; Ross, C.A. PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD). Cell Cycle 2015, 14, 1716–1729. [Google Scholar] [CrossRef]
- Banasavadi-Siddegowda, Y.K.; Russell, L.; Frair, E.; Karkhanis, V.A.; Relation, T.; Yoo, J.Y.; Zhang, J.; Sif, S.; Imitola, J.; Baiocchi, R.; et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene 2017, 36, 263–274. [Google Scholar] [CrossRef]
- Braun, C.J.; Stanciu, M.; Boutz, P.L.; Patterson, J.C.; Calligaris, D.; Higuchi, F.; Neupane, R.; Fenoglio, S.; Cahill, D.P.; Wakimoto, H.; et al. Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer Cell 2017, 32, 411–426.e411. [Google Scholar] [CrossRef] [PubMed]
- Sachamitr, P.; Ho, J.C.; Ciamponi, F.E.; Ba-Alawi, W.; Coutinho, F.J.; Guilhamon, P.; Kushida, M.M.; Cavalli, F.M.G.; Lee, L.; Rastegar, N.; et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 2021, 12, 979. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.; Benavides-Serrato, A.; Saunders, J.T.; Landon, K.A.; Schreck, A.J.; Nishimura, R.N.; Gera, J. The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma. J. Neurooncol. 2019, 145, 11–22. [Google Scholar] [CrossRef]
- Du, C.; Hansen, L.J.; Singh, S.X.; Wang, F.; Sun, R.; Moure, C.J.; Roso, K.; Greer, P.K.; Yan, H.; He, Y. A PRMT5-RNF168-SMURF2 Axis Controls H2AX Proteostasis. Cell Rep. 2019, 28, 3199–3211.e3195. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, X.O.; Rozen, E.J.; Sun, X.; Sallis, B.; Verdejo-Torres, O.; Wigglesworth, K.; Moon, D.; Huang, T.; Cavaretta, J.P.; et al. PRMT5 activates AKT via methylation to promote tumor metastasis. Nat. Commun. 2022, 13, 3955. [Google Scholar] [CrossRef]
- Chen, H.; Lorton, B.; Gupta, V.; Shechter, D. A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 2017, 36, 373–386. [Google Scholar] [CrossRef]
- Fu, T.; Lv, X.; Kong, Q.; Yuan, C. A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget 2017, 8, 54809–54820. [Google Scholar] [CrossRef]
- Jing, P.; Zhao, N.; Ye, M.; Zhang, Y.; Zhang, Z.; Sun, J.; Wang, Z.; Zhang, J.; Gu, Z. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett. 2018, 427, 38–48. [Google Scholar] [CrossRef]
- Zhou, H.; Chang, J.; Zhang, J.; Zheng, H.; Miao, X.; Mo, H.; Sun, J.; Jia, Q.; Qi, G. PRMT5 activates KLF5 by methylation to facilitate lung cancer. J. Cell Mol. Med. 2023, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhu, Y.; Zhou, Z.; Xu, J.; Jin, S.; Xu, K.; Zhang, H.; Sun, Q.; Wang, J.; Xu, J. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018, 7, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, J.; Liu, X.; Li, S.; Wang, Q.; Di, C.; Hu, Z.; Yu, T.; Ding, J.; Li, J.; et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 2018, 9, 1572. [Google Scholar] [CrossRef]
- Zhu, K.; Peng, Y.; Hu, J.; Zhan, H.; Yang, L.; Gao, Q.; Jia, H.; Luo, R.; Dai, Z.; Tang, Z.; et al. Metadherin-PRMT5 complex enhances the metastasis of hepatocellular carcinoma through the WNT-beta-catenin signaling pathway. Carcinogenesis 2020, 41, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Hing, Z.A.; Walker, J.S.; Whipp, E.C.; Brinton, L.; Cannon, M.; Zhang, P.; Sher, S.; Cempre, C.B.; Brown, F.; Smith, P.L.; et al. Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter’s transformation. Nat. Commun. 2023, 14, 97. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hu, Q.; Xu, J.; Ji, S.; Dai, W.; Liu, W.; Xu, W.; Sun, Q.; Zhang, Z.; Ni, Q.; et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun. Signal. 2019, 17, 30. [Google Scholar] [CrossRef]
- Orben, F.; Lankes, K.; Schneeweis, C.; Hassan, Z.; Jakubowsky, H.; Krauss, L.; Boniolo, F.; Schneider, C.; Schafer, A.; Murr, J.; et al. Epigenetic drug screening defines a PRMT5 inhibitor-sensitive pancreatic cancer subtype. JCI Insight 2022, 7, e151353. [Google Scholar] [CrossRef]
- Dong, Y.; Song, C.; Wang, Y.; Lei, Z.; Xu, F.; Guan, H.; Chen, A.; Li, F. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell Signal. 2017, 34, 55–65. [Google Scholar] [CrossRef]
- Kota, S.K.; Roening, C.; Patel, N.; Kota, S.B.; Baron, R. PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone 2018, 117, 37–46. [Google Scholar] [CrossRef]
- Mounir, Z.; Korn, J.M.; Westerling, T.; Lin, F.; Kirby, C.A.; Schirle, M.; McAllister, G.; Hoffman, G.; Ramadan, N.; Hartung, A.; et al. ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the Androgen Receptor. eLife 2016, 5, e13964. [Google Scholar] [CrossRef]
- Beketova, E.; Fang, S.; Owens, J.L.; Liu, S.; Chen, X.; Zhang, Q.; Asberry, A.M.; Deng, X.; Malola, J.; Huang, J.; et al. Protein Arginine Methyltransferase 5 Promotes pICln-Dependent Androgen Receptor Transcription in Castration-Resistant Prostate Cancer. Cancer Res. 2020, 80, 4904–4917. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Zhao, S.; Liu, T.; Liu, Y.; Liu, Y.; Yang, X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem. 2013, 61, 206–217. [Google Scholar] [CrossRef]
- Wang, Z.; Kong, J.; Wu, Y.; Zhang, J.; Wang, T.; Li, N.; Fan, J.; Wang, H.; Zhang, J.; Ling, R. PRMT5 determines the sensitivity to chemotherapeutics by governing stemness in breast cancer. Breast Cancer Res. Treat. 2018, 168, 531–542. [Google Scholar] [CrossRef]
- Chiang, K.; Zielinska, A.E.; Shaaban, A.M.; Sanchez-Bailon, M.P.; Jarrold, J.; Clarke, T.L.; Zhang, J.; Francis, A.; Jones, L.J.; Smith, S.; et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep. 2017, 21, 3498–3513. [Google Scholar] [CrossRef]
- Yin, S.; Liu, L.; Brobbey, C.; Palanisamy, V.; Ball, L.E.; Olsen, S.K.; Ostrowski, M.C.; Gan, W. PRMT5-mediated arginine methylation activates AKT kinase to govern tumorigenesis. Nat. Commun. 2021, 12, 3444. [Google Scholar] [CrossRef]
- Hu, D.; Gur, M.; Zhou, Z.; Gamper, A.; Hung, M.C.; Fujita, N.; Lan, L.; Bahar, I.; Wan, Y. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat. Commun. 2015, 6, 8419. [Google Scholar] [CrossRef]
- Rengasamy, M.; Zhang, F.; Vashisht, A.; Song, W.M.; Aguilo, F.; Sun, Y.; Li, S.; Zhang, W.; Zhang, B.; Wohlschlegel, J.A.; et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017, 45, 11106–11120. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.A.; Fay, M.M.; Factor, R.E.; Welm, A.L.; Ullman, K.S. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011, 71, 5579–5587. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wu, J. Epigenetic regulation in medulloblastoma. Mol. Cell Neurosci. 2018, 87, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Roussel, M.F.; Stripay, J.L. Epigenetic Drivers in Pediatric Medulloblastoma. Cerebellum 2018, 17, 28–36. [Google Scholar] [CrossRef]
- Dubuc, A.M.; Remke, M.; Korshunov, A.; Northcott, P.A.; Zhan, S.H.; Mendez-Lago, M.; Kool, M.; Jones, D.T.; Unterberger, A.; Morrissy, A.S.; et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013, 125, 373–384. [Google Scholar] [CrossRef]
- Alimova, I.; Venkataraman, S.; Harris, P.; Marquez, V.E.; Northcott, P.A.; Dubuc, A.; Taylor, M.D.; Foreman, N.K.; Vibhakar, R. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int. J. Cancer 2012, 131, 1800–1809. [Google Scholar] [CrossRef]
- Favia, A.; Salvatori, L.; Nanni, S.; Iwamoto-Stohl, L.K.; Valente, S.; Mai, A.; Scagnoli, F.; Fontanella, R.A.; Totta, P.; Nasi, S.; et al. The Protein Arginine Methyltransferases 1 and 5 affect Myc properties in glioblastoma stem cells. Sci. Rep. 2019, 9, 15925. [Google Scholar] [CrossRef]
- Chittka, A.; Nitarska, J.; Grazini, U.; Richardson, W.D. Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 2012, 287, 42995–43006. [Google Scholar] [CrossRef]
- Zhou, Z.; Feng, Z.; Hu, D.; Yang, P.; Gur, M.; Bahar, I.; Cristofanilli, M.; Gradishar, W.J.; Xie, X.Q.; Wan, Y. A novel small-molecule antagonizes PRMT5-mediated KLF4 methylation for targeted therapy. EBioMedicine 2019, 44, 98–111. [Google Scholar] [CrossRef]
- Wu, Q.; Schapira, M.; Arrowsmith, C.H.; Barsyte-Lovejoy, D. Protein arginine methylation: From enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021, 20, 509–530. [Google Scholar] [CrossRef] [PubMed]
- Feustel, K.; Falchook, G.S. Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors in Oncology Clinical Trials: A review. J. Immunother. Precis. Oncol. 2022, 5, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Millar, H.J.; Brehmer, D.; Verhulst, T.; Haddish-Berhane, N.; Greway, T.; Gaffney, D.; Boeckx, A.; Heerde, E.V.; Nys, T.; Portale, J.; et al. In vivo efficacy and pharmacodynamic modulation of JNJ-64619178, a selective PRMT5 inhibitor, in human lung and hematologic preclinical models [abstract]. In Proceedings of the American Association for Cancer Research Annual Meeting 2019, Atlanta, GA, USA, 29 March–3 April 2019; AACR: Philadelphia, PA, USA, 2019. Volume 79 (Suppl 13), Abstract nr 950. [Google Scholar]
- Jensen-Pergakes, K.; Tatlock, J.; Maegley, K.A.; McAlpine, I.J.; McTigue, M.; Xie, T.; Dillon, C.P.; Wang, Y.; Yamazaki, S.; Spiegel, N.; et al. SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance. Mol. Cancer Ther. 2022, 21, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.W.; Rioux, N.; Boriack-Sjodin, P.A.; Munchhof, M.J.; Reiter, L.A.; Majer, C.R.; Jin, L.; Johnston, L.D.; Chan-Penebre, E.; Kuplast, K.G.; et al. Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666. ACS Med. Chem. Lett. 2016, 7, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Gulla, A.; Hideshima, T.; Bianchi, G.; Fulciniti, M.; Kemal Samur, M.; Qi, J.; Tai, Y.T.; Harada, T.; Morelli, E.; Amodio, N.; et al. Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia 2018, 32, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, X.; Han, Y.; Wang, P. Protein arginine methyltransferase 5 promotes bladder cancer growth through inhibiting NF-kB dependent apoptosis. EXCLI J. 2018, 17, 1157–1166. [Google Scholar] [CrossRef]
- Kaushik, S.; Liu, F.; Veazey, K.J.; Gao, G.; Das, P.; Neves, L.F.; Lin, K.; Zhong, Y.; Lu, Y.; Giuliani, V.; et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 2018, 32, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Serio, J.; Ropa, J.; Chen, W.; Mysliwski, M.; Saha, N.; Chen, L.; Wang, J.; Miao, H.; Cierpicki, T.; Grembecka, J.; et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene 2018, 37, 450–460. [Google Scholar] [CrossRef] [PubMed]
- AbuHammad, S.; Cullinane, C.; Martin, C.; Bacolas, Z.; Ward, T.; Chen, H.; Slater, A.; Ardley, K.; Kirby, L.; Chan, K.T.; et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl. Acad. Sci. USA 2019, 116, 17990–18000. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, S.V.; Kellner, W.A.; Thompson, C.; Pappalardi, M.B.; Zhang, X.P.; Montes de Oca, R.; Penebre, E.; Duncan, K.; Boriack-Sjodin, A.; Le, B.; et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci. Rep. 2018, 8, 9711. [Google Scholar] [CrossRef] [PubMed]
- Vinet, M.; Suresh, S.; Maire, V.; Monchecourt, C.; Nemati, F.; Lesage, L.; Pierre, F.; Ye, M.; Lescure, A.; Brisson, A.; et al. Protein arginine methyltransferase 5: A novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019, 8, 2414–2428. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Guo, H.; Bates, P.D.; Zhang, S.; Zhang, H.; Nomie, K.J.; Li, Y.; Lu, L.; Seibold, K.R.; Wang, F.; et al. PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia 2019, 33, 2898–2911. [Google Scholar] [CrossRef]
- Villalona-Calero, M.A.; Patnaik, A.; Maki, R.G.; O’Neil, B.; Abbruzzese, J.L.; Dagogo-Jack, I.; Devarakonda, S.; Wahlroos, S.; Lin, C.-C.; Fujiwara, Y.; et al. Design and rationale of a phase 1 dose-escalation study of AMG 193, a methylthioadenosine (MTA)-cooperative PRMT5 inhibitor, in patients with advanced methylthioadenosine phosphorylase (MTAP)-null solid tumors. J. Clin. Oncol. 2022, 40, TPS3167. [Google Scholar] [CrossRef]
- Bhagwat, N.; Zhang, Y.; Lin, H.; Wang, M.; Rominger, D.; Emm, T.; Chugani-Mahtani, D.; Angelis, D.; Shetty, R.; Leal, R.; et al. Preclinical Characterization of PRT543, a Potent and Selective Inhibitor of Protein Arginine Methyltransferase 5 (PRMT5), with Broad Antitumor Activity in In vitro and In vivo Models. In Proceedings of the Annual Meeting of the American Association for Cancer Research, Philadelphia, PA, USA, 27–28 April 2020. [Google Scholar]
- Falchook, G. A phase 1 dose-escalation study of protein arginine methyltransferase 5 (PRMT5) brain-penetrant inhibitor PRT811 in patients with advanced solid tumors, including recurrent high-grade gliomas. In Proceedings of the AACR-NCI-EORTC Virtual Conference, Virtual, 7–10 October 2021; Available online: www.aacr.org/meeting/aacr-nci-eortc-international (accessed on 17 November 2023).
- Zhang, M.; Tsai, A.; Cottrell, K.M.; Haines, B.B.; Wilker, E.; DiBenedetto, H.; Weitzman, R.; Huang, A.; Davis, C.B.; Maxwell, J.P.; et al. TNG908, a brain-penetrant MTA-cooperative PRMT5 inhibitor, is efficacious in preclinical glioblastoma models. In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023; Part 1 (Regular and Invited Abstracts). AACR: Philadelphia, PA, USA, 2023; Volume 83. [Google Scholar]
- Vegar, L.; Aranda, R.; Waters, L.; Moya, K.; Hebbert, A.; Smith, C.S.; Hallin, J.; David, B.M.; Engstrom, L.D.; Vanderpool, D.; et al. A novel MTA-cooperative PRMT5 inhibitor, MRTX1719, stabilizes the ternary MTA-PRMT5 complex and leads to synthetic lethality in MTAP deleted cancers [abstract]. In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023; Part 1 (Regular and Invited Abstracts). AACR: Philadelphia, PA, USA, 2023; Volume 83. [Google Scholar]
- Engstrom, L.D.; Aranda, R.; Waters, L.; Moya, K.; Bowcut, V.; Vegar, L.; Trinh, D.; Hebbert, A.; Smith, C.R.; Kulyk, S.; et al. MRTX1719 is an MTA-cooperative PRMT5 inhibitor that exhibits synthetic lethality in preclinical models and patients with MTAP deleted cancer. Cancer Discov. 2023, 13, 2412–2431. [Google Scholar] [CrossRef]
- Bonday, Z.Q.; Cortez, G.S.; Grogan, M.J.; Antonysamy, S.; Weichert, K.; Bocchinfuso, W.P.; Li, F.; Kennedy, S.; Li, B.; Mader, M.M.; et al. LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity. ACS Med. Chem. Lett. 2018, 9, 612–617. [Google Scholar] [CrossRef]
- Snyder, K.J.; Zitzer, N.C.; Gao, Y.; Choe, H.K.; Sell, N.E.; Neidemire-Colley, L.; Ignaci, A.; Kale, C.; Devine, R.D.; Abad, M.G.; et al. PRMT5 regulates T cell interferon response and is a target for acute graft-versus-host disease. JCI Insight 2020, 5, e131099. [Google Scholar] [CrossRef]
- Palte, R.L.; Schneider, S.E.; Altman, M.D.; Hayes, R.P.; Kawamura, S.; Lacey, B.M.; Mansueto, M.S.; Reutershan, M.; Siliphaivanh, P.; Sondey, C.; et al. Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5). ACS Med. Chem. Lett. 2020, 11, 1688–1693. [Google Scholar] [CrossRef] [PubMed]
- Sivanandhan, D.; Gajendran, C.; Mohammed, Z.; Gosu, R.; Sher, D.; Mansur, S.; Friedmann-Morvinski, D.; Rajagopal, S.; Rastelli, L. JBI-778, a novel brain-penetrant small molecule PRMT5 inhibitor for treatment of cancer [abstract]. In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023; Part 1 (Regular and Invited Abstracts). AACR: Philadelphia, PA, USA, 2023; Volume 83. [Google Scholar]
- Zheng, J.; Li, B.; Wu, Y.; Wu, X.; Wang, Y. Targeting Arginine Methyltransferase PRMT5 for Cancer Therapy: Updated Progress and Novel Strategies. J. Med. Chem. 2023, 66, 8407–8427. [Google Scholar] [CrossRef] [PubMed]
- NCT03573310. A Phase 1, First-In-Human, Open-Label Study of the Safety, Pharmacokinetics, and Pharmacodynamics of JNJ64619178 in Subjects with Advanced Cancers. ClinicalTrials.gov Identifier: NCT03573310. Available online: https://clinicaltrials.gov/study/NCT03573310 (accessed on 17 November 2023).
- NIH. JNJ-64619178. NCI Drug Dictionary, National Institutes of Health–National Cancer Institute. Available online: www.cancer.gov/publications/dictionaries/cancer-drug/def/onametostat (accessed on 17 November 2021).
- Vieito Villar, M.; Spreafico, A.; Brana, I.; Hernandez, T.; Razak, A.A.; Wang, J.; Haddish-Berhane, N.; Mehta, J.; Johnson, A.; Maes, A.; et al. Firstin-human study of JNJ-64619178, a protein arginine methyltransferase 5 (PRMT5) inhibitor, in patients with advanced cancers. Ann. Oncol. 2020, 31, S462–S504. [Google Scholar]
- Vieito, M.; Moreno, V.; Spreafico, A.; Brana, I.; Wang, J.S.; Preis, M.; Hernandez-Guerrero, T.; Genta, S.; Hansen, A.R.; Doger, B.; et al. Phase 1 Study of JNJ-64619178, a Protein Arginine Methyltransferase 5 Inhibitor, in Advanced Solid Tumors. Clin. Cancer Res. 2023, 29, 3592–3602. [Google Scholar] [CrossRef] [PubMed]
- NCT03854227. A Phase 1 Study to Evaluate the Safety, Pharmacokinetics, and Pharmacodynamics of Escalating Doses of PF-0693999 (PRMT5 Inhibitor) in Participants with Advanced or Metastatic Non-Small Cell Lung Cancer, Head and Neck Squamous Cell Carcinoma, Esophageal Cancer, Endometrial Cancer, Cervical Cancer and Bladder Cancer. ClinicalTrials.gov identifier: NCT03854227. Available online: https://clinicaltrials.gov/study/NCT03854227 (accessed on 17 November 2023).
- Rodon Ahnert, J.; Perez, C.A.; Wong, K.M.; Maitland, M.L.; Tsai, F.; Berlin, J.; Liao, K.H.; Wang, I.-M.; Markovtsova, L.; Jacobs, I.A.; et al. PF-06939999, a potent and selective PRMT5 inhibitor, in patients with advanced or metastatic solid tumors: A phase 1 dose escalation study. J. Clin. Oncol. 2021, 29, 3019. [Google Scholar] [CrossRef]
- Ernzen, K.; Melvin, C.; Yu, L.; Phelps, C.; Niewiesk, S.; Green, P.L.; Panfil, A.R. The PRMT5 inhibitor EPZ015666 is effective against HTLV-1-transformed T-cell lines in vitro and in vivo. Front. Microbiol. 2023, 14, 1101544. [Google Scholar] [CrossRef]
- Chen, Y.; Shao, X.; Zhao, X.; Ji, Y.; Liu, X.; Li, P.; Zhang, M.; Wang, Q. Targeting protein arginine methyltransferase 5 in cancers: Roles, inhibitors and mechanisms. Biomed. Pharmacother. 2021, 144, 112252. [Google Scholar] [CrossRef]
- NCT02783300. Dose Escalation and Expansion Study of GSK3326595 in Subjects with Advanced or Recurrent Solid Tumors as well as Clinical Activity in Participants with a Subset of Solid Tumors and Non-Hodgkin’s Lymphoma. Clinical Trials.gov Identifier: NCT02783300. Available online: https://clinicaltrials.gov/study/nct02783300 (accessed on 17 November 2023).
- Watts, J.M.; Bradley, T.J.; Thomassen, A.; Brunner, A.M.; Minden, M.D.; Papadantonakis, N.; Abedin, S.; Baines, A.J.; Barbash, O.; Gorman, S.; et al. A phase I/II study to investigate the safety and clinical activity of the protein arginine methyltransferase 5 inhibitor GSK3326595 in subjects with myelodysplastic syndrome and acute myeloid leukemia. Blood 2019, 134, 2656. [Google Scholar] [CrossRef]
- Siu, L.L.; Rasco, D.W.; Vinay, S.P.; Romano, P.M.; Menis, J.; Opdam, F.L.; Heinhuis, K.M.; Egger, J.L.; Gorman, S.A.; Parasrampuria, R.; et al. 3410–METEOR-1: A phase I study of GSK3326595, a first-in-class protein arginine methyltransferase 5 (PRMT5) inhibitor, in advanced solid tumors. Ann. Oncol. 2019, 30, 2656. [Google Scholar] [CrossRef]
- NCT03614728. Safety, Tolerability and Clinical Activity of GSK3326595 in Subjects with Refractory MDS, CMML, and AML. ClinicalTrials.gov Identifier: NCT03614728. Available online: https://clinicaltrials.gov/study/NCT03614728 (accessed on 17 November 2023).
- NCT04676516. Randomized Window of Opportunity Study of GSK3326595 in Subjects with Early Stage Breast Cancers. ClinicalTrials.gov Identifier: NCT04676516. Available online: https://clinicaltrials.gov/study/NCT04676516 (accessed on 17 November 2023).
- NCT05094336. Dose Escalation and Expansion Study of AMG 193 in Subjects with Metastatic or Locally Advanced Methylthioadenosine Phosphorylase (MTAP)-Null Solid Tumors. Available online: https://clinicaltrials.gov/study/NCT05094336 (accessed on 17 November 2023).
- NIH. PRT543. NCI Drug Dictionary, National Institutes of Health–National Cancer Institute. Available online: https://www.cancer.gov/publications/dictionaries/cancer-drug/def/protein-arginine-methyltransferase-5-inhibitor-prt543 (accessed on 17 November 2021).
- NCT04089449. A Phase 1, Open-Label, Multicenter, Dose Escalation and Expansion Study of PRT811 in Subjects with Advanced Solid Tumors, CNS Lymphoma, and Recurrent High-Grade Gliomas. ClinicalTrials.gov Identifier: NCT04089449. Available online: https://clinicaltrials.gov/study/NCT04089449 (accessed on 17 November 2023).
- Briggs, K.J.; Cottrell, K.M.; Tonini, M.R.; Wilker, E.W.; Gu, L.; Davis, C.B.; Zhang, M.; Whittington, D.; Gotur, D.; Goldstein, M.J.; et al. TNG908 is an MTAPnull-selective PRMT5 inhibitor that drives tumor regressions in MTAP-deleted xenograft models across multiple histologies [abstract]. In Proceedings of the American Association for Cancer Research Annual Meeting 2022, New Orleans, LA, USA, 8–13 April 2022; Volume 82. [Google Scholar]
- Malamas, M.S.; Erdei, J.; Gunawan, I.; Turner, J.; Hu, Y.; Wagner, E.; Fan, K.; Chopra, R.; Olland, A.; Bard, J.; et al. Design and synthesis of 5,5’-disubstituted aminohydantoins as potent and selective human beta-secretase (BACE1) inhibitors. J. Med. Chem. 2010, 53, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Ohguchi, H.; Hideshima, T.; Anderson, K.C. The biological significance of histone modifiers in multiple myeloma: Clinical applications. Blood Cancer J. 2018, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Banasavadi-Siddegowda, Y.K.; Welker, A.M.; An, M.; Yang, X.; Zhou, W.; Shi, G.; Imitola, J.; Li, C.; Hsu, S.; Wang, J.; et al. PRMT5 as a druggable target for glioblastoma therapy. Neuro Oncol. 2018, 20, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, K.J.; McDonald, E.R., 3rd; Schlabach, M.R.; Billy, E.; Hoffman, G.R.; deWeck, A.; Ruddy, D.A.; Venkatesan, K.; Yu, J.; McAllister, G.; et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 2016, 351, 1208–1213. [Google Scholar] [CrossRef]
- Liu, F.; Cheng, G.; Hamard, P.J.; Greenblatt, S.; Wang, L.; Man, N.; Perna, F.; Xu, H.; Tadi, M.; Luciani, L.; et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Investig. 2015, 125, 3532–3544. [Google Scholar] [CrossRef] [PubMed]
- Fromm, M.F. Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol. Sci. 2004, 25, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Endicott, J.A.; Ling, V. The biochemistry of P-glycoprotein-mediated multidrug resistance. Annu. Rev. Biochem. 1989, 58, 137–171. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.A. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 2020, 12, eaaz5683. [Google Scholar] [CrossRef]
- Hu, R.; Zhou, B.; Chen, Z.; Chen, S.; Chen, N.; Shen, L.; Xiao, H.; Zheng, Y. PRMT5 Inhibition Promotes PD-L1 Expression and Immuno-Resistance in Lung Cancer. Front. Immunol. 2021, 12, 722188. [Google Scholar] [CrossRef]
- Carter, J.; Hulse, M.; Sivakumar, M.; Burtell, J.; Thodima, V.; Wang, M.; Agarwal, A.; Vykuntam, K.; Spruance, J.; Bhagwat, N.; et al. PRMT5 Inhibitors Regulate DNA Damage Repair Pathways in Cancer Cells and Improve Response to PARP Inhibition and Chemotherapies. Cancer Res. Commun. 2023, 3, 2233–2243. [Google Scholar] [CrossRef]
- Dakroub, R.; Huard, S.; Hajj-Younes, Y.; Suresh, S.; Badran, B.; Fayyad-Kazan, H.; Dubois, T. Therapeutic Advantage of Targeting PRMT5 in Combination with Chemotherapies or EGFR/HER2 Inhibitors in Triple-Negative Breast Cancers. Breast Cancer 2023, 15, 785–799. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).