Disturbances in Nitric Oxide Cycle and Related Molecular Pathways in Clear Cell Renal Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. The Disruption of NO Homeostasis in ccRCC
2.1. The Disruption of NO Biosynthesis in ccRCC
2.2. The Disruption of NO Signaling in ccRCC
2.3. Bimodal Effects of NO in ccRCC
2.4. Quantitative Determination of NO Metabolites in Biological Samples
- Spectrophotometry: Utilizing azoic dyes and the Griess test.
- Fluorescence: Employing reagents such as Diaminofluorescein (DAF-2).
- Luminescence: Using luciferin–luciferase assays.
- Electrochemical: Employing amperometric NO microelectrodes.
- Tandem Mass Spectrometry: Including mass spectrometry in tandem (MS/MS) and electrospray ionization mass spectrometry (ESI-MS/MS).
- Liquid Chromatography-Mass Spectrometry (LC-MS): Combining liquid chromatography with mass spectrometry
- Electron Paramagnetic Resonance (EPR): Employed for NO detection.
- HPLC: High-Performance Liquid Chromatography is another technique.
- Antibody-Based Methods: These encompass immunohistochemical, immunoblotting, and enzyme-linked immunosorbent assays (ELISAs).
- Chemiluminescence: A method based on the detection of light emission.
- UV-Visible Absorption Spectrum: Measuring the absorption of UV-visible light.
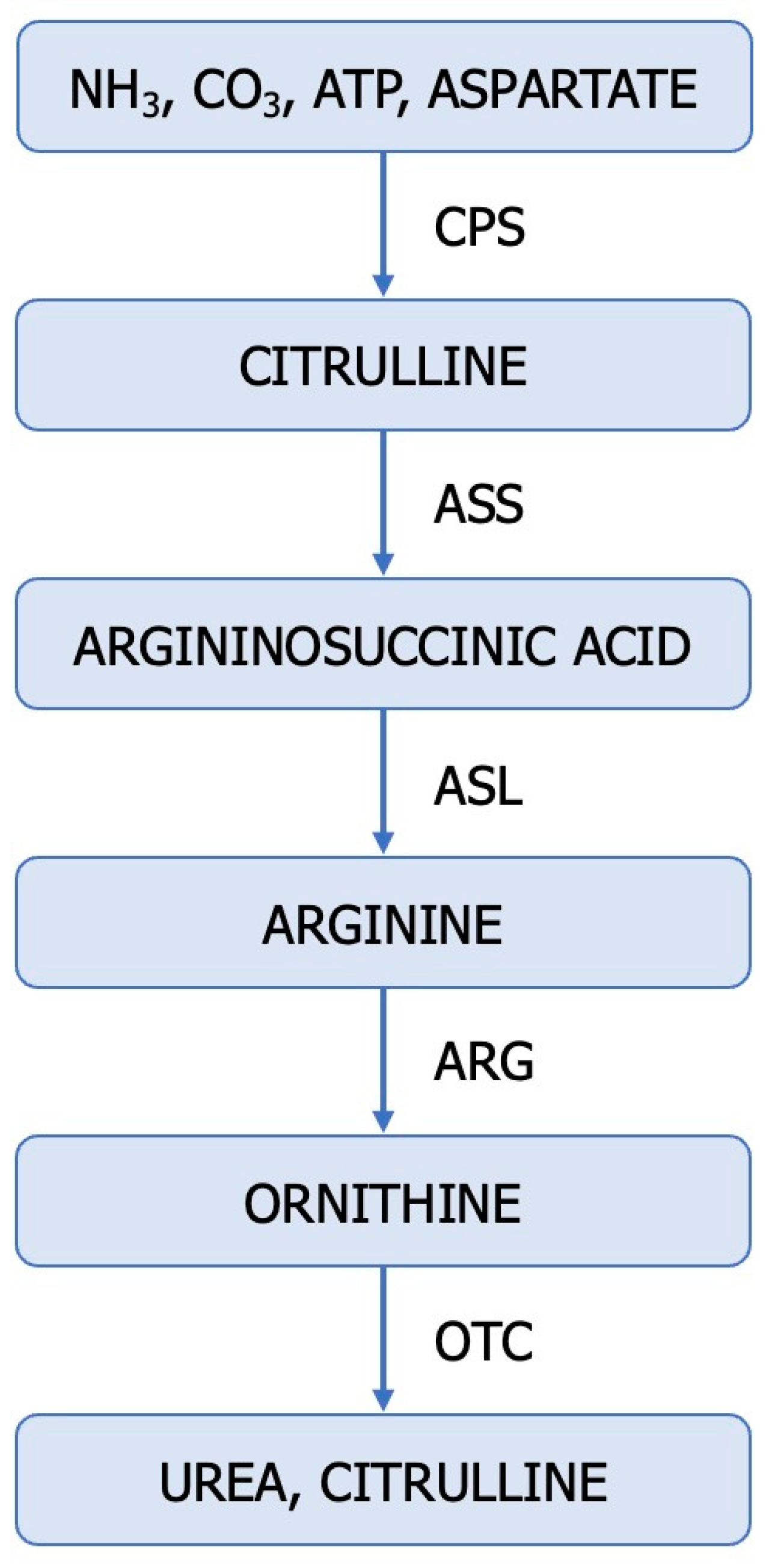
3. Dysregulated Ureagenic Cycle—A Distinctive Sign in ccRCC
4. The Upregulation of Glutamine: An Alternative Source of Nitrogen for ccRCC
5. Cellular Arginine Depletion—A Proliferation Strategy in ccRCC
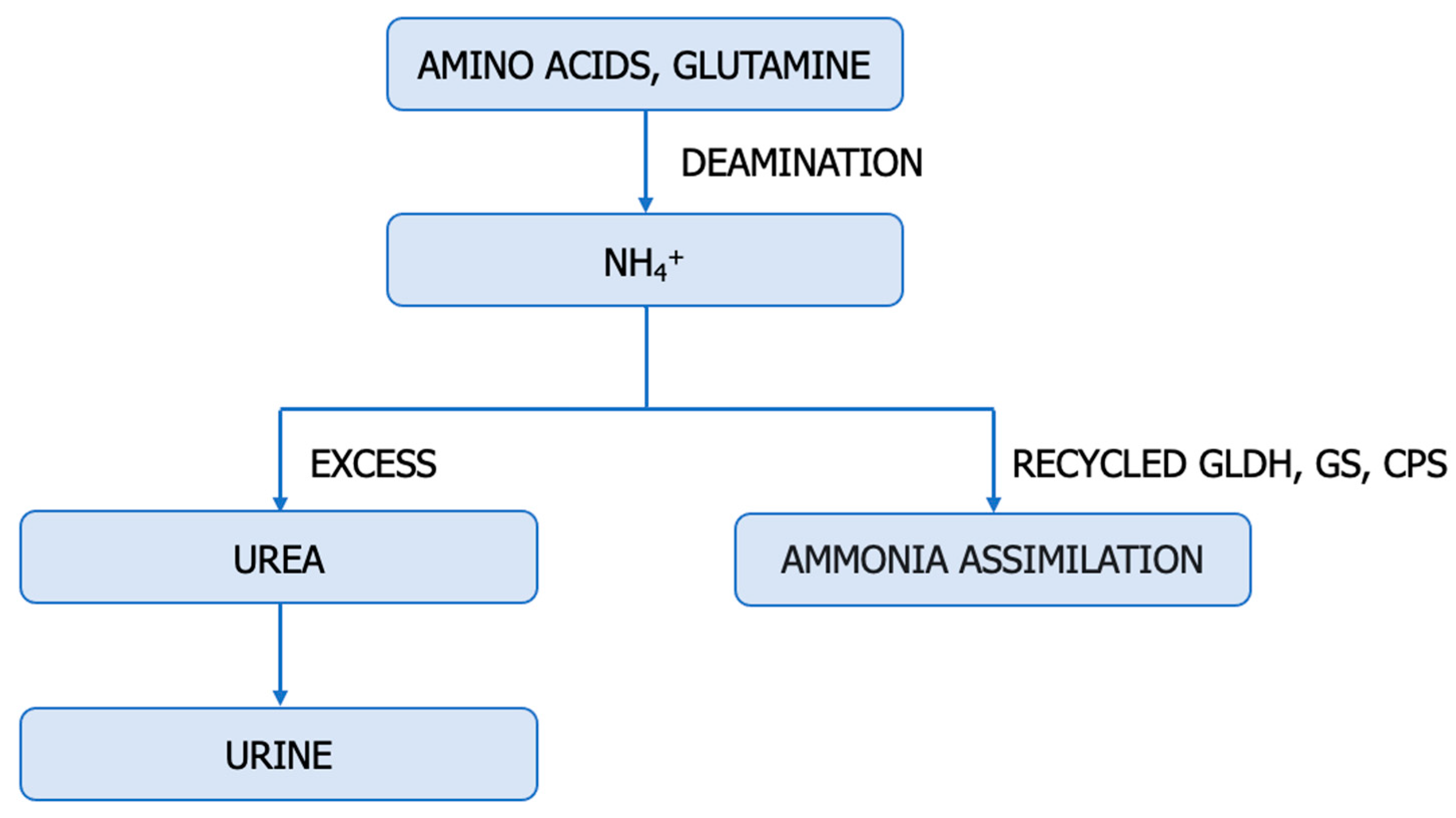
6. Hyperammonemia in ccRCC
7. The Reduction in BCAA Catabolism in ccRCC
8. Endogenous Inhibitors of NO Synthesis
9. The Inactivation of VHL and the Accumulation of HIFs—Essential Characteristics of ccRCC
10. NO-Based Therapy for ccRCC
11. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sciacovelli, M.; Dugourd, A.; Jimenez, L.V.; Yang, M.; Nikitopoulou, E.; Costa, A.S.H.; Tronci, L.; Caraffini, V.; Rodrigues, P.; Schmidt, C.; et al. Nitrogen partitioning between branched-chain amino acids and urea cycle enzymes sustains renal cancer progression. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ene, C.D.; Nicolae, I. The Inflammatory Profile Orchestrated by Inducible Nitric Oxide Synthase in Systemic Lupus Erythematosus. J. Pers. Med. 2023, 13, 934. [Google Scholar] [CrossRef] [PubMed]
- Ene, C.D.; Penescu, M.N.; Georgescu, S.R.; Tampa, M.; Nicolae, I. Posttranslational modifications pattern in clear cell renal cell carcinoma. Metabolites 2021, 11, 10. [Google Scholar] [CrossRef]
- Ene, C.D.; Nicolae, I. Hypoxia-Nitric Oxide Axis and the Associated Damage Molecular Pattern in Cutaneous Melanoma. J. Pers. Med. 2022, 12, 1646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, L.; Chen, J.; Liu, L.; Yang, X.; Cui, H.; Yue, G. Role of Metabolic Reprogramming of Long non-coding RNA in Clear Cell Renal Cell Carcinoma. J. Cancer 2022, 13, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Czyzyk-Krzeska, M.F.; Landero Figueroa, J.A.; Gulati, S.; Cunningham, J.T.; Meller, J.; Shamsaei, B.; Vemuri, B.; Plas, D.R. Molecular and metabolic subtypes in sporadic and inherited clear cell renal cell carcinoma. Genes 2021, 12, 388. [Google Scholar] [CrossRef] [PubMed]
- Ochocki, J.D.; Khare, S.; Hess, M.; Ackerman, D.; Qiu, B.; Daisak, J.I.; Worth, A.J.; Lin, N.; Lee, P.; Xie, H.; et al. Arginase 2 Suppresses Renal Carcinoma Progression via Biosynthetic Cofactor Pyridoxal Phosphate Depletion and Increased Polyamine Toxicity. Cell Metab. 2018, 27, 1263–1280.e6. [Google Scholar] [CrossRef]
- Hu, Y.; Xiang, J.; Su, L.; Tang, X. The regulation of nitric oxide in tumor progression and therapy. J. Int. Med. Res. 2020, 48, 0300060520905985. [Google Scholar] [CrossRef]
- Kurmi, K.; Haigis, M.C. Nitrogen Metabolism in Cancer and Immunity. Trends Cell Biol. 2020, 30, 408–424. [Google Scholar] [CrossRef]
- Lee, J.S.; Adler, L.; Karathia, H.; Carmel, N.; Rabinovich, S.; Auslander, N.; Keshet, R.; Stettner, N.; Silberman, A.; Agemy, L.; et al. Urea Cycle Dysregulation Generates Clinically Relevant Genomic and Biochemical Signatures. Cell 2018, 174, 1559–1570.e22. [Google Scholar] [CrossRef]
- Fukumura, D.; Kashiwagi, S.; Jain, R.K. The role of nitric oxide in tumour progression. Nat. Rev. Cancer 2006, 6, 521–534. [Google Scholar] [CrossRef]
- Hulin, J.A.; Gubareva, E.A.; Jarzebska, N.; Rodionov, R.N.; Mangoni, A.A.; Tommasi, S. Inhibition of Dimethylarginine Dimethylaminohydrolase (DDAH) Enzymes as an Emerging Therapeutic Strategy to Target Angiogenesis and Vasculogenic Mimicry in Cancer. Front. Oncol. 2020, 9, 1455. [Google Scholar] [CrossRef]
- Ene, C.V.; Nicolae, I.; Geavlete, B.; Geavlete, P.; Ene, C.D. IL-6 Signaling Link between Inflammatory Tumor Microenvironment and Prostatic Tumorigenesis. Anal. Cell. Pathol. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2021, 8, 603837. [Google Scholar] [CrossRef]
- Somasundaram, V.; Basudhar, D.; Bharadwaj, G.; No, J.H.; Ridnour, L.A.; Cheng, R.Y.S.; Fujita, M.; Thomas, D.D.; Anderson, S.K.; McVicar, D.W.; et al. Molecular mechanisms of nitric oxide in cancer progression, signal transduction, and metabolism. Antioxidants Redox Signal. 2019, 30, 1124–1143. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, W.; Zhou, J.; Wang, Y.; Wang, H.; Wang, Y. Nitrate Metabolism and Ischemic Cerebrovascular Disease: A Narrative Review. Front. Neurol. 2022, 13, 735181. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, A. Quantitative aspects of nitric oxide production from nitrate and nitrite. Excli J. 2022, 21, 470–486. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.O.; Weitzberg, E.; Gladwin, M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008, 7, 156–167. [Google Scholar] [CrossRef]
- Di Meo, N.A.; Lasorsa, F.; Rutigliano, M.; Loizzo, D.; Ferro, M.; Stella, A.; Bizzoca, C.; Vincenti, L.; Pandolfo, S.D.; Autorino, R.; et al. Renal Cell Carcinoma as a Metabolic Disease: An Update on Main Pathways, Potential Biomarkers, and Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 14360. [Google Scholar] [CrossRef] [PubMed]
- Trott, J.F.; Hwang, V.J.; Ishimaru, T.; Chmiel, K.J.; Zhou, J.X.; Shim, K.; Stewart, B.J.; Mahjoub, M.R.; Jen, K.Y.; Barupal, D.K.; et al. Arginine reprogramming in ADPKD results in arginine-dependent cystogenesis. Am. J. Physiol. Ren. Physiol. 2018, 315, F1855–F1868. [Google Scholar] [CrossRef]
- Keshet, R.; Erez, A. Arginine and the metabolic regulation of nitric oxide synthesis in cancer. DMM Dis. Model. Mech. 2018, 11, dmm033332. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, V.L.; Pinzon-Guzman, C.; Barbul, A. Arginine—Dual roles as an onconutrient and immunonutrient. J. Surg. Oncol. 2017, 115, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, X.; Jia, P.; Liu, C.; Wei, Y.; Song, Y.; Li, S.; Liu, L.; Wang, B.; Shi, H. Association between serum arginine levels and cancer risk: A community-based nested case-control study. Front. Nutr. 2022, 9, 1069113. [Google Scholar] [CrossRef] [PubMed]
- Baylis, C. Nitric oxide deficiency in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2008, 294, F1–F9. [Google Scholar] [CrossRef] [PubMed]
- Morou-Bermúdez, E.; Torres-Colón, J.E.; Bermúdez, N.S.; Patel, R.P.; Joshipura, K.J. Pathways Linking Oral Bacteria, Nitric Oxide Metabolism, and Health. J. Dent. Res. 2022, 101, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Piechowicz, J.; Gamian, A.; Chukwu, O.; Polak-Jonkisz, D. Nitric Oxide Synthesis Metabolites—As Potential Markers in Chronic Kidney Disease in Children. Curr. Issues Mol. Biol. 2022, 44, 3518–3532. [Google Scholar] [CrossRef] [PubMed]
- Carlström, M. Nitric oxide signalling in kidney regulation and cardiometabolic health. Nat. Rev. Nephrol. 2021, 17, 575–590. [Google Scholar] [CrossRef]
- Khan, F.H.; Dervan, E.; Bhattacharyya, D.D.; McAuliffe, J.D.; Miranda, K.M.; Glynn, S.A. The role of nitric oxide in cancer: Master regulator or not? Int. J. Mol. Sci. 2020, 21, 9393. [Google Scholar] [CrossRef]
- Krishnan, S.M.; Kraehling, J.R.; Eitner, F.; Bénardeau, A.; Sandner, P. The impact of the nitric oxide (no)/soluble guanylyl cyclase (sGC) signaling cascade on kidney health and disease: A preclinical perspective. Int. J. Mol. Sci. 2018, 19, 1712. [Google Scholar] [CrossRef]
- Mishra, D.; Patel, V.; Banerjee, D. Nitric Oxide and S-Nitrosylation in Cancers: Emphasis on Breast Cancer. Breast Cancer Basic Clin. Res. 2020, 14, 1178223419882688. [Google Scholar] [CrossRef]
- Hara, N.; Bilim, V.; Kasahara, T.; Obara, K.; Saito, K.; Takahashi, K.; Tomita, Y. Inducible Nitric Oxide Synthase in Renal Cell Carcinoma: Expression in Tumor Thrombi and Induction under Hypoxic Conditions. Anticancer. Res. 2003, 23, 4641–4649. [Google Scholar]
- Perske, C.; Lahat, N.; Levin, S.S.; Bitterman, H.; Hemmerlein, B.; Rahat, M.A. Loss of inducible nitric oxide synthase expression in the mouse renal cell carcinoma cell line RENCA is mediated by MicroRNA miR-146a. Am. J. Pathol. 2010, 177, 2046–2054. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Oh, C.K.; Zhang, X.; Tannenbaum, S.R.; Lipton, S.A. Protein transnitrosylation signaling networks contribute to inflammaging and neurodegenerative disorders. Antioxidants Redox Signal. 2021, 35, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Faienza, F.; Rasola, A.; Filomeni, G. Nitric oxide-based regulation of metabolism: Hints from TRAP1 and SIRT3 crosstalk. Front. Mol. Biosci. 2022, 9, 942729. [Google Scholar] [CrossRef]
- Yoon, S.; Eom, G.H.; Kang, G. Nitrosative stress and human disease: Therapeutic potential of denitrosylation. Int. J. Mol. Sci. 2021, 22, 9794. [Google Scholar] [CrossRef]
- Gu, Y.R.; Kim, J.; Na, J.C.; Han, W.K. Mitochondrial metabolic reprogramming by SIRT3 regulation ameliorates drug resistance in renal cell carcinoma. PLoS ONE 2022, 17, e0269432. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martin, C.; Serapian, S.A.; Colombo, G.; Rasola, A. Dynamically Shaping Chaperones. Allosteric Modulators of HSP90 Family as Regulatory Tools of Cell Metabolism in Neoplastic Progression. Front. Oncol. 2020, 10, 1177. [Google Scholar] [CrossRef] [PubMed]
- Poderoso, J.J.; Helfenberger, K.; Poderoso, C. The effect of nitric oxide on mitochondrial respiration. Nitric Oxide Biol. Chem. 2019, 88, 61–72. [Google Scholar] [CrossRef]
- Lowenstein, C.J. Metabolism reprogrammed by the nitric oxide signalling molecule. Nature 2019, 565, 33–34. [Google Scholar] [CrossRef]
- Zhang, S.X.; Marzluff, E.M.; Lindgren, C.A. Quantitative determination of nitric oxide from tissue samples using liquid chromatography—Mass spectrometry. MethodsX 2021, 8, 101412. [Google Scholar] [CrossRef]
- Möller, M.N.; Rios, N.; Trujillo, M.; Radi, R.; Denicola, A.; Alvarez, B. Detection and quantification of nitric oxide-derived oxidants in biological systems. J. Biol. Chem. 2019, 294, 14776–14802. [Google Scholar] [CrossRef] [PubMed]
- Jansson, O.T.; Morcos, E.; Brundin, L.; Beroerheim, U.S.R.; Adolfsson, J.; Wiklund, N.P. Nitric oxide synthase activity in human renal cell carcinoma. J. Urol. 1998, 160, 556–560. [Google Scholar] [CrossRef]
- Zequi, C.; de, S.; Fregnani, J.H.G.T.; Favaretto, R.L.; Costa, W.H.; Madeira Campos, R.S.; Fonseca, F.P.; Guimaraes, G.C.; Soares, F.A.; da Cunha, I.W.; et al. The impact of immunohistochemical expression of nitric oxide synthases on clinical and pathological features of renal cell carcinoma. World J. Urol. 2013, 31, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Renaudin, K.; Denis, M.G.; Karam, G.; Vallette, G.; Buzelin, F.; Laboisse, C.L.; Jarry, A. Loss of NOSI expression in high-grade renal cell carcinoma associated with a shift of NO signalling. Br. J. Cancer 2004, 90, 2364–2369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ali, M.A.; Akhmedkhanov, A.; Zeleniuch-Jaquotte, A.; Toniolo, P.; Frenkel, K.; Huang, X. Reliability of serum iron, ferritin, nitrite, and association with risk of renal cancer in women. Cancer Detect. Prev. 2003, 27, 116–121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khare, S.; Kim, L.C.; Lobel, G.; Doulias, P.-T.; Ischiropoulos, H.; Nissim, I.; Keith, B.; Simon, M.C. ASS1 and ASL suppress growth in clear cell renal cell carcinoma via altered nitrogen metabolism. Cancer Metab. 2021, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, G.; Ferro, M.; Ditonno, P.; Battaglia, M. The urea cycle enzymes act as metabolic suppressors in clear cell renal cell carcinoma. Transl. Cancer Res. 2018, 7, S766–S769. [Google Scholar] [CrossRef]
- Carney, E.F. Altered ammonia metabolism in ccRCC. Nat. Rev. Nephrol. 2018, 14, 476. [Google Scholar] [CrossRef]
- Minoru, M.; Masao, O.; Toshihiko, K.; Takao, S.; Kenji, S.; Teruo, N. Concentrations of polyamines in renal cell carcinoma. Clin. Chim. Acta 1978, 87, 93–99. [Google Scholar] [CrossRef]
- Koida, T. The clinical significance of tissue, blood and urine polyamine in renal cell carcinoma. Japanese J. Urol. 1992, 83, 1228–1237. [Google Scholar] [CrossRef]
- Dallmann, K.; Junker, H.; Balabanov, S.; Zimmermann, U.; Giebel, J.; Walther, R. Human agmatinase is diminished in the clear cell type of renal cell carcinoma. Int. J. Cancer 2004, 108, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, L.; Huang, D.; Yue, W.; Chen, J.; Liu, C. A nitric oxide-releasing prodrug promotes apoptosis in human renal carcinoma cells: Involvement of reactive oxygen species. Open Chem. 2021, 19, 635–645. [Google Scholar] [CrossRef]
- Morbidelli, L.; Donnini, S.; Ziche, M. Role of Nitric Oxide in the Modulation of Angiogenesis. Curr. Pharm. Des. 2005, 9, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Said Abasse, K.; Essien, E.E.; Abbas, M.; Yu, X.; Xie, W.; Jinfang, S.; Akter, L.; Cote, A. Association between Dietary Nitrate, Nitrite Intake, and Site-Specific Cancer Risk: A Systematic Review and Meta-Analysis. Nutrients 2022, 14, 666. [Google Scholar] [CrossRef]
- Kamal, N.; Ilowefah, M.A.; Hilles, A.R.; Anua, N.A.; Awin, T.; Alshwyeh, H.A.; Aldosary, S.K.; Jambocus, N.G.S.; Alosaimi, A.A.; Rahman, A.; et al. Genesis and Mechanism of Some Cancer Types and an Overview on the Role of Diet and Nutrition in Cancer Prevention. Molecules 2022, 27, 1794. [Google Scholar] [CrossRef]
- Hord, N.G.; Tang, Y.; Bryan, N.S. Food sources of nitrates and nitrites: The physiologic context for potential health benefits. Am. J. Clin. Nutr. 2009, 90, 1–10. [Google Scholar] [CrossRef]
- Mirvish, S.S. Experimental evidence for inhibition of N-nitroso compound formation as a factor in the negative correlation between vitamin C consumption and the incidence of certain cancers. Cancer Res. 1994, 54, 1948s–1951s. [Google Scholar]
- Bartsch, H.; Ohshima, H.; Pignatelli, B. Inhibitors of endogenous nitrosation mechanisms and implications in human cancer prevention. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1988, 202, 307–324. [Google Scholar] [CrossRef]
- Bogovski, P.; Bogovski, S. Special report animal species in which n-nitroso compounds induce cancer. Int. J. Cancer 1981, 27, 471–474. [Google Scholar] [CrossRef]
- Brock, K.E.; Gridley, G.; Chiu, B.C.H.; Ershow, A.G.; Lynch, C.F.; Cantor, K.P. Dietary fat and risk of renal cell carcinoma in the USA: A case-control study. Br. J. Nutr. 2009, 101, 1228–1238. [Google Scholar] [CrossRef]
- Alexander, D.D.; Cushing, C.A. Quantitative assessment of red meat or processed meat consumption and kidney cancer. Cancer Epidemiol. 2009, 32, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Q.; He, J. Correction: Intake of red and processed meat and risk of renal cell carcinoma: A meta-analysis of observational studies. Oncotarget 2018, 9, 29018. [Google Scholar] [CrossRef]
- Liao, Z.; Fang, Z.; Gou, S.; Luo, Y.; Liu, Y.; He, Z.; Li, X.; Peng, Y.; Fu, Z.; Li, D.; et al. The role of diet in renal cell carcinoma incidence: An umbrella review of meta-analyses of observational studies. BMC Med. 2022, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Hu, L.; Feng, X.; Wang, S. Nitrate and nitrite in health and disease. Aging Dis. 2018, 9, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Sözen, S.; Coskun, U.; Sancak, B.; Bukan, N.; Günel, N.; Tunc, L.; Bozkirli, I. Serum levels of interleukin-18 and nitrite+nitrate in renal cell carcinoma patients with different tumor stage and grade. Neoplasma 2004, 51, 25–29. [Google Scholar]
- Tate, D.J.; Vonderhaar, D.J.; Caldas, Y.A.; Metoyer, T.; Patterson IV, J.R.; Aviles, D.H.; Zea, A.H. Effect of arginase II on L-arginine depletion and cell growth in murine cell lines of renal cell carcinoma. J. Hematol. Oncol. 2008, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Zea, A.H.; Rodriguez, P.C.; Atkins, M.B.; Hernandez, C.; Signoretti, S.; Zabaleta, J.; McDermott, D.; Quiceno, D.; Youmans, A.; O’Neill, A.; et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: A mechanism of tumor evasion. Cancer Res. 2005, 65, 3044–3048. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhanasekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; da Veiga Leprevost, F.; Reva, B.; Lih, T.S.M.; Chang, H.Y.; et al. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 2019, 179, 964–983.e31. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef]
- Pandey, N.; Lanke, V.; Vinod, P.K. Network-based metabolic characterization of renal cell carcinoma. Sci. Rep. 2020, 10, 5955. [Google Scholar] [CrossRef]
- Liu, H.; Li, S.; Liu, X.; Chen, Y.; Deng, H. SIRT3 Overexpression Inhibits Growth of Kidney Tumor Cells and Enhances Mitochondrial Biogenesis. J. Proteome Res. 2018, 17, 3143–3152. [Google Scholar] [CrossRef]
- Morigi, M.; Perico, L.; Benigni, A. Sirtuins in Renal Health and Disease. J. Am. Soc. Nephrol. 2018, 29, 1799–1809. [Google Scholar] [CrossRef]
- Costa-Machado, L.F.; Fernandez-Marcos, P.J. The sirtuin family in cancer. Cell Cycle 2019, 18, 2164–2196. [Google Scholar] [CrossRef]
- Galiniak, S.; Biesiadecki, M.; Mołoń, M.; Olech, P.; Balawender, K. Serum Oxidative and Nitrosative Stress Markers in Clear Cell Renal Cell Carcinoma. Cancers 2023, 15, 3995. [Google Scholar] [CrossRef] [PubMed]
- Poillet-Perez, L.; Xie, X.; Zhan, L.; Yang, Y.; Sharp, D.W.; Hu, Z.S.; Su, X.; Maganti, A.; Jiang, C.; Lu, W.; et al. Autophagy maintains tumour growth through circulating arginine. Nature 2018, 563, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Hajaj, E.; Sciacovelli, M.; Frezza, C.; Erez, A. The context-specific roles of urea cycle enzymes in tumorigenesis. Mol. Cell 2021, 81, 3749–3759. [Google Scholar] [CrossRef]
- Nagamani, S.C.S.; Erez, A. A metabolic link between the urea cycle and cancer cell proliferation. Mol. Cell. Oncol. 2016, 3, e1127314. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Wang, H.; Dong, D.; Li, T.; Yu, Z.; Guo, J.; Zhou, W.; Li, D.; Yan, R.; Wang, L.; et al. Urea as a By-Product of Ammonia Metabolism Can Be a Potential Serum Biomarker of Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 677. [Google Scholar] [CrossRef]
- Hoerner, C.R.; Chen, V.J.; Fan, A.C. The “achilles heel” of metabolism in renal cell carcinoma: Glutaminase inhibition as a rational treatment strategy. Kidney Cancer 2019, 3, 15–29. [Google Scholar] [CrossRef]
- Okazaki, A.; Gameiro, P.; Stephanopoulos, G.; Iliopoulos, O. Abstract 1123: Glutaminase inhibitors suppress pyrimidine synthesis and promote DNA replication stress in VHL -deficient human renal cancer cells. Cancer Res. 2015, 75, 1123. [Google Scholar] [CrossRef]
- Kaushik, A.K.; Tarangelo, A.; Boroughs, L.K.; Ragavan, M.; Zhang, Y.; Wu, C.Y.; Li, X.; Ahumada, K.; Chiang, J.C.; Tcheuyap, V.T.; et al. In vivo characterization of glutamine metabolism identifies therapeutic targets in clear cell renal cell carcinoma. Sci. Adv. 2022, 8, eabp8293. [Google Scholar] [CrossRef] [PubMed]
- Nabi, S.; Kessler, E.R.; Bernard, B.; Flaig, T.W.; Lam, E.T. Renal cell carcinoma: A review of biology and pathophysiology. F1000Research 2018, 7, 307. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Neri, I.; Ramírez-Bermúdez, J.; Ojeda-López, C.; Montes, S.; Soto-Hernández, J.L.; Ríos, C. Glutamine and citrulline concentrations reflect nitric oxide synthesis in the human nervous system. Neurologia 2020, 35, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Bryk, J.; Ochoa, J.B.; Correia, M.I.T.D.; Munera-Seeley, V.; Popovic, P.J. Effect of citrulline and glutamine on nitric oxide production in RAW 264.7 cells in an arginine-depleted environment. J. Parenter. Enter. Nutr. 2008, 32, 377–383. [Google Scholar] [CrossRef]
- Chen, C.L.; Hsu, S.C.; Ann, D.K.; Yen, Y.; Kung, H.J. Arginine signaling and cancer metabolism. Cancers 2021, 13, 3541. [Google Scholar] [CrossRef] [PubMed]
- Ochocki, J.; Lin, N.; Qiu, B.; Simon, M.C. Abstract C32: Metabolic advantages of urea cycle misregulation in renal cell carcinoma. Cancer Res. 2013, 73, C32. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Yoon, H.; Ringel, A.E.; Jeanfavre, S.; Clish, C.B.; Haigis, M.C. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 2017, 358, 941–946. [Google Scholar] [CrossRef]
- Kimura, S.; Fujisaki, Y.; Onizuka, C.; Hasuike, S.; Sato, Y.; Mukai, S.; Kamoto, T. A case of hyperammonemia occurring during treatment of metastatic renal cell carcinoma with axitinib. IJU Case Reports 2023, 6, 206–210. [Google Scholar] [CrossRef]
- Milewski, K.; Bogacińska-Karaś, M.; Frȩśko, I.; Hilgier, W.; Jaźwiec, R.; Albrecht, J.; Zielińska, M. Ammonia reduces intracellular asymmetric dimethylarginine in cultured astrocytes stimulating its y+LAT2 carrier-mediated loss. Int. J. Mol. Sci. 2017, 18, 2308. [Google Scholar] [CrossRef]
- Peng, H.; Wang, Y.; Luo, W. Multifaceted role of branched-chain amino acid metabolism in cancer. Oncogene 2020, 39, 6747–6756. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Wilkinson, A.C. Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.J.; Lee, J.H.; Kim, D.H.; Kim, K.T.; Lee, G.W.; Choi, S.J.; Chang, P.S.; Paik, H.D. Antioxidative and nitric oxide scavenging activity of branched-chain amino acids. Food Sci. Biotechnol. 2015, 24, 1555–1558. [Google Scholar] [CrossRef]
- Chachaj, A.; Wiśniewski, J.; Rybka, J.; Butrym, A.; Biedroń, M.; Krzystek-Korpacka, M.; Fleszar, M.G.; Karczewski, M.; Wróbel, T.; Mazur, G.; et al. Asymmetric and symmetric dimethylarginines and mortality in patients with hematological malignancies—A prospective study. PLoS ONE 2018, 13, e0197148. [Google Scholar] [CrossRef]
- Li, Z.L.; Wang, B.; Wen, Y.; Wu, Q.L.; Lv, L.L.; Liu, B.C. Disturbance of Hypoxia Response and Its Implications in Kidney Diseases. Antioxidants Redox Signal. 2022, 37, 936–955. [Google Scholar] [CrossRef]
- Lasorsa, F.; Rutigliano, M.; Milella, M.; Ferro, M.; Pandolfo, S.D.; Crocetto, F.; Autorino, R.; Battaglia, M.; Ditonno, P.; Lucarelli, G. Cancer Stem Cells in Renal Cell Carcinoma: Origins and Biomarkers. Int. J. Mol. Sci. 2023, 24, 13179. [Google Scholar] [CrossRef]
- Lasorsa, F.; Rutigliano, M.; Milella, M.; Ferro, M.; Pandolfo, S.D.; Crocetto, F.; Tataru, O.S.; Autorino, R.; Battaglia, M.; Ditonno, P.; et al. Cellular and Molecular Players in the Tumor Microenvironment of Renal Cell Carcinoma. J. Clin. Med. 2023, 12, 3888. [Google Scholar] [CrossRef]
- Sinha, B.K. Can Nitric Oxide-Based Therapy Be Improved for the Treatment of Cancers? A Perspective. Int. J. Mol. Sci. 2023, 24, 13611. [Google Scholar] [CrossRef]
- Zhao, Y.; Ouyang, X.; Peng, Y.; Peng, S. Stimuli responsive nitric oxide-based nanomedicine for synergistic therapy. Pharmaceutics 2021, 13, 1917. [Google Scholar] [CrossRef]






| NO Parameters (References) | Biological Systems | Results | Conclusions |
|---|---|---|---|
| Calcium-dependent and calcium-independent NO synthetase [42] | Human kidney/RCC, proximal tubular cell lines HN4, HN51. | Calcium-dependent NOS activity, identified in all the samples studied, was downregulated in RCC compared to non-malignant renal tissues studied; calcium-independent NOS activity was inconsistently expressed in the renal tissue. | NO exerted cytostatic effects on cultured renal cells. |
| NOS1, NOS2, NOS3 [43] | Non-neoplastic renal tissues and RCC | In non-neoplastic tissues, NOS3 immunoreactivity was increased and NOS2 was reduced compared to RCC. The NOS expression was correlated with tumour size and a poor prognosis. | NOS3 as a predictive factor in RCC |
| NOS, sGC, nitrotirosine [44] | Normal and tumoural renal tissue (benign and malignant tumours). | NOS1 is downregulated in malignant tissues and associated with the tumour grade; sGC is present in all renal tumours; nitrotyrosine is present in normal renal parenchyma and tumour tissues. | Autocrine signalling of NO is similar in normal and non-malignant renal tissues and altered in malignant tissues |
| Nitrites [45] | Serum (apparently healthy women diagnosed with RCC) | Elevated serum level in patients with RCC | Elevated serum nitrite levels are associated with a low risk of renal cancer. |
| ASS1, ASL, Arg2 [46,47] | RCC tissue samples and control | mRNA and ASS1, ASL, Arg2 activity are reduced in RCC vs. normal kidney Altered urea cycle-metabolic pathway in RCC. | Attenuation of the cytotoxic effects of NO. ASS1, ASL, Arg2 -metabolic suppressors in RCC. |
| NOSi-ARN [31] | RCC and control tissue samples | mRNA and iNOS protein present in tumour thrombi in patients with RCC and in A498 and A704 cells under hypoxic conditions. | It mediates the formation of tumour thrombi and hypoxic adaptation. |
| Arg2, ASS1 [46,48] | Normal and malignant renal cell lines | The expression of enzymes in the urea cycle is downregulated in RCC compared to the control. Deficiency of enzymes in the urea cycle disrupts polyamine synthesis, conservation of pyridoxal phosphate, arginine auxotrophy, infiltration of cytotoxic T cells in the tumour tissue, and immunosuppression in the tumour microenvironment. | Arg2 and ASS1 are potential metabolic suppressors of renal tumourigenesis. |
| ASS1, ADI (E.C.3.5.3.6) [35] | Biopsy samples, animal models, cell lines. | Low or undetectable ASS1 in RCC, present in normal proximal tubule epithelium. Exogenous ADI (arginine deiminase) determines antiproliferative and antiangiogenic effects in vivo on RENCA tumour cells and extends the survival of tumour-bearing mice. | Arginine deprivation via ADI—an antitumour strategy in RCC |
| Spermine, spermidine [49] | Normal and malignant human renal tissue | Spermidine levels and spermidine/spermine ratio increase; normal tissue < differentiated RCC < poorly differentiated RCC. The other polyamines do not show differences between normal tissues, tumours, and metastases. | Polyamines—biochemical markers for the malignancy of RCC |
| Diamine, spermidine, spermine [50] | Tissue, urine, blood | Elevated levels are correlated with the progression of RCC | Polyamines—tumour markers in RCC |
| Agmatinase (E. C.) [51] | Normal and malignant renal tissue | The expression and mRNA of agmatinase are decreased in RCC compared to benign renal tumours. Accumulated agmatine stimulates NOS3 and NOS2, leading to NO synthesis. | Reduced agmatinase increases the cytotoxic activity of NO in RCC |
| RNS, NO2− [52] | Cell cultures | JS-K, a NO donor, stimulates the increase in ROS (Reactive Oxygen Species) and RNS (Reactive Nitrogen Species), the reduction in GSH/GSSG (glutathione redox status), the increase in pro-apoptotic proteins (Bak, Bax), and the reduction in anti-apoptotic proteins (Bcl-2) in RCC (Renal Cell Carcinoma). JS-K induces apoptosis in cancer cells by modulating the production and signalling of NO, MAPK (Mitogen-Activated Protein Kinase), the ubiquitin–proteasome pathway, and the β-catenin/T-cell factor (TCF) signalling pathway | NO released by JS-K induces apoptosis in renal carcinoma cells by increasing the levels of ROS/RNS and induces chemosensitivity of tumour cells to doxorubicin |
| NOS, cGMP [53] | Experimental and human tumours. | The NOS enzymes are overexpressed in tumour tissues. | NO metabolites correlate with angiogenesis and tumour aggressiveness. |
| Dietary nitrite, nitrate intake [54] | Evaluation of nitrates and nitrites in food sources (41 articles, 13 types of cancer). | Nitrates from plant sources and nitrates in general do not affect the development of renal cancer. Nitrites from processed meat are associated with an increased risk of renal cancer < pancreatic cancer < thyroid cancer, stomach cancer, glioma, and others. | The nitrites/nitrates intake has specific effects on the type and site-specific risk of cancer |
| Phytonutrients [55,56,57,58,59,60] | Epidemiological studies on the role of dietary factors. | Phytonutrients play an essential role in cancer prevention. Dietary sources of nitrites and nitrates have a role in immunity and vascular function. Dietary sources of nitrites include vegetables, fruits, and processed meat. | Vitamin C inhibits endogenous nitrosation. |
| Red and processed meat [61] | Meta-analysis (12 case-control studies, 16 cohorts) | No statistically significant data were obtained between the consumption of red and processed meat, individual variables (BMI, smoking, total energy intake), and the development of renal cancer. | An independent relationship between meat consumption and the risk of renal cancer was not evident. |
| Red and processed meat [62] | Meta−analysis (23 eligible publications) on the association and impact of red meat consumption on RCC | Positive relationship between consumption of beef, salami, ham, bacon, sausages, hamburgers, and renal cancer. | Statistically significant positive association between red meat consumption and RCC |
| Dietary factors [63,64] | Report on 22 meta−analyses (566 publications). | No suggestive or convincing evidence between the consumption of foods, beverages, alcohol, macronutrients, micronutrients, and the incidence of RCC | The intake of vegetables and vitamin C is associated with the risk of RCC |
| Serum NO2−, NO3− [65] | RCC patients and control patients | No significant differences between patients and controls. Variations depending on the tumour grade. | NO exerts immunoregulatory effects in RCC |
| Arginase 2 [66] | Murin renal cell lines, normal and neoplastic. | Arginase 2 rapidly metabolizes L-arginine, suppresses tumour growth, and reduces the expression of CD3zeta. | Arginase 2 modulates the function of T cells, depleting arginine. |
| Arginase 2 [67] | Peripheral blood of metastatic RCC and control patients | Myeloid suppressor cells producing arginase present in patients with metastatic ccRCC | Arginase 2 regulates the availability of arginine. |
| Arginase 2 [7] | Arginase 2 supports the growth of ccRCC | ||
| BCAA, BCAT, ASS1 [1,21,68,69,70] | Cultured primary and metastatic renal cancer cells (omic study) | Transcriptionally suppressed BCAA catabolism, overexpressed BCAT, urea cycle, glutathione, cysteine/methionine, arginine, glutamine, tryptophan, reactivated polyamines in ccRCC show metabolic flexibility during tumour progression and offer invasive potential. | Altered metabolic advantage for cancer cell survival |
| SIRTs [34,36,71,72,73] | ccRCC cells, cell lines, genetic models, pharmacological models, omic studies, computational | SIRTs-1,3,6,7 maintain renal homeostasis. SIRT1 regulates NOSe in glomerular cells. NO regulates the stability of the SIRT3/TRAP1 complex. SIRT3 has dual effects on tumour growth. SIRT3 has antioxidant and anti-inflammatory effects in kidney disease. SIRT4 inhibits glutamine metabolism | Activating SIRTs before tumour initiation is a preventive strategy. NO/SIRT3 regulates mitochondrial biogenesis in ccRCC and cell sensitivity to antitumour therapy. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ene, C.D.; Tampa, M.; Georgescu, S.R.; Matei, C.; Leulescu, I.M.T.; Dogaru, C.I.; Penescu, M.N.; Nicolae, I. Disturbances in Nitric Oxide Cycle and Related Molecular Pathways in Clear Cell Renal Cell Carcinoma. Cancers 2023, 15, 5797. https://doi.org/10.3390/cancers15245797
Ene CD, Tampa M, Georgescu SR, Matei C, Leulescu IMT, Dogaru CI, Penescu MN, Nicolae I. Disturbances in Nitric Oxide Cycle and Related Molecular Pathways in Clear Cell Renal Cell Carcinoma. Cancers. 2023; 15(24):5797. https://doi.org/10.3390/cancers15245797
Chicago/Turabian StyleEne, Corina Daniela, Mircea Tampa, Simona Roxana Georgescu, Clara Matei, Iulia Maria Teodora Leulescu, Claudia Ioana Dogaru, Mircea Nicolae Penescu, and Ilinca Nicolae. 2023. "Disturbances in Nitric Oxide Cycle and Related Molecular Pathways in Clear Cell Renal Cell Carcinoma" Cancers 15, no. 24: 5797. https://doi.org/10.3390/cancers15245797
APA StyleEne, C. D., Tampa, M., Georgescu, S. R., Matei, C., Leulescu, I. M. T., Dogaru, C. I., Penescu, M. N., & Nicolae, I. (2023). Disturbances in Nitric Oxide Cycle and Related Molecular Pathways in Clear Cell Renal Cell Carcinoma. Cancers, 15(24), 5797. https://doi.org/10.3390/cancers15245797








