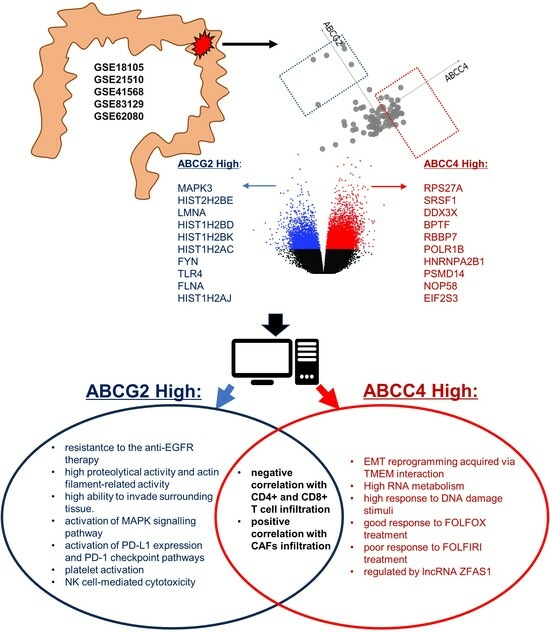
Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC—A New Opportunity to Predict Therapy Response
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Microarray Data Processing and Analysis
2.2. Survival Probability Analysis
2.3. Enrichment Analysis
2.4. Hierarchical Clustering
2.5. Protein–Protein Interaction Network
2.6. Analysis of Immune Cell Tumour Infiltration
2.7. Statistics
3. Results
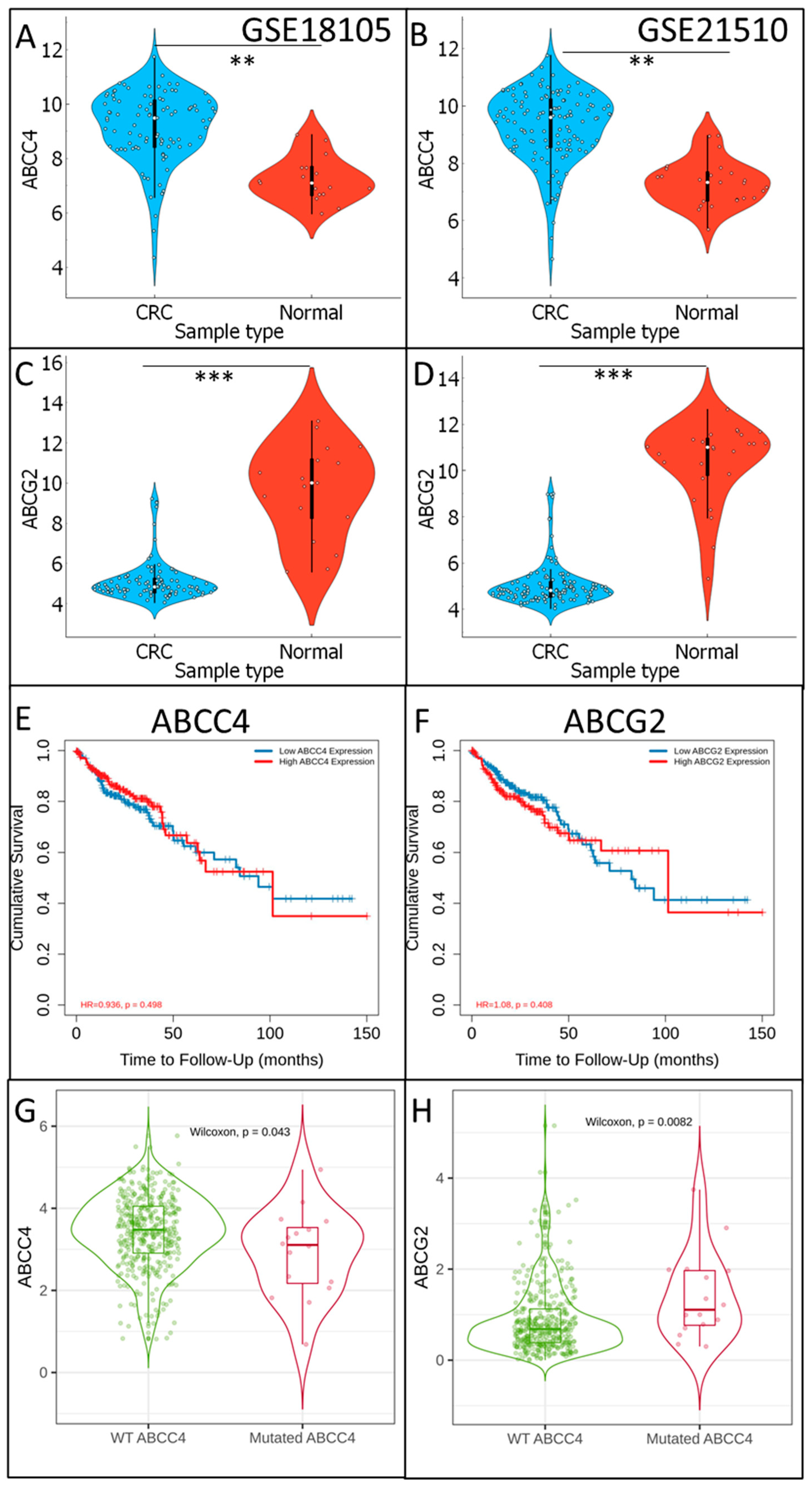
3.1. Analysis of ABCC4 and ABCG2 Expression Level in CRC
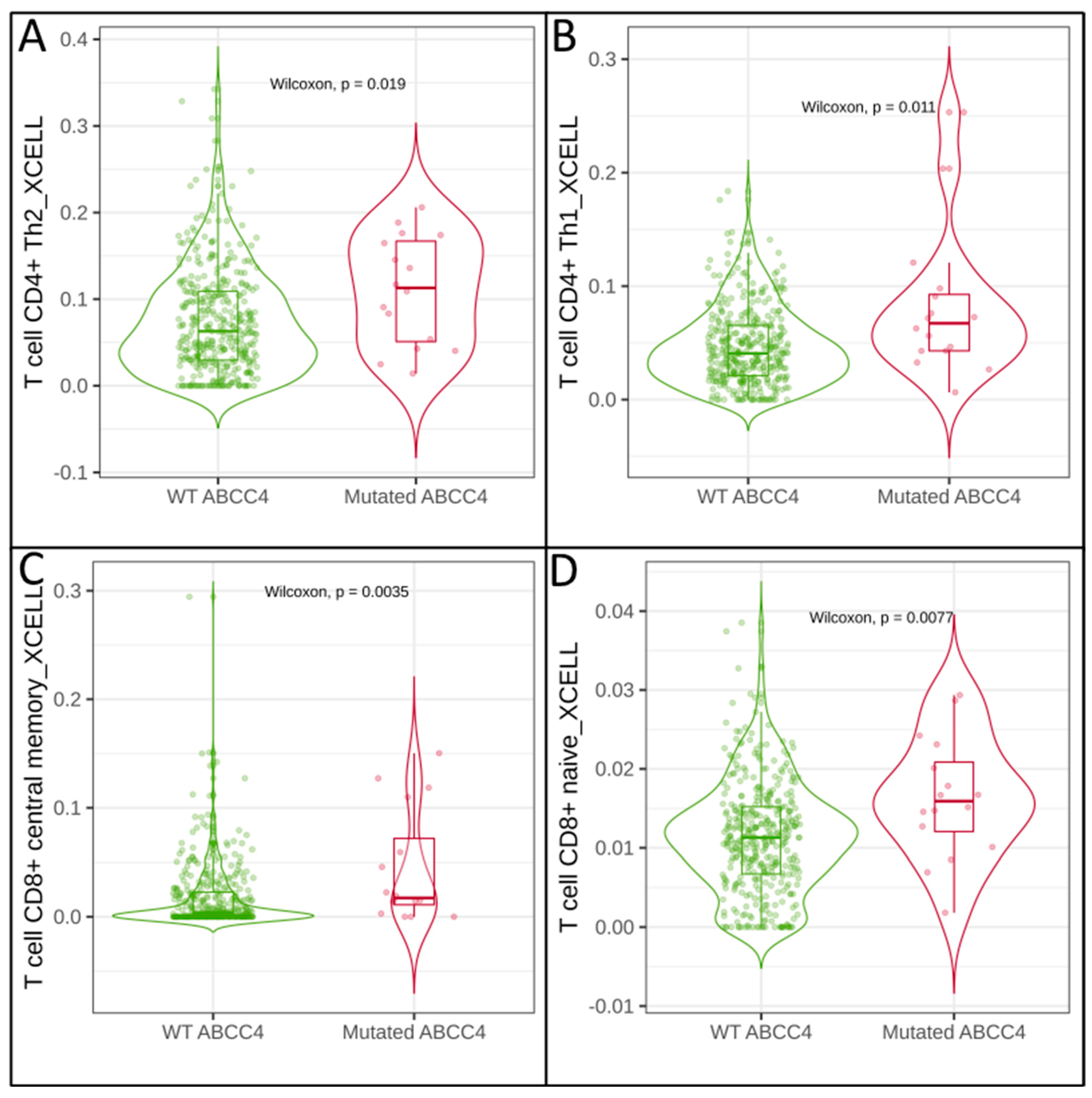
3.2. Correlation of Immune Cell Infiltration with ABCG2 and ABCC4 Expression Levels in CRC
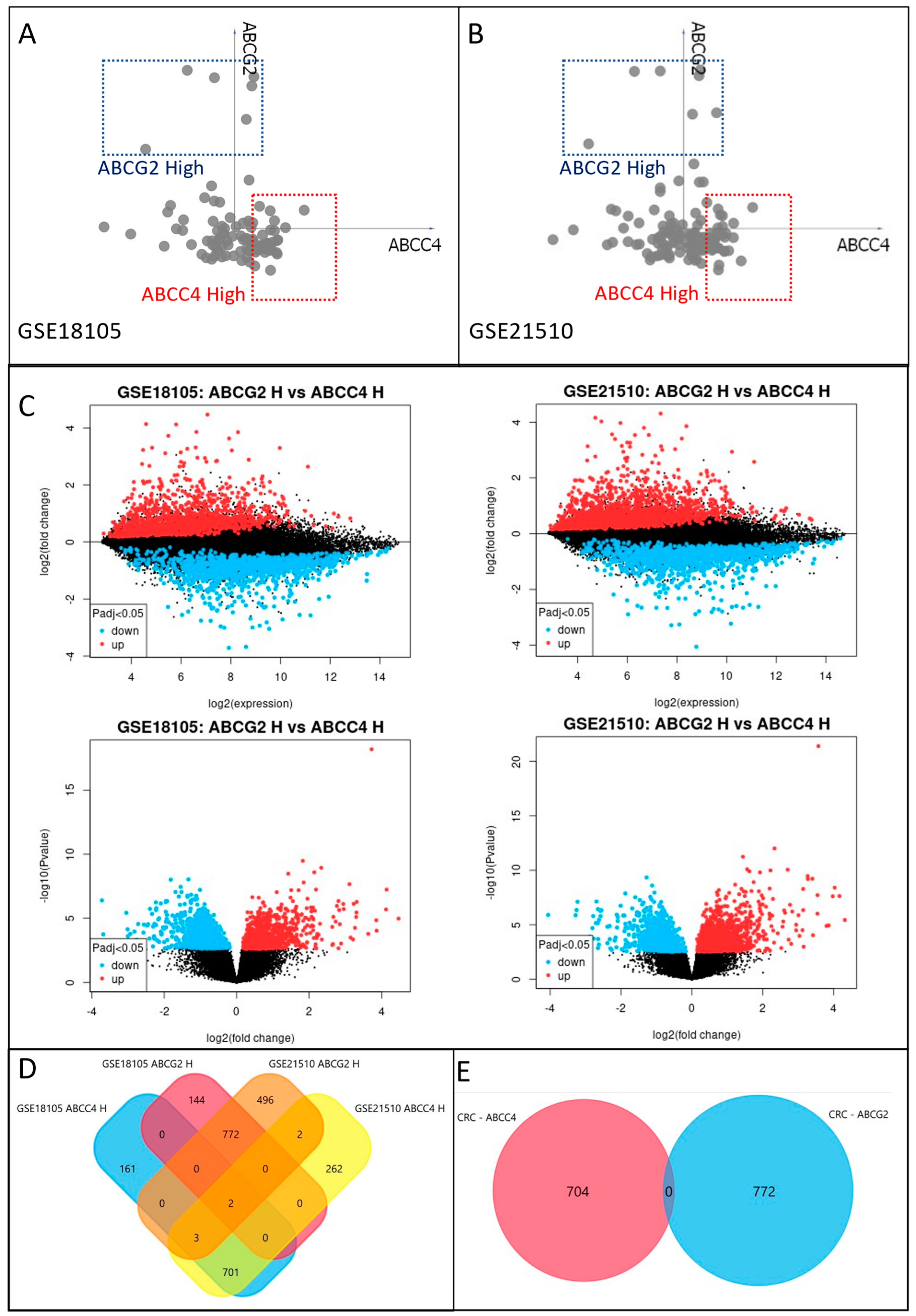
3.3. Identification of ABCC4 and ABCG2 High Expression CRC Subsets
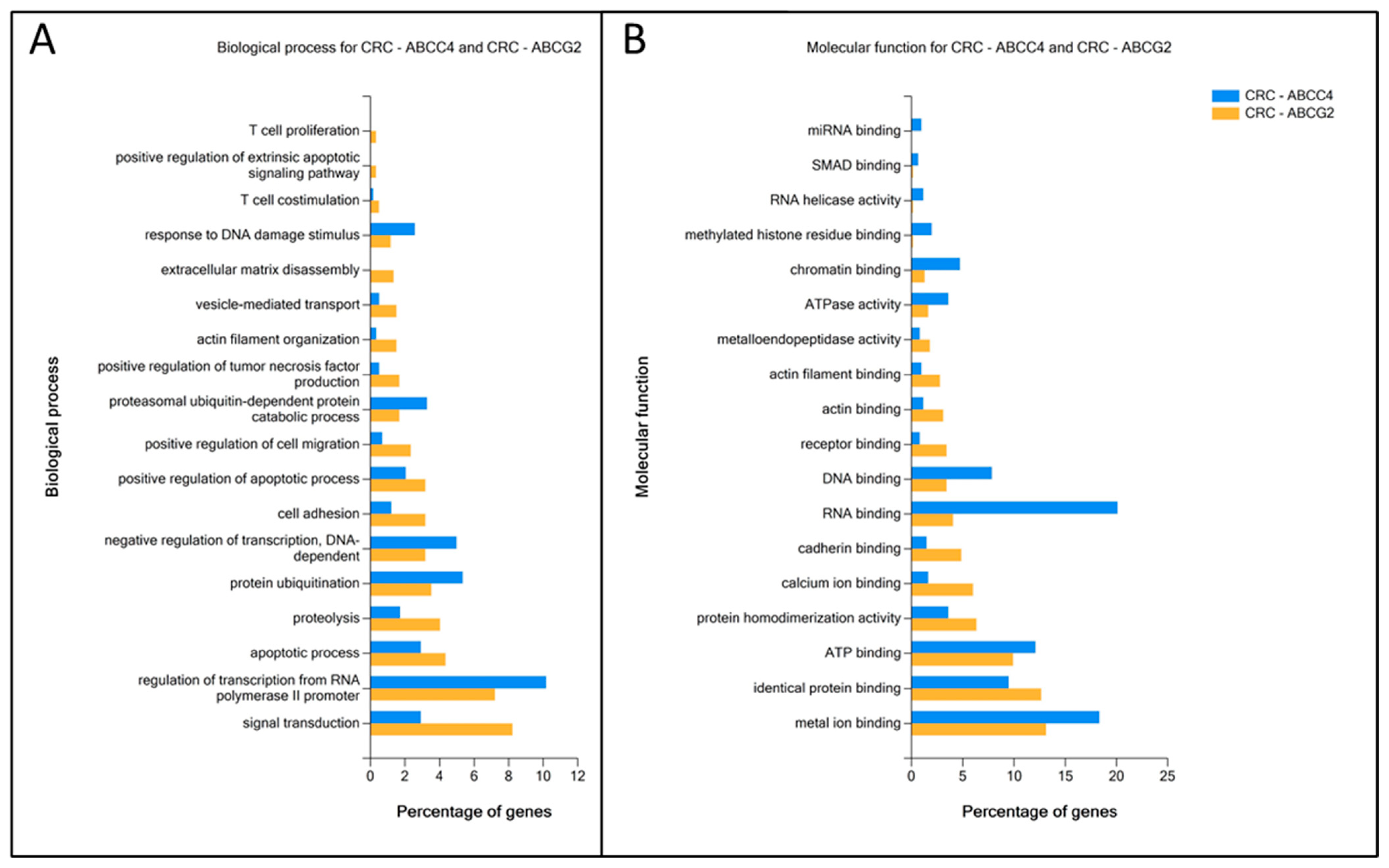
3.4. Enrichment Analysis of DEGs Unique to ABCC4 High or ABCG2 High CRC Subsets
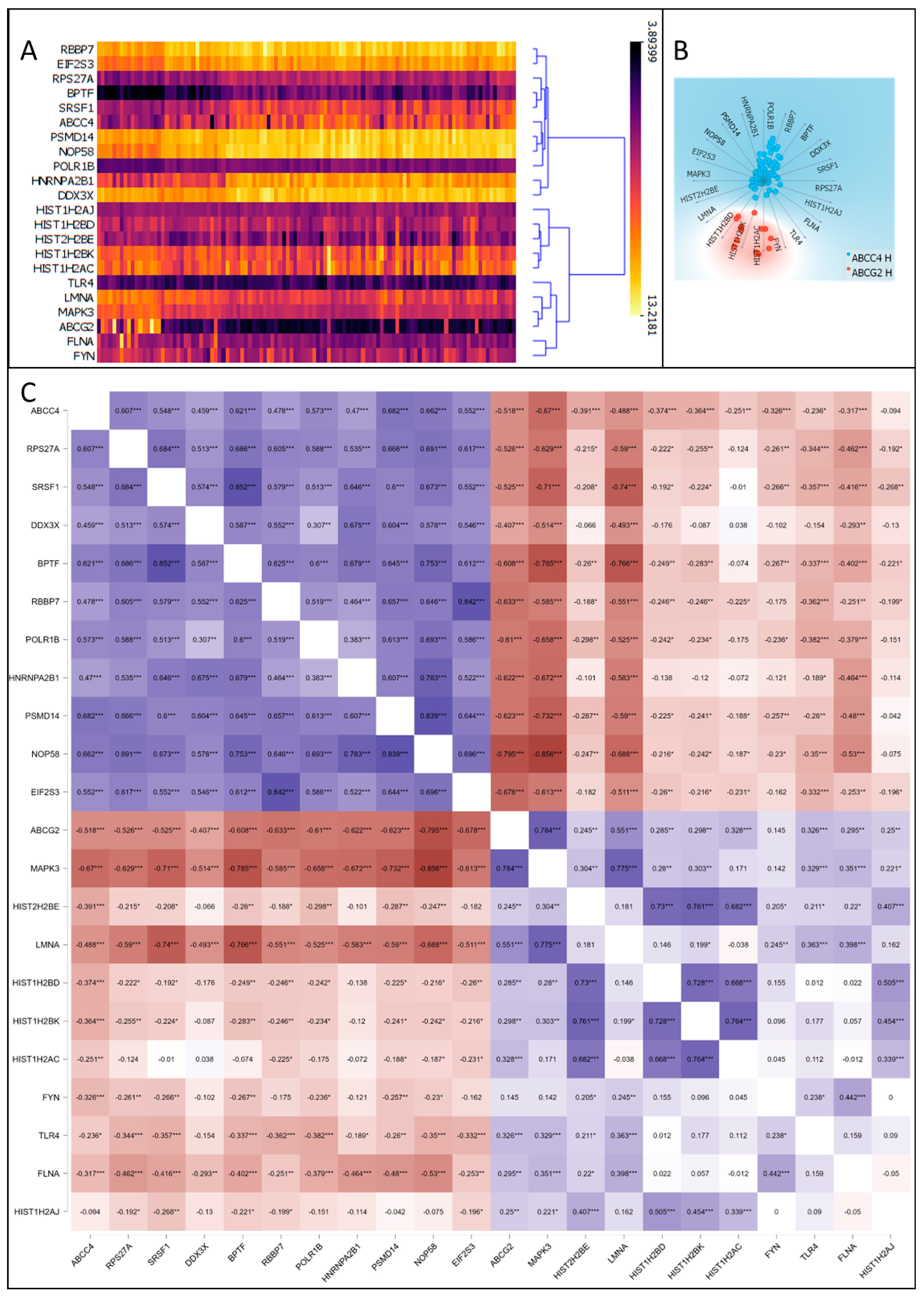
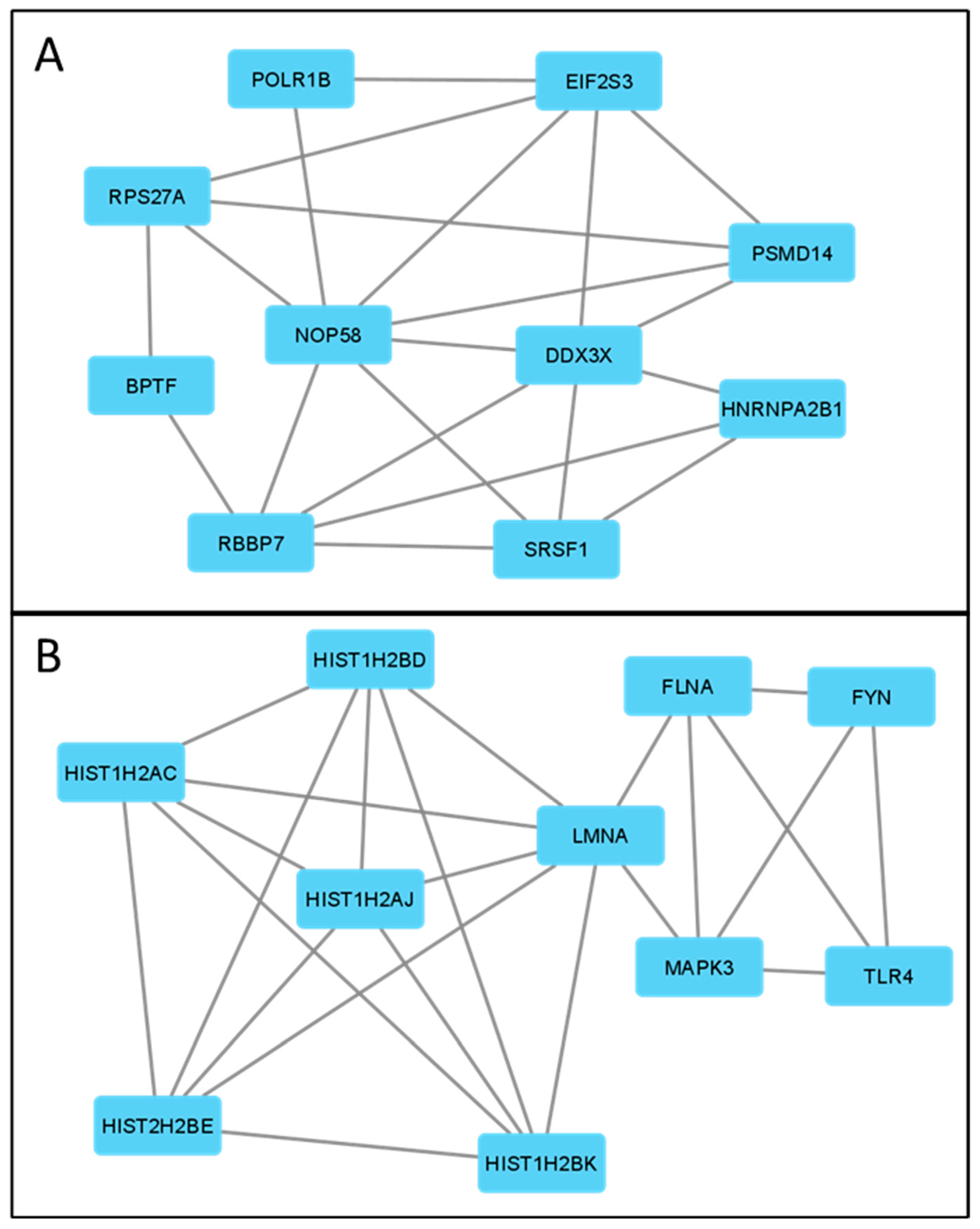
3.5. Correlation of ABCC4 and ABCG2 Expression Levels with Major Dysregulated Protein Hubs
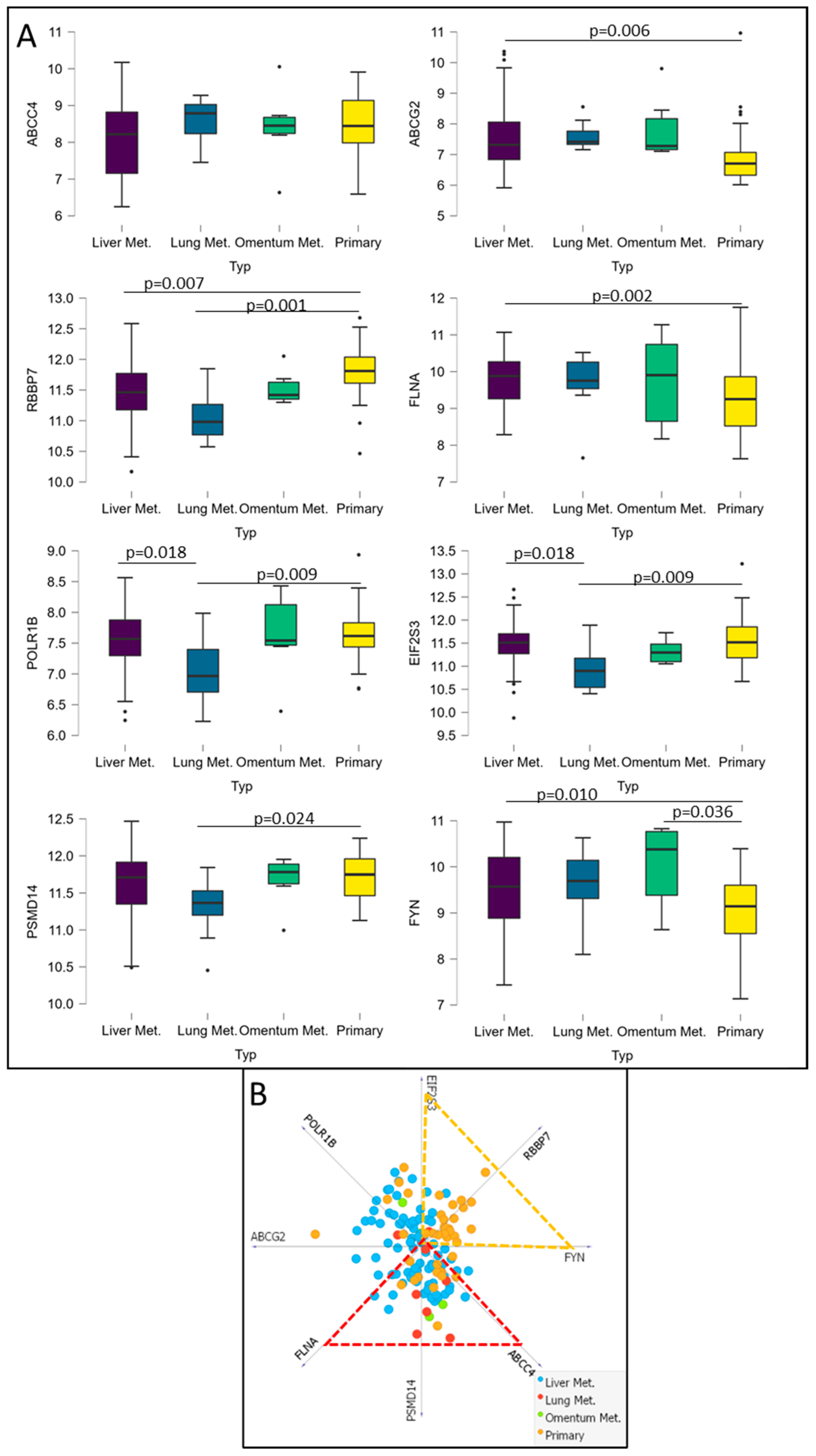
3.6. Analysis of Potential CRC Metastatic Organotropism Biomarkers
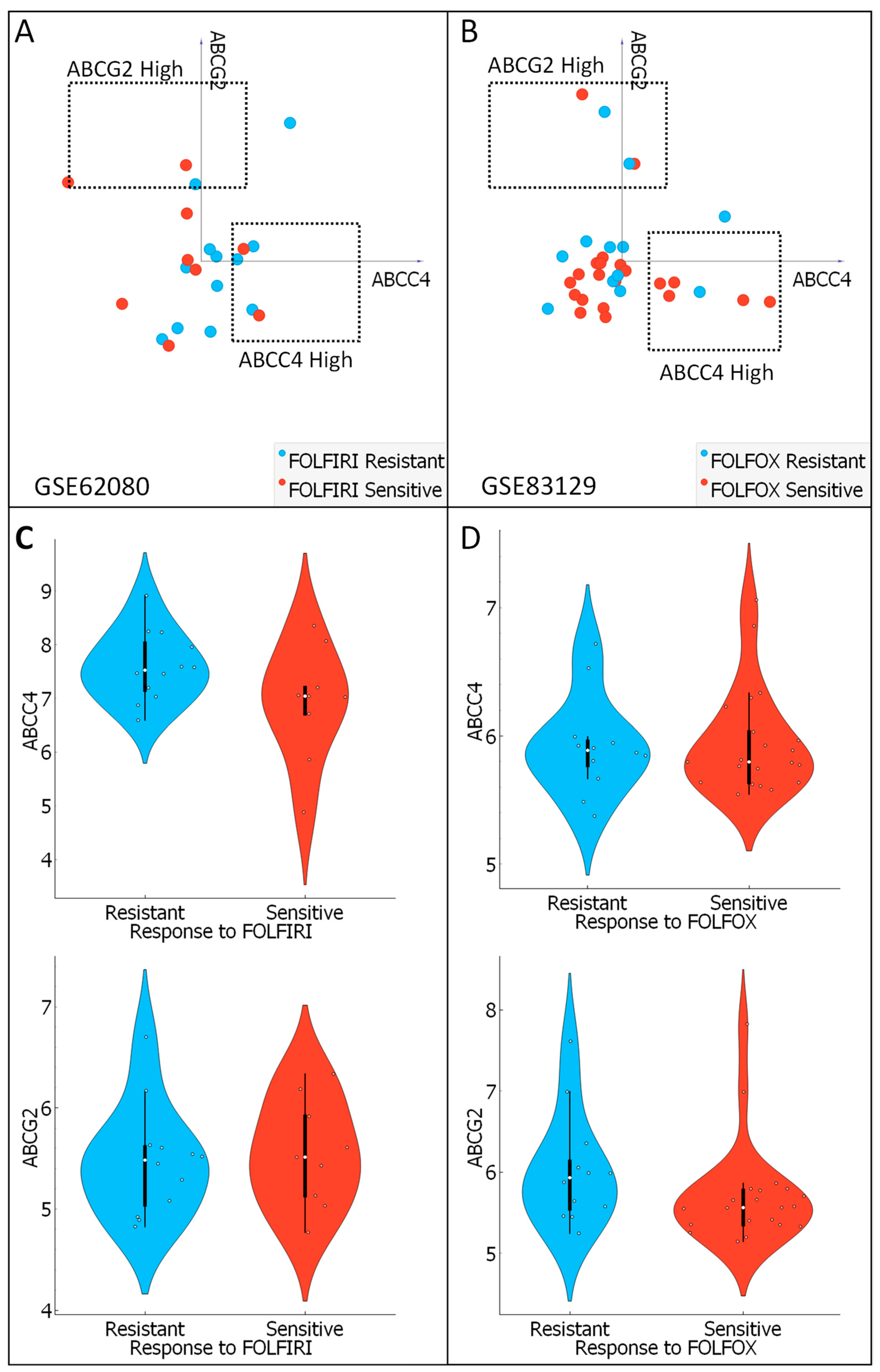
3.7. Analysis of ABCC4 and ABCG2 Potential Chemotherapy Response Predictive Capabilities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henry, J.T.; Johnson, B. Current and evolving biomarkers for precision oncology in the management of metastatic colorectal cancer. Chin. Clin. Oncol. 2019, 8, 49. [Google Scholar] [CrossRef]
- Xie, Y.-H.; Chen, Y.-X.; Fang, J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef]
- Vatandoust, S.; Price, T.J.; Karapetis, C.S. Colorectal cancer: Metastases to a single organ. World J. Gastroenterol. 2015, 21, 11767–11776. [Google Scholar] [CrossRef]
- Baran, B.; Mert Ozupek, N.; Yerli Tetik, N.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol. Res. 2018, 11, 264–273. [Google Scholar] [CrossRef]
- Hsu, H.-H.; Chen, M.-C.; Baskaran, R.; Lin, Y.-M.; Day, C.H.; Lin, Y.-J.; Tu, C.-C.; Vijaya Padma, V.; Kuo, W.-W.; Huang, C.-Y. Oxaliplatin resistance in colorectal cancer cells is mediated via activation of ABCG2 to alleviate ER stress induced apoptosis. J. Cell. Physiol. 2018, 233, 5458–5467. [Google Scholar] [CrossRef] [PubMed]
- Blondy, S.; David, V.; Verdier, M.; Mathonnet, M.; Perraud, A.; Christou, N. 5-Fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacol. Res. 2019, 148, 104398. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.M.; Deeley, R.G.; Cole, S.P.C. Multidrug resistance proteins: Role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol. Appl. Pharmacol. 2005, 204, 216–237. [Google Scholar] [CrossRef] [PubMed]
- Hlavata, I.; Mohelnikova-Duchonova, B.; Vaclavikova, R.; Liska, V.; Pitule, P.; Novak, P.; Bruha, J.; Vycital, O.; Holubec, L.; Treska, V.; et al. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis 2012, 27, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.L.; Palshof, J.A.; Brünner, N.; Stenvang, J.; Viuff, B.M. Implications of ABCG2 Expression on Irinotecan Treatment of Colorectal Cancer Patients: A Review. Int. J. Mol. Sci. 2017, 18, 1926. [Google Scholar] [CrossRef]
- Kryczka, J.; Sochacka, E.; Papiewska-Pająk, I.; Boncela, J. Implications of ABCC4–Mediated cAMP Eflux for CRC Migration. Cancers 2020, 12, 3547. [Google Scholar] [CrossRef]
- Przygodzka, P.; Papiewska-Pajak, I.; Bogusz, H.; Kryczka, J.; Sobierajska, K.; Kowalska, M.A.; Boncela, J. Neuromedin U is upregulated by Snail at early stages of EMT in HT29 colon cancer cells. Biochim. Biophys. Acta 2016, 1860, 2445–2453. [Google Scholar] [CrossRef]
- Fletcher, J.I.; Haber, M.; Henderson, M.J.; Norris, M.D. ABC transporters in cancer: More than just drug efflux pumps. Nat. Rev. Cancer 2010, 10, 147–156. [Google Scholar] [CrossRef]
- Muriithi, W.; Macharia, L.W.; Heming, C.P.; Echevarria, J.L.; Nyachieo, A.; Filho, P.N.; Neto, V.M. ABC transporters and the hallmarks of cancer: Roles in cancer aggressiveness beyond multidrug resistance. Cancer Biol. Med. 2020, 17, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.I.; Williams, R.T.; Henderson, M.J.; Norris, M.D.; Haber, M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 2016, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kryczka, J.; Boncela, J. Integrated Bioinformatics Analysis of the Hub Genes Involved in Irinotecan Resistance in Colorectal Cancer. Biomedicines 2022, 10, 1720. [Google Scholar] [CrossRef] [PubMed]
- Makondi, P.T.; Chu, C.-M.; Wei, P.-L.; Chang, Y.-J. Prediction of novel target genes and pathways involved in irinotecan-resistant colorectal cancer. PLoS ONE 2017, 12, e0180616. [Google Scholar] [CrossRef] [PubMed]
- Chmielewska-Kassassir, M.; Sobierajska, K.; Ciszewski, W.M.; Kryczka, J.; Zieleniak, A.; Wozniak, L.A. Evening Primrose Extract Modulates TYMS Expression via SP1 Transcription Factor in Malignant Pleural Mesothelioma. Cancers 2023, 15, 5003. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Baran, K.; Waśko, J.; Kryczka, J.; Boncela, J.; Jabłoński, S.; Kolesińska, B.; Brzeziańska-Lasota, E.; Kordiak, J. The Comparison of Serum Exosome Protein Profile in Diagnosis of NSCLC Patients. Int. J. Mol. Sci. 2023, 24, 13669. [Google Scholar] [CrossRef] [PubMed]
- Kryczka, J.; Kryczka, J.; Czarnecka-Chrebelska, K.H.; Brzeziańska-Lasota, E. Molecular Mechanisms of Chemoresistance Induced by Cisplatin in NSCLC Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 8885. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Neophytou, C.M.; Panagi, M.; Stylianopoulos, T.; Papageorgis, P. The Role of Tumor Microenvironment in Cancer Metastasis: Molecular Mechanisms and Therapeutic Opportunities. Cancers 2021, 13, 2053. [Google Scholar] [CrossRef]
- Shokati, E.; Safari, E. The immunomodulatory role of exosomal microRNA networks in the crosstalk between tumor-associated myeloid-derived suppressor cells and tumor cells. Int. Immunopharmacol. 2023, 120, 110267. [Google Scholar] [CrossRef]
- Kuwahara, T.; Hazama, S.; Suzuki, N.; Yoshida, S.; Tomochika, S.; Nakagami, Y.; Matsui, H.; Shindo, Y.; Kanekiyo, S.; Tokumitsu, Y.; et al. Intratumoural-infiltrating CD4+ and FOXP3 + T cells as strong positive predictive markers for the prognosis of resectable colorectal cancer. Br. J. Cancer 2019, 121, 659–665. [Google Scholar] [CrossRef]
- Ito, A.; Yamada, N.; Yoshida, Y.; Morino, S.; Yamamoto, O. Myofibroblastic differentiation in atypical fibroxanthomas occurring on sun-exposed skin and in a burn scar: An ultrastructural and immunohistochemical study. J. Cutan. Pathol. 2011, 38, 670–676. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Xu, J.-Y.; Shi, X.-Y.; Huang, W.; Ruan, T.-Y.; Xie, P.; Ding, J.-L. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. J. Tech. Methods Pathol. 2013, 93, 844–854. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Leban, G.; Zupan, B.; Vidmar, G.; Bratko, I. VizRank: Data Visualization Guided by Machine Learning. Data Min. Knowl. Discov. 2006, 13, 119–136. [Google Scholar] [CrossRef]
- Mu, Q.; Luo, G.; Wei, J.; Zheng, L.; Wang, H.; Yu, M.; Xu, N. Apolipoprotein M promotes growth and inhibits apoptosis of colorectal cancer cells through upregulation of ribosomal protein S27a. EXCLI J. 2021, 20, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Qin, W.; Lu, S.; Wang, X.; Zhang, J.; Sun, T.; Hu, X.; Li, Y.; Chen, Q.; Wang, Y.; et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2′-O-methylation via NOP58 recruitment in colorectal cancer. Mol. Cancer 2020, 19, 95. [Google Scholar] [CrossRef]
- Lu, M.; Zessin, A.S.; Glover, W.; Hsu, D.S. Activation of the mTOR Pathway by Oxaliplatin in the Treatment of Colorectal Cancer Liver Metastasis. PLoS ONE 2017, 12, e0169439. [Google Scholar] [CrossRef]
- Ooi, Z.S.; Pang, S.W.; Teow, S.Y. RAS and BRAF genes as biomarkers and target for personalised colorectal cancer therapy: An update. Malays. J. Pathol. 2022, 44, 415–428. [Google Scholar] [PubMed]
- Ooki, A.; Shinozaki, E.; Yamaguchi, K. Immunotherapy in Colorectal Cancer: Current and Future Strategies. J. Anus Rectum Colon 2021, 5, 11–24. [Google Scholar] [CrossRef]
- Giddings, E.L.; Champagne, D.P.; Wu, M.-H.; Laffin, J.M.; Thornton, T.M.; Valenca-Pereira, F.; Culp-Hill, R.; Fortner, K.A.; Romero, N.; East, J.; et al. Mitochondrial ATP fuels ABC transporter-mediated drug efflux in cancer chemoresistance. Nat. Commun. 2021, 12, 2804. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, R.; Scartozzi, M.; Loretelli, C.; Piva, F.; Mandolesi, A.; Lezoche, G.; Prete, M.D.; Bittoni, A.; Faloppi, L.; Bianconi, M.; et al. Cancer Stem Cell Gene Profile as Predictor of Relapse in High Risk Stage II and Stage III, Radically Resected Colon Cancer Patients. PLoS ONE 2013, 8, e72843. [Google Scholar] [CrossRef]
- Ween, M.P.; Armstrong, M.A.; Oehler, M.K.; Ricciardelli, C. The role of ABC transporters in ovarian cancer progression and chemoresistance. Crit. Rev. Oncol. Hematol. 2015, 96, 220–256. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef]
- Saxena, M.; Stephens, M.A.; Pathak, H.; Rangarajan, A. Transcription factors that mediate epithelial–mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011, 2, e179. [Google Scholar] [CrossRef]
- Shi, X.; Wei, K.; Wu, Y.; Mao, L.; Pei, W.; Zhu, H.; Shi, Y.; Zhang, S.; Tao, S.; Wang, J.; et al. Exosome-derived miR-372-5p promotes stemness and metastatic ability of CRC cells by inducing macrophage polarization. Cell. Signal. 2023, 111, 110884. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Zhang, P.; Wang, L.; He, X.; Yang, S.; Lu, H. RBBP7 is a prognostic biomarker in patients with esophageal squamous cell carcinoma. Oncol. Lett. 2018, 16, 7204–7211. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zou, R.; Gu, Q.; Wang, X.; Zhang, J.; Ma, R.; Wang, T.; Wu, J.; Feng, J.; Zhang, Y. CRNDE mediated hnRNPA2B1 stability facilitates nuclear export and translation of KRAS in colorectal cancer. Cell Death Dis. 2023, 14, 611. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol.J Hematol Oncol 2020, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Anczuków, O.; Rosenberg, A.Z.; Akerman, M.; Das, S.; Zhan, L.; Karni, R.; Muthuswamy, S.K.; Krainer, A.R. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 2012, 19, 220–228. [Google Scholar] [CrossRef]
- Guo, P.; Zu, S.; Han, S.; Yu, W.; Xue, G.; Lu, X.; Lin, H.; Zhao, X.; Lu, H.; Hua, C.; et al. BPTF inhibition antagonizes colorectal cancer progression by transcriptionally inactivating Cdc25A. Redox Biol. 2022, 55, 102418. [Google Scholar] [CrossRef]
- Wang, J.; Huang, R.; Huang, Y.; Chen, Y.; Chen, F. Overexpression of NOP58 as a Prognostic Marker in Hepatocellular Carcinoma: A TCGA Data-Based Analysis. Adv. Ther. 2021, 38, 3342–3361. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Qin, W.; Sun, T.; Lu, S.; Li, Y.; Wang, Y.; Hu, X.; Xu, D.; Wu, Y.; et al. Long non-coding RNA ZFAS1 promotes colorectal cancer tumorigenesis and development through DDX21-POLR1B regulatory axis. Aging 2020, 12, 22656–22687. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J.; Fiechter, C.; Burton, J.; Hallion, J.; Paas, M.; Patel, A.; Patel, A.; Rochet, A.; Scheurlen, K.; Gardner, S.; et al. Long non-coding RNA ZFAS1 is a major regulator of epithelial-mesenchymal transition through miR-200/ZEB1/E-cadherin, vimentin signaling in colon adenocarcinoma. Cell Death Discov. 2021, 7, 61. [Google Scholar] [CrossRef]
- Wang, R.; Huang, Z.; Lin, Z.; Chen, B.; Pang, X.; Du, C.; Fan, H. Hypoxia-induced RBBP7 promotes esophagus cancer progression by inducing CDK4 expression. Acta Biochim. Biophys. Sin. 2022, 54, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, J.; Xu, M.; Zheng, Y.; Wang, M.; Yang, S.; Qin, B.; Li, S.; Dong, L.; Dai, F. DDX3 acts as a tumor suppressor in colorectal cancer as loss of DDX3 in advanced cancer promotes tumor progression by activating the MAPK pathway. Int. J. Biol. Sci. 2022, 18, 3918–3933. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, Y.; Zhang, J.; Jin, L.; Cao, P. A feedback loop between GATA2-AS1 and GATA2 promotes colorectal cancer cell proliferation, invasion, epithelial-mesenchymal transition and stemness via recruiting DDX3X. J. Transl. Med. 2022, 20, 287. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-T.; Huang, C.-S.; Yao, C.-T.; Su, S.-L.; Terng, H.-J.; Chou, H.-L.; Chou, Y.-C.; Chen, K.-H.; Shih, Y.-W.; Lu, C.-Y.; et al. Gene expression profile of peripheral blood in colorectal cancer. World J. Gastroenterol. 2014, 20, 14463–14471. [Google Scholar] [CrossRef]
- Lin, X.-H.; Li, D.-P.; Liu, Z.-Y.; Zhang, S.; Tang, W.-Q.; Chen, R.-X.; Weng, S.-Q.; Tseng, Y.-J.; Xue, R.-Y.; Dong, L. Six immune-related promising biomarkers may promote hepatocellular carcinoma prognosis: A bioinformatics analysis and experimental validation. Cancer Cell Int. 2023, 23, 52. [Google Scholar] [CrossRef]
- Liu, W.; Xu, Z.; Zhou, J.; Xing, S.; Li, Z.; Gao, X.; Feng, S.; Xiao, Y. High Levels of HIST1H2BK in Low-Grade Glioma Predicts Poor Prognosis: A Study Using CGGA and TCGA Data. Front. Oncol. 2020, 10, 627. [Google Scholar] [CrossRef]
- Kim, J.C.; Ha, Y.J.; Park, I.J.; Kim, C.W.; Yoon, Y.S.; Lee, J.L.; Tak, K.H.; Cho, D.-H.; Park, S.H.; Kim, S.-K.; et al. Tumor immune microenvironment of primary colorectal adenocarcinomas metastasizing to the liver or lungs. J. Surg. Oncol. 2021, 124, 1136–1145. [Google Scholar] [CrossRef]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.G.; Lannon, W.A.; Grotzinger, C.; Del Rio, M.; et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef]
- Ju, H.; Hu, Z.; Lu, Y.; Wu, Y.; Zhang, L.; Wei, D.; Guo, W.; Xia, W.; Liu, S.; Ren, G.; et al. TLR4 activation leads to anti-EGFR therapy resistance in head and neck squamous cell carcinoma. Am. J. Cancer Res. 2020, 10, 454–472. [Google Scholar] [PubMed]
- Lu, K.V.; Zhu, S.; Cvrljevic, A.; Huang, T.T.; Sarkaria, S.; Ahkavan, D.; Dang, J.; Dinca, E.B.; Plaisier, S.B.; Oderberg, I.; et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009, 69, 6889–6898. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. A novel strategy for the study on molecular mechanism of prostate injury induced by 4,4′-sulfonyldiphenol based on network toxicology analysis. J. Appl. Toxicol. JAT 2023, 1–13. [Google Scholar] [CrossRef]
- Ye, B.; Chen, P.; Lin, C.; Zhang, C.; Li, L. Study on the material basis and action mechanisms of sophora davidii (Franch.) skeels flower extract in the treatment of non-small cell lung cancer. J. Ethnopharmacol. 2023, 317, 116815. [Google Scholar] [CrossRef] [PubMed]
| Infiltrating Cells | Correlation Rho | p | |
|---|---|---|---|
| ABCC4 H | |||
| CAFs | 0.188 | 1.72 × 10−3 | |
| Neutrophils | 0.226 | 8.01 × 10−6 | |
| NK | −0.181 | 2.54 × 10−3 | |
| Macrophage | 0.326 | 3.26 × 10−8 | |
| Macrophage M1 | 0.358 | 9.51 × 10−10 | |
| Macrophage M2 | 0.304 | 2.86 × 10−7 | |
| CD8+ T-cell effector memory | −0.119 | 4,87 × 10−2 | |
| CD8+ T-cell-naive | −0.143 | 1.75 × 10−2 | |
| CD4+ T cell Th1 | −0.207 | 5.41 × 10−4 | |
| CD4+ central memory | −0.127 | 3.50 × 10−2 | |
| ABCG2 H | |||
| CAFs | 0.182 | 2.48 × 10−3 | |
| Neutrophils | 0.119 | 4.89 × 10−2 | |
| Class-switched memory B cells | −0.228 | 1.35 × 10−4 | |
| CD8+ T-cell central memory | −0.14 | 2.04 × 10−2 | |
| CD4+ T-cell effector memory | −0.153 | 1.12 × 10−2 | |
| CD4+ T cell Th2 | −0.143 | 1.75 × 10−2 | |
| CD4+ T cell non-regulatory | −0.138 | 2.20 × 10−2 |
| Gene | Protein | Number of Edges | |
|---|---|---|---|
| ABCC4 H | |||
| RPS27A | ribosomal protein S27a | 87 | |
| SRSF1 | Serine/arginine-rich splicing factor 1 | 56 | |
| DDX3X | DEAD-box helicase family member | 50 | |
| BPTF | bromodomain PHD finger transcription factor | 44 | |
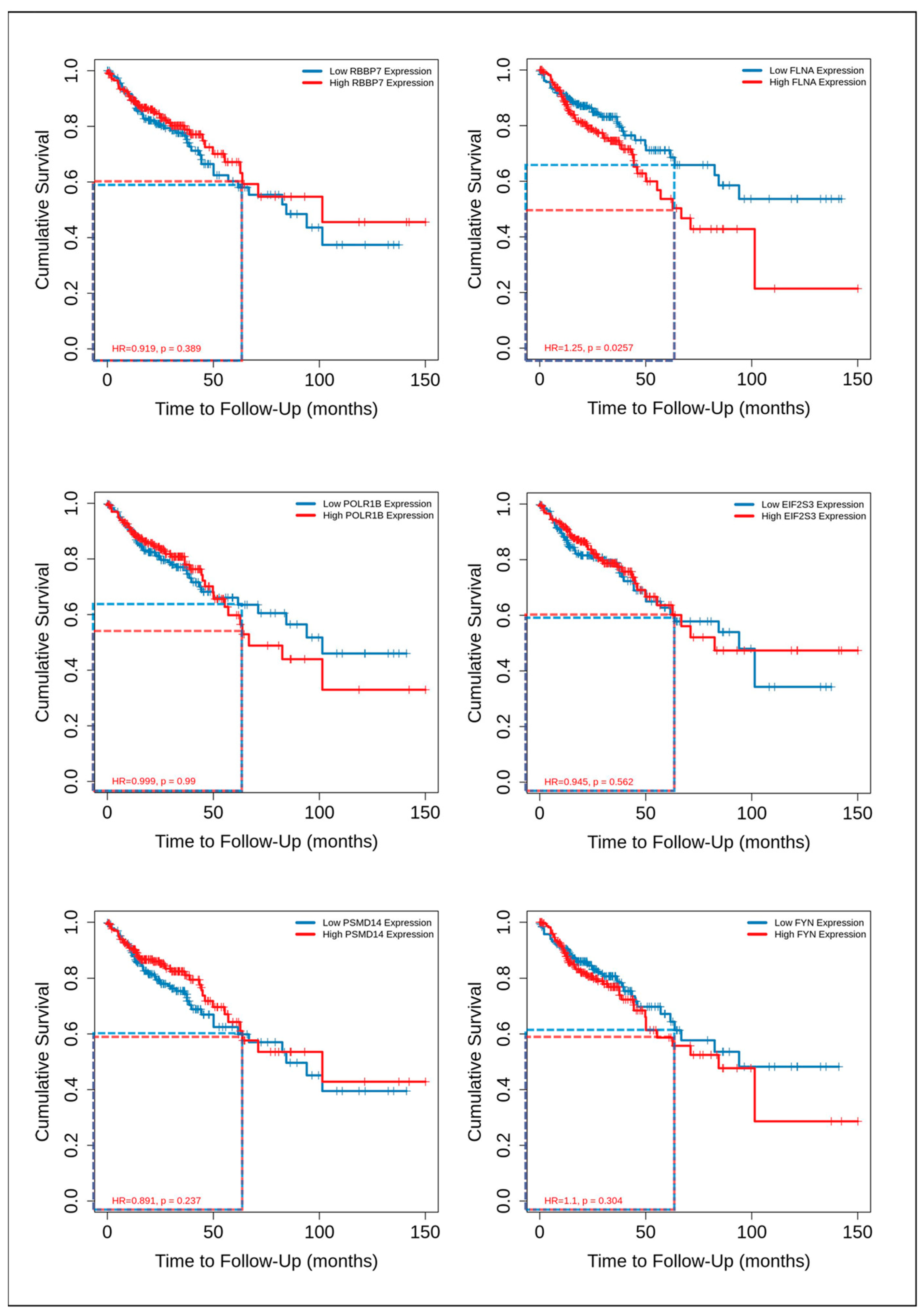
| RBBP7 | RB Binding Protein 7, Chromatin Remodeling Factor | 44 | |
| POLR1B | RNA Polymerase I Subunit B | 44 | |
| HNRNPA2B1 | heterogeneous nuclear ribonucleoprotein A2/B1 | 43 | |
| PSMD14 | proteasome 26S subunit, non-ATPase 14 | 42 | |
| NOP58 | ribonucleoprotein | 42 | |
| EIF2S3 | eukaryotic translation initiation factor 2 subunit gamma | 41 | |
| ABCG2 H | |||
| MAPK3 | mitogen-activated protein kinase 3 | 53 | |
| HIST2H2BE | histone cluster 2 H2B family member E (H2B clustered histone 21) | 34 | |
| LMNA | Lamin A/C | 29 | |
| HIST1H2BD | histone cluster 1 H2B family member D (H2B clustered histone 5) | 29 | |
| HIST1H2BK | histone cluster 1 H2B family member K (H2B clustered histone 12) | 29 | |
| HIST1H2AC | histone cluster 1 H2A family member C (H2A clustered histone 6) | 29 | |
| FYN | FYN proto-oncogene, Src family tyrosine kinase | 28 | |
| TLR4 | toll-like receptor 4 | 28 | |
| FLNA | filamin A | 28 | |
| HIST1H2AJ | histone cluster 1 H2B family member J (H2A clustered histone 14) | 27 |
| KEGG ID | Description | Strength | False Discovery Rate | |
|---|---|---|---|---|
| ABCC4 H | ||||
| hsa03050 | Proteasome | 2.14 | 2.26 × 10−9 | |
| hsa05012 | Parkinson’s disease | 1.46 | 5.28 × 10−7 | |
| hsa05017 | Spinocerebellar ataxia | 1.64 | 5.28 × 10−7 | |
| hsa05014 | Amyotrophic lateral sclerosis | 1.29 | 3.48 × 10−6 | |
| hsa05020 | Prion disease | 1.35 | 1.47 × 10−5 | |
| hsa05016 | Huntington’s disease | 1.29 | 2.41 × 10−5 | |
| hsa05010 | Alzheimer’s disease | 1.22 | 5.63 × 10−5 | |
| hsa05169 | Epstein–Barr virus infection | 1.31 | 0.0018 | |
| hsa03020 | RNA polymerase | 1.8 | 0.0192 | |
| ABCG2 H | ||||
| hsa05034 | Alcoholism | 1.74 | 2.90 × 10−7 | |
| hsa05133 | Pertussis | 1.95 | 4.60 × 10−7 | |
| hsa05322 | Systemic lupus erythematosus | 1.85 | 9.16 × 10−7 | |
| hsa04620 | Toll-like receptor signalling pathway | 1.81 | 1.02 × 10−6 | |
| hsa05132 | Salmonella infection | 1.49 | 2.75 × 10−5 | |
| hsa04064 | NF-kappa B signalling pathway | 1.71 | 5.70 × 10−5 | |
| hsa04217 | Necroptosis | 1.54 | 0.00022 | |
| hsa05203 | Viral carcinogenesis | 1.46 | 0.00042 | |
| hsa05205 | Proteoglycans in cancer | 1.43 | 0.00049 | |
| hsa05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1.65 | 0.0014 | |
| hsa05142 | Chagas disease | 1.6 | 0.0018 | |
| hsa05145 | Toxoplasmosis | 1.57 | 0.0020 | |
| hsa05135 | Yersinia infection | 1.5 | 0.0030 | |
| hsa05161 | Hepatitis B | 1.39 | 0.0056 | |
| hsa05152 | Tuberculosis | 1.37 | 0.0059 | |
| hsa05164 | Influenza A | 1.38 | 0.0059 | |
| hsa04621 | NOD-like receptor signalling pathway | 1.35 | 0.0060 | |
| hsa05130 | Pathogenic Escherichia coli infection | 1.32 | 0.0070 | |
| hsa04510 | Focal adhesion | 1.3 | 0.0078 | |
| hsa05131 | Shigellosis | 1.25 | 0.0098 | |
| hsa05134 | Legionellosis | 1.68 | 0.0137 | |
| hsa04010 | MAPK signalling pathway | 1.13 | 0.0185 | |
| hsa04520 | Adherens junction | 1.59 | 0.0185 | |
| hsa04664 | Fc epsilon RI signalling pathway | 1.6 | 0.0185 | |
| hsa05140 | Leishmaniasis | 1.57 | 0.0185 | |
| hsa05221 | Acute myeloid leukemia | 1.6 | 0.0185 | |
| hsa05220 | Chronic myeloid leukemia | 1.54 | 0.0193 | |
| hsa04012 | ErbB signalling pathway | 1.5 | 0.0226 | |
| hsa04660 | T-cell-receptor signalling pathway | 1.41 | 0.0313 | |
| hsa05146 | Amoebiasis | 1.42 | 0.0313 | |
| hsa04066 | HIF-1 signalling pathway | 1.39 | 0.0327 | |
| hsa04725 | Cholinergic synapse | 1.38 | 0.0341 | |
| hsa04071 | Sphingolipid signalling pathway | 1.35 | 0.0366 | |
| hsa04380 | Osteoclast differentiation | 1.33 | 0.0385 | |
| hsa04611 | Platelet activation | 1.33 | 0.0385 | |
| hsa04650 | Natural-killer-cell-mediated cytotoxicity | 1.33 | 0.0385 | |
| hsa04210 | Apoptosis | 1.3 | 0.0418 | |
| hsa04910 | Insulin signalling pathway | 1.29 | 0.0418 | |
| hsa04145 | Phagosome | 1.26 | 0.0456 | |
| hsa04072 | Phospholipase D signalling pathway | 1.25 | 0.0475 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryczka, J.; Boncela, J. Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC—A New Opportunity to Predict Therapy Response. Cancers 2023, 15, 5623. https://doi.org/10.3390/cancers15235623
Kryczka J, Boncela J. Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC—A New Opportunity to Predict Therapy Response. Cancers. 2023; 15(23):5623. https://doi.org/10.3390/cancers15235623
Chicago/Turabian StyleKryczka, Jakub, and Joanna Boncela. 2023. "Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC—A New Opportunity to Predict Therapy Response" Cancers 15, no. 23: 5623. https://doi.org/10.3390/cancers15235623
APA StyleKryczka, J., & Boncela, J. (2023). Characteristics of ABCC4 and ABCG2 High Expression Subpopulations in CRC—A New Opportunity to Predict Therapy Response. Cancers, 15(23), 5623. https://doi.org/10.3390/cancers15235623