Spatial Distribution of Non-Immune Cells Expressing Glycoprotein A Repetitions Predominant in Human and Murine Metastatic Lymph Nodes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Single-Cell RNA Sequencing of Human LN Cells
2.2. Single-Cell RNA Sequencing of Mouse LN Cells
2.3. In Vitro Cell Culture
2.4. Western Blot
2.5. Flow Cytometry
2.6. Multiplexed Immunofluorescence on Human LN Sections
2.7. Ear Sponge Assay
2.8. In Situ RNA Hybridization
2.9. Slide Scanning and Image Analysis with Olyvia and QuPath
2.10. Statistics
2.11. Study Approval
3. Results
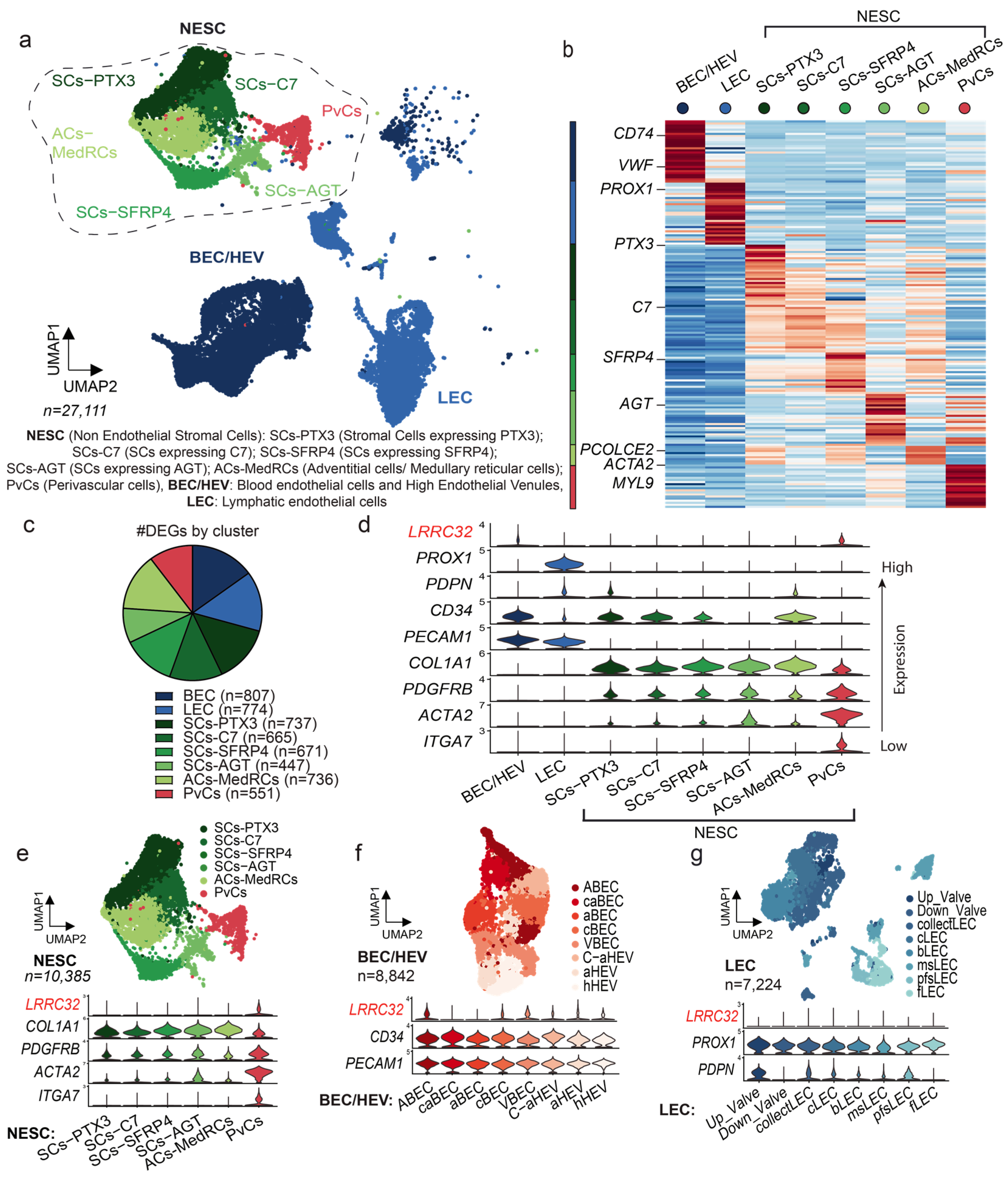
3.1. Single-Cell RNA Sequencing Analysis of Human LNs Uncovers the LRRC32 Gene Expression by Subpopulations of Endothelial and Perivascular Cells in Human LNs
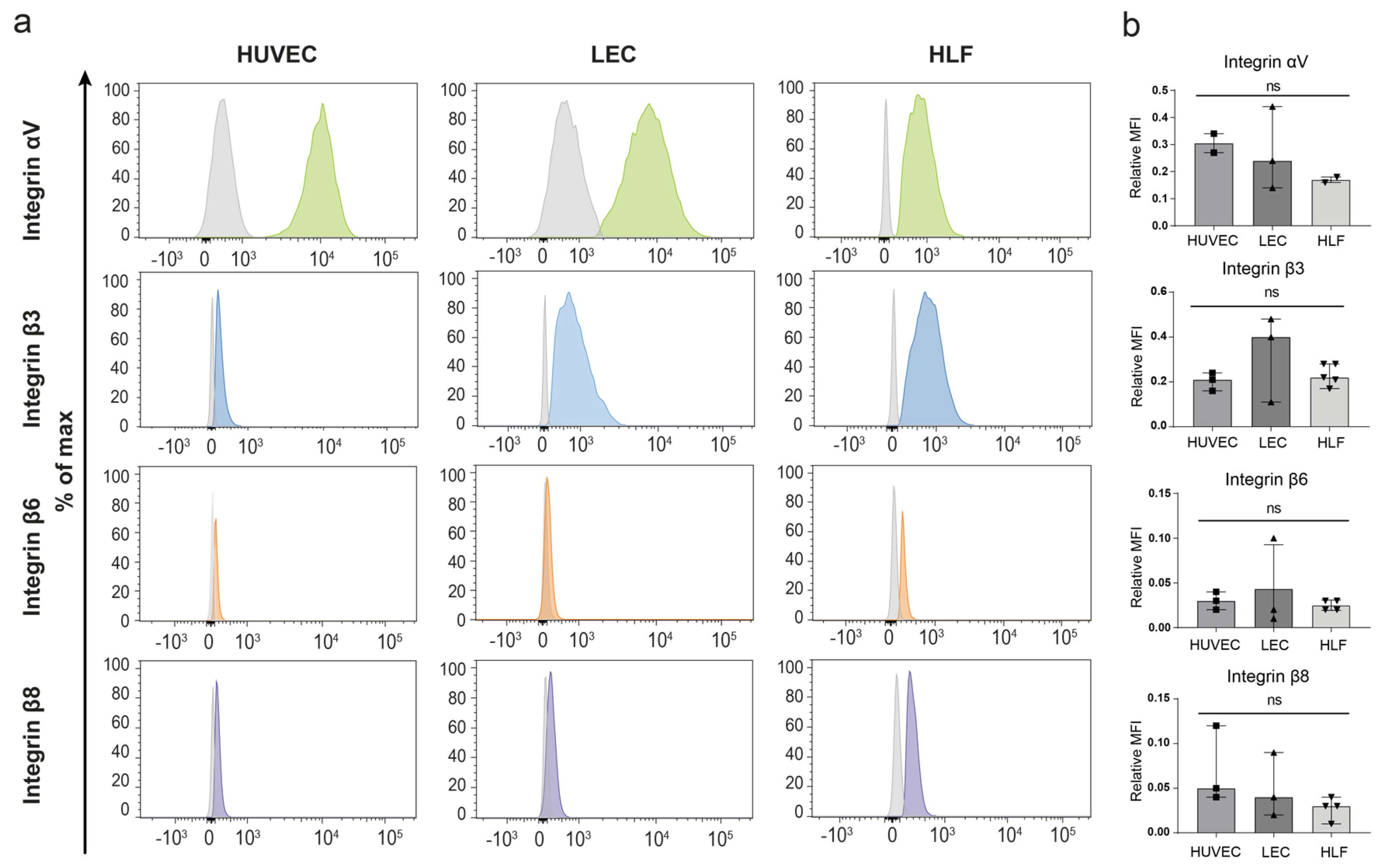
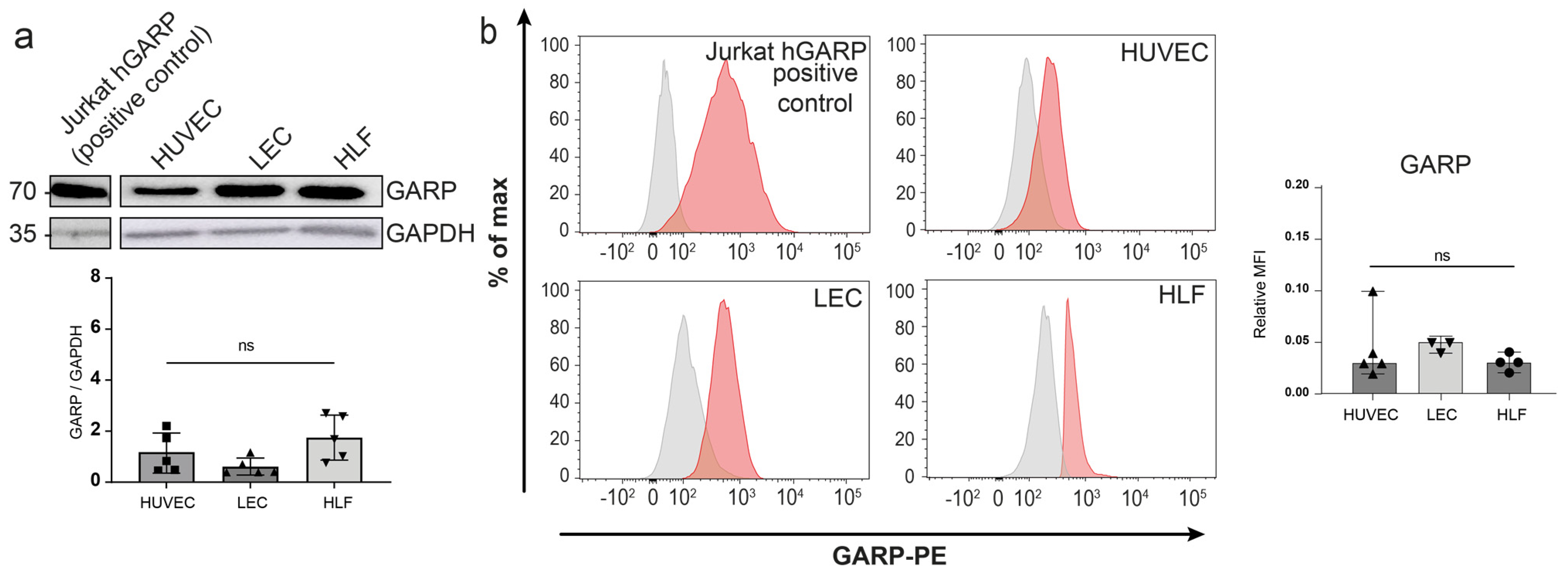
3.2. Expression of GARP and Integrins Are Produced In Vitro by Human Endothelial Cells and LN Fibroblasts
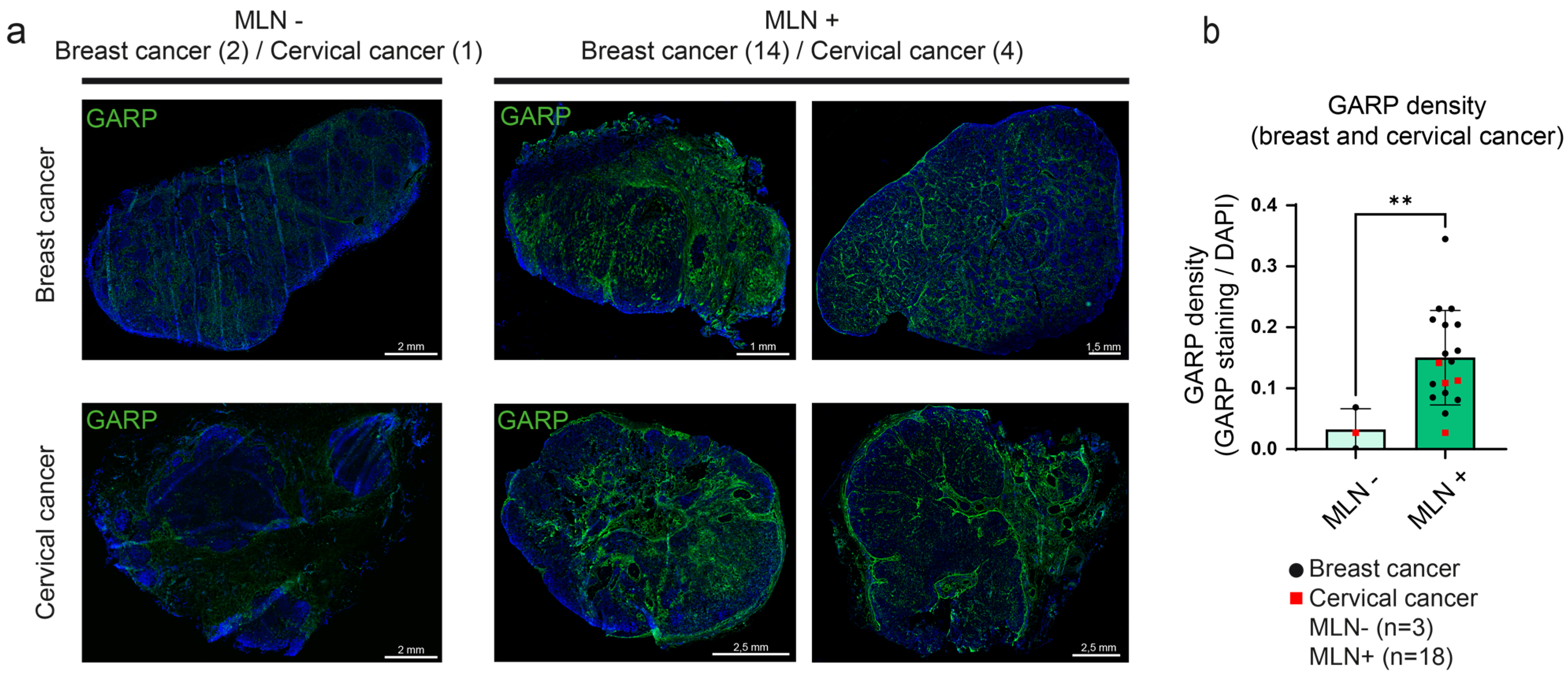
3.3. Detection and Mapping of GARP in Human Metastatic LN Samples
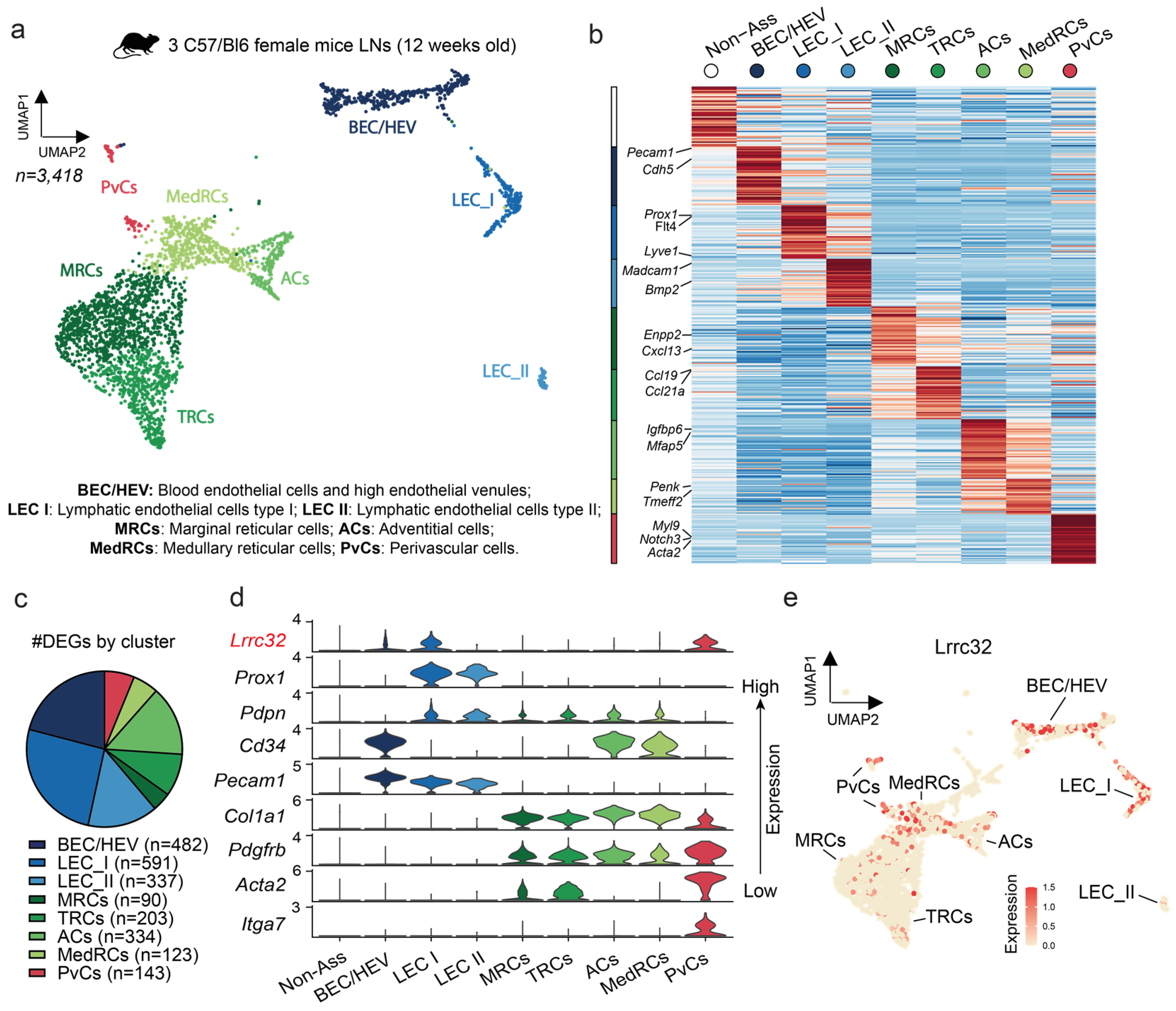
3.4. Single-Cell RNAseq Analysis Uncovers the Lrrc32 Gene Expression in Endothelial and Peri-Vascular Cells of Murine LNs
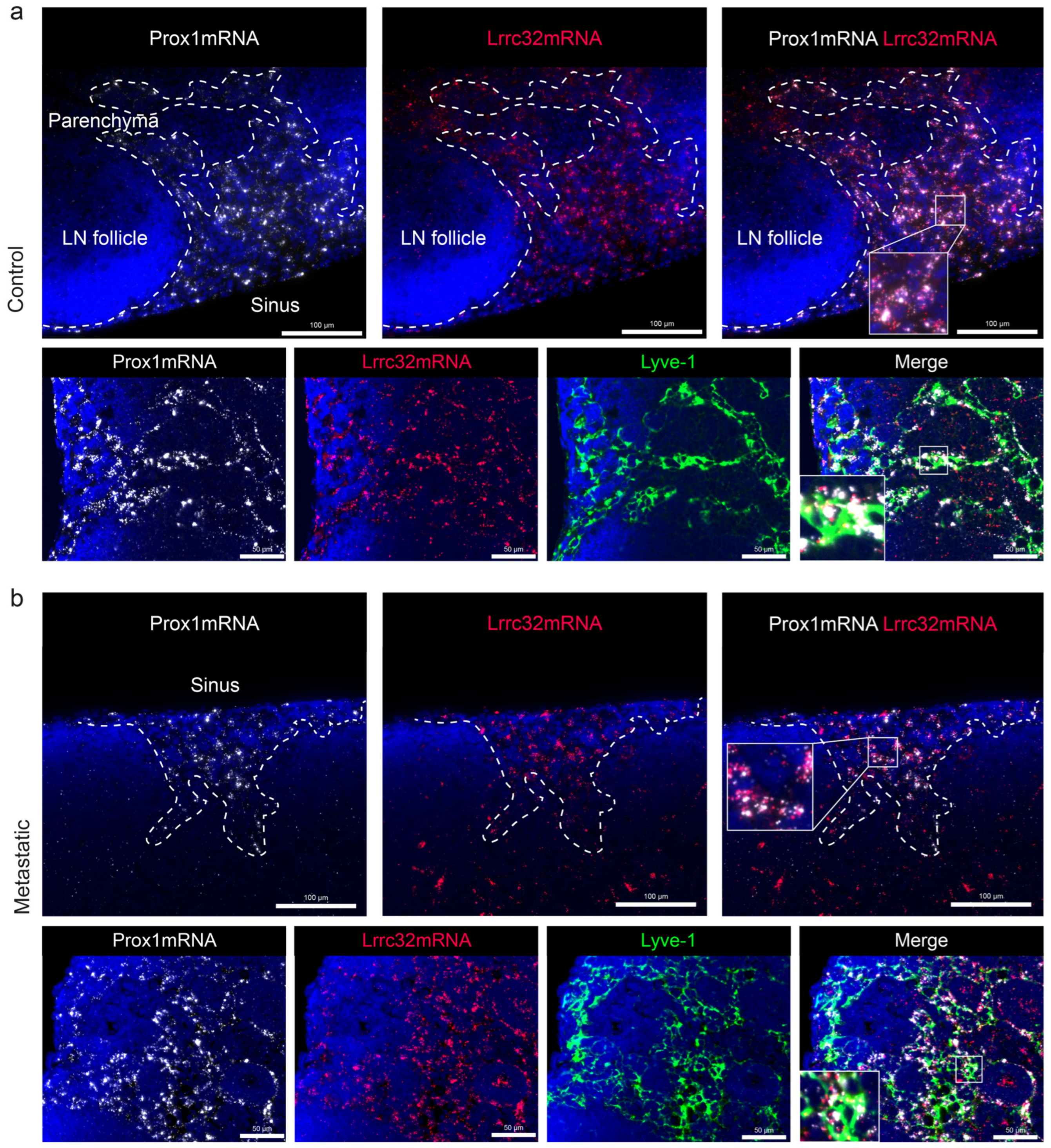
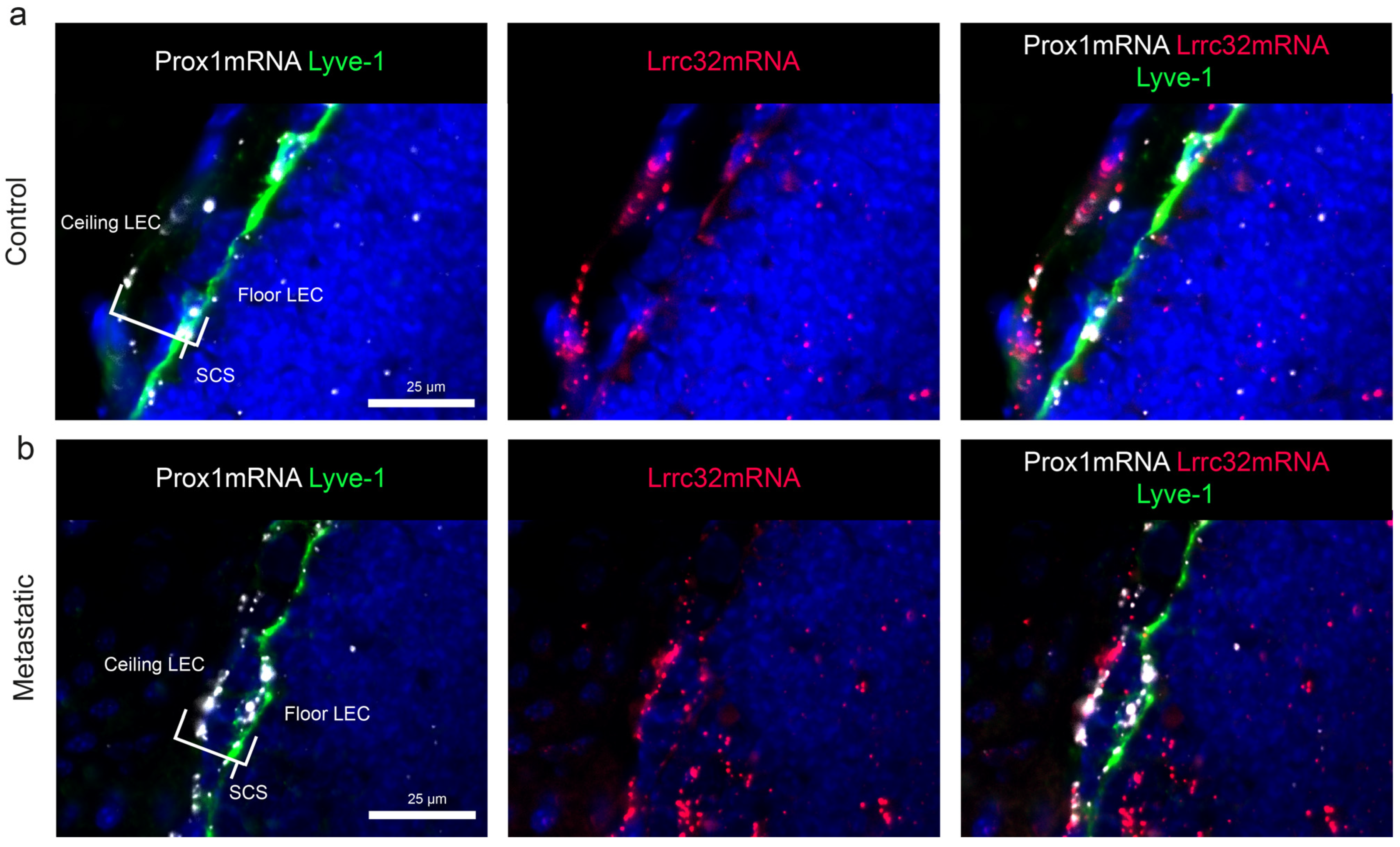
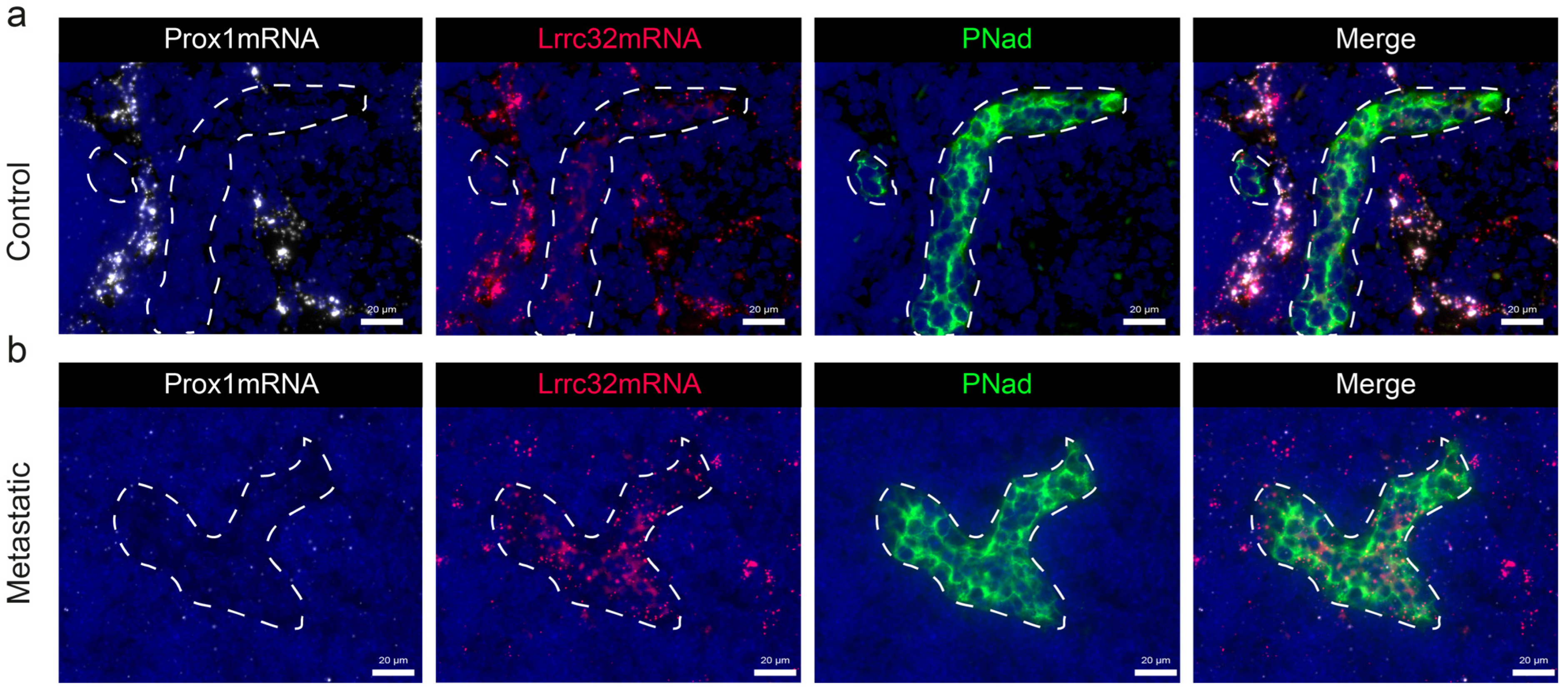
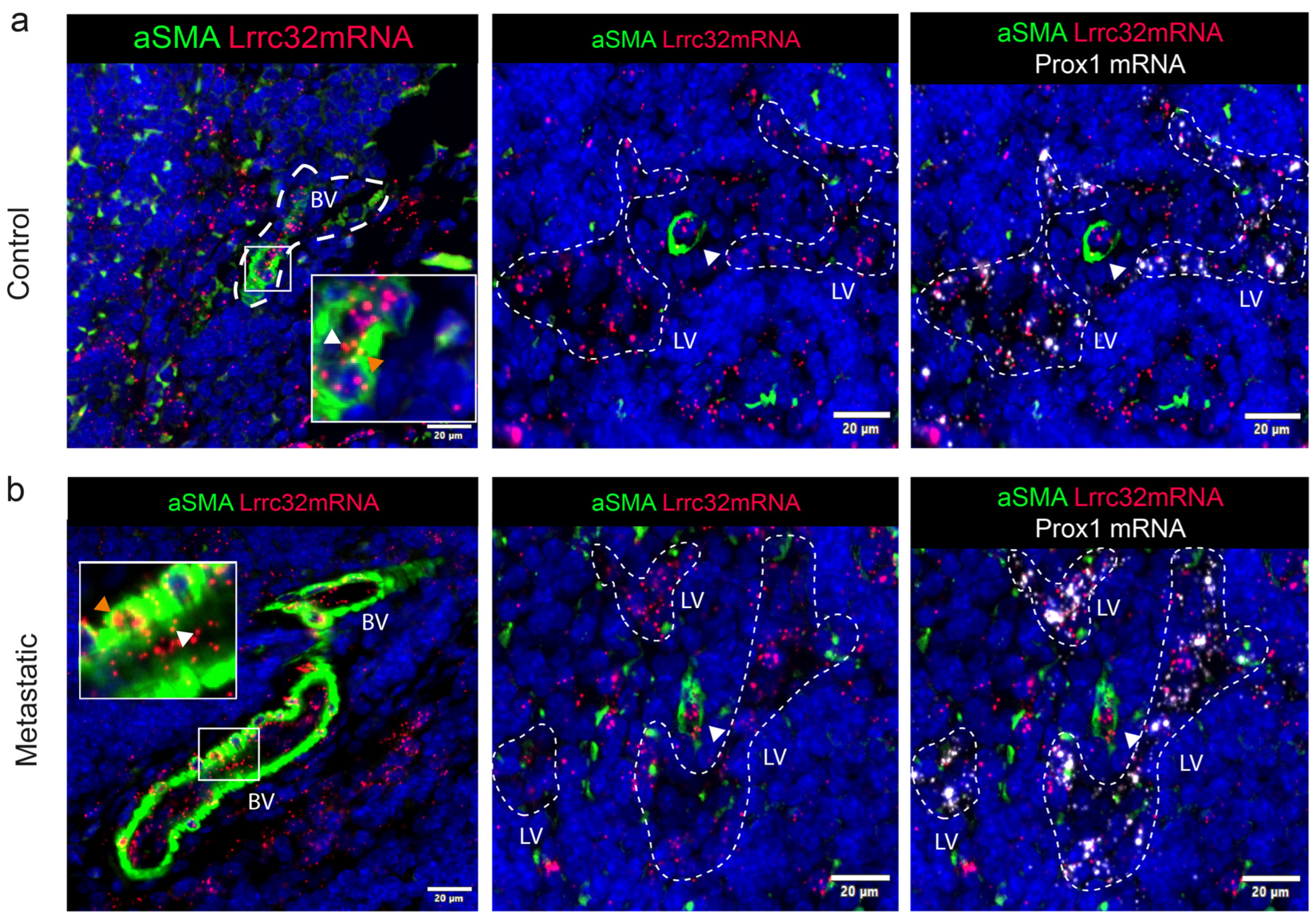
3.5. Mapping of Lrrc32 mRNA Expression in Mouse LNs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Chatterjee, G.; Pai, T.; Hardiman, T.; Avery-Kiejda, K.; Scott, R.J.; Spencer, J.; Pinder, S.E.; Grigoriadis, A. Molecular Patterns of Cancer Colonisation in Lymph Nodes of Breast Cancer Patients. Breast Cancer Res. 2018, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Balsat, C.; Blacher, S.; Herfs, M.; Van de Velde, M.; Signolle, N.; Sauthier, P.; Pottier, C.; Gofflot, S.; De Cuypere, M.; Delvenne, P.; et al. A Specific Immune and Lymphatic Profile Characterizes the Pre-Metastatic State of the Sentinel Lymph Node in Patients with Early Cervical Cancer. Oncoimmunology 2017, 6, e1265718. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka, N.; Hasegawa, Y.; Yoshimoto, S.; Miura, K.; Shiotani, A.; Yokoyama, J.; Sugasawa, M.; Moriyama-Kita, M.; Endo, K.; Yoshizaki, T. Primary Tumor-Secreted Lymphangiogenic Factors Induce Pre-Metastatic Lymphvascular Niche Formation at Sentinel Lymph Nodes in Oral Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0144056. [Google Scholar] [CrossRef] [PubMed]
- Tammela, T.; Alitalo, K. Lymphangiogenesis: Molecular Mechanisms and Future Promise. Cell 2010, 140, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Maus, R.L.G.; Jakub, J.W.; Hieken, T.J.; Nevala, W.K.; Christensen, T.A.; Sutor, S.L.; Flotte, T.J.; Markovic, S.N. Identification of Novel, Immune-Mediating Extracellular Vesicles in Human Lymphatic Effluent Draining Primary Cutaneous Melanoma. OncoImmunology 2019, 8, e1667742. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.K.; Hyun, S.H.; Choi, N.; Kim, M.J.; Padera, T.P.; Choi, J.Y.; Jeong, H.S. Significance of Lymph Node Metastasis in Cancer Dissemination of Head and Neck Cancer. Transl. Oncol. 2015, 8, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Stacker, S.A.; Williams, S.P.; Karnezis, T.; Shayan, R.; Fox, S.B.; Achen, M.G. Lymphangiogenesis and Lymphatic Vessel Remodelling in Cancer. Nat. Rev. Cancer 2014, 14, 159–172. [Google Scholar] [CrossRef]
- Brown, M.; Assen, F.P.; Leithner, A.; Abe, J.; Schachner, H.; Asfour, G.; Bago-Horvath, Z.; Stein, J.V.; Uhrin, P.; Sixt, M.; et al. Lymph Node Blood Vessels Provide Exit Routes for Metastatic Tumor Cell Dissemination in Mice. Science 2018, 359, 1408–1411. [Google Scholar] [CrossRef]
- Padera, T.P.; Meijer, E.F.J.; Munn, L.L. The Lymphatic System in Disease Processes and Cancer Progression. Annu. Rev. Biomed. Eng. 2016, 18, 125–158. [Google Scholar] [CrossRef]
- Sleeman, J.P. The Lymph Node Pre-Metastatic Niche. J. Mol. Med. 2015, 93, 1173–1184. [Google Scholar] [CrossRef]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-Positive Haematopoietic Bone Marrow Progenitors Initiate the Pre-Metastatic Niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-Metastatic Niches: Organ-Specific Homes for Metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Psaila, B.; Lyden, D. The Metastatic Niche: Adapting the Foreign Soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, S.; Kodama, S.; Kunstfeld, R.; Kajiya, K.; Brown, L.F.; Detmar, M. VEGF-A Induces Tumor and Sentinel Lymph Node Lymphangiogenesis and Promotes Lymphatic Metastasis. J. Exp. Med. 2005, 201, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, S.; Brown, L.F.; Kodama, S.; Paavonen, K.; Alitalo, K.; Detmar, M. VEGF-C–Induced Lymphangiogenesis in Sentinel Lymph Nodes Promotes Tumor Metastasis to Distant Sites. Blood 2007, 109, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Gillot, L.; Lebeau, A.; Baudin, L.; Pottier, C.; Louis, T.; Durré, T.; Longuespée, R.; Mazzucchelli, G.; Nizet, C.; Blacher, S.; et al. Periostin in Lymph Node Pre-Metastatic Niches Governs Lymphatic Endothelial Cell Functions and Metastatic Colonization. Cell. Mol. Life Sci. 2022, 79, 295. [Google Scholar] [CrossRef] [PubMed]
- De Streel, G.; Lucas, S. Targeting Immunosuppression by TGF-Β1 for Cancer Immunotherapy. Biochem. Pharmacol. 2021, 192, 114697. [Google Scholar] [CrossRef]
- Travis, M.A.; Sheppard, D. TGF-β Activation and Function in Immunity. Annu. Rev. Immunol. 2014, 32, 51–82. [Google Scholar] [CrossRef]
- Liénart, S.; Merceron, R.; Vanderaa, C.; Lambert, F.; Colau, D.; Stockis, J.; Van Der Woning, B.; De Haard, H.; Saunders, M.; Coulie, P.G.; et al. Structural Basis of Latent TGF-Β1 Presentation and Activation by GARP on Human Regulatory T Cells. Science 2018, 362, 952–956. [Google Scholar] [CrossRef]
- Cuende, J.; Liénart, S.; Dedobbeleer, O.; Van Der Woning, B.; De Boeck, G.; Stockis, J.; Huygens, C.; Colau, D.; Somja, J.; Delvenne, P.; et al. Monoclonal Antibodies against GARP/TGF-Β1 Complexes Inhibit the Immunosuppressive Activity of Human Regulatory T Cells In Vivo. Sci. Transl. Med. 2015, 7, 284ra56. [Google Scholar] [CrossRef]
- Zimmer, N.; Trzeciak, E.R.; Graefen, B.; Satoh, K.; Tuettenberg, A. GARP as a Therapeutic Target for the Modulation of Regulatory T Cells in Cancer and Autoimmunity. Front. Immunol. 2022, 13, 928450. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.Q.; Andersson, J.; Wang, R.; Ramsey, H.; Unutmaz, D.; Shevach, E.M. GARP (LRRC32) Is Essential for the Surface Expression of Latent TGF-Beta on Platelets and Activated FOXP3+ Regulatory T Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 13445–13450. [Google Scholar] [CrossRef] [PubMed]
- Dedobbeleer, O.; Stockis, J.; Van Der Woning, B.; Coulie, P.G.; Lucas, S. Cutting Edge: Active TGF-Β1 Released from GARP/TGF-Β1 Complexes on the Surface of Stimulated Human B Lymphocytes Increases Class-Switch Recombination and Production of IgA. J. Immunol. 2017, 199, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sharma, P.; Maschmeyer, P.; Hu, Y.; Lou, M.; Kim, J.; Fujii, H.; Unutmaz, D.; Schwabe, R.F.; Winau, F. GARP on Hepatic Stellate Cells Is Essential for the Development of Liver Fibrosis. J. Hepatol. 2023, 79, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, C.; Van Meerbeeck, P.; De Streel, G.; Vaherto-Bleeckx, N.; Benhaddi, F.; Rouaud, L.; Noël, A.; Coulie, P.G.; Van Baren, N.; Lucas, S. Combined Blockade of GARP:TGF-Β1 and PD-1 Increases Infiltration of T Cells and Density of Pericyte-Covered GARP+ Blood Vessels in Mouse MC38 Tumors. Front. Immunol. 2021, 12, 704050. [Google Scholar] [CrossRef] [PubMed]
- Vermeersch, E.; Denorme, F.; Maes, W.; De Meyer, S.F.; Vanhoorelbeke, K.; Edwards, J.; Shevach, E.M.; Unutmaz, D.; Fujii, H.; Deckmyn, H.; et al. The Role of Platelet and Endothelial GARP in Thrombosis and Hemostasis. PLoS ONE 2017, 12, e0173329. [Google Scholar] [CrossRef] [PubMed]
- Gillot, L.; Baudin, L.; Rouaud, L.; Kridelka, F.; Noël, A. The Pre-Metastatic Niche in Lymph Nodes: Formation and Characteristics. Cell. Mol. Life Sci. 2021, 78, 5987–6002. [Google Scholar] [CrossRef]
- Miyasaka, M.; Hata, E.; Tohya, K.; Hayasaka, H. Lymphocyte Recirculation. In Encyclopedia of Immunobiology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 486–492. ISBN 978-0-08-092152-5. [Google Scholar]
- Acton, S.E.; Onder, L.; Novkovic, M.; Martinez, V.G.; Ludewig, B. Communication, Construction, and Fluid Control: Lymphoid Organ Fibroblastic Reticular Cell and Conduit Networks. Trends Immunol. 2021, 42, 782–794. [Google Scholar] [CrossRef]
- Rodda, L.B.; Lu, E.; Bennett, M.L.; Sokol, C.L.; Wang, X.; Luther, S.A.; Barres, B.A.; Luster, A.D.; Ye, C.J.; Cyster, J.G. Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 2018, 48, 1014–1028.e6. [Google Scholar] [CrossRef]
- Abe, Y.; Sakata-Yanagimoto, M.; Fujisawa, M.; Miyoshi, H.; Suehara, Y.; Hattori, K.; Kusakabe, M.; Sakamoto, T.; Nishikii, H.; Nguyen, T.B.; et al. A Single-Cell Atlas of Non-Haematopoietic Cells in Human Lymph Nodes and Lymphoma Reveals a Landscape of Stromal Remodelling. Nat. Cell Biol. 2022, 24, 565–578. [Google Scholar] [CrossRef]
- Takeda, A.; Hollmén, M.; Dermadi, D.; Pan, J.; Brulois, K.F.; Kaukonen, R.; Lönnberg, T.; Boström, P.; Koskivuo, I.; Irjala, H.; et al. Single-Cell Survey of Human Lymphatics Unveils Marked Endothelial Cell Heterogeneity and Mechanisms of Homing for Neutrophils. Immunity 2019, 51, 561–572.e5. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Grosso, R.A.; Takeda, A.; Pan, J.; Bekkhus, T.; Brulois, K.; Dermadi, D.; Nordling, S.; Vanlandewijck, M.; Jalkanen, S.; et al. A Single-Cell Transcriptional Roadmap of the Mouse and Human Lymph Node Lymphatic Vasculature. Front. Cardiovasc. Med. 2020, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, N.; He, Y.; D’Addio, M.; Tacconi, C.; Detmar, M.; Dieterich, L.C. Single-Cell Mapping Reveals New Markers and Functions of Lymphatic Endothelial Cells in Lymph Nodes. PLoS Biol. 2020, 18, e3000704. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shirkey, M.W.; Zhang, T.; Piao, W.; Li, X.; Zhao, J.; Mei, Z.; Guo, Y.; Saxena, V.; Kensiski, A.; et al. Lymph Node Fibroblastic Reticular Cells Preserve a Tolerogenic Niche in Allograft Transplantation through Laminin A4. J. Clin. Investig. 2022, 132, e156994. [Google Scholar] [CrossRef] [PubMed]
- Stockis, J.; Colau, D.; Coulie, P.G.; Lucas, S. Membrane Protein GARP Is a Receptor for Latent TGF-β on the Surface of Activated Human Treg: Cellular Immune Response. Eur. J. Immunol. 2009, 39, 3315–3322. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.E.; Turley, S.J. Stromal Infrastructure of the Lymph Node and Coordination of Immunity. Trends Immunol. 2015, 36, 30–39. [Google Scholar] [CrossRef]
- Van De Velde, M.; García-Caballero, M.; Durré, T.; Kridelka, F.; Noël, A. Ear Sponge Assay: A Method to Investigate Angiogenesis and Lymphangiogenesis in Mice. In Proteases and Cancer; Cal, S., Obaya, A.J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1731, pp. 223–233. ISBN 978-1-4939-7594-5. [Google Scholar]
- De Streel, G.; Bertrand, C.; Chalon, N.; Liénart, S.; Bricard, O.; Lecomte, S.; Devreux, J.; Gaignage, M.; De Boeck, G.; Mariën, L.; et al. Selective Inhibition of TGF-Β1 Produced by GARP-Expressing Tregs Overcomes Resistance to PD-1/PD-L1 Blockade in Cancer. Nat. Commun. 2020, 11, 4545. [Google Scholar] [CrossRef]
- Lahimchi, M.R.; Eslami, M.; Yousefi, B. New Insight into GARP Striking Role in Cancer Progression: Application for Cancer Therapy. Med. Oncol. 2022, 40, 33. [Google Scholar] [CrossRef]
- Jalkanen, S.; Salmi, M. Lymphatic Endothelial Cells of the Lymph Node. Nat. Rev. Immunol. 2020, 20, 566–578. [Google Scholar] [CrossRef]
- Gerli, M.F.M.; Moyle, L.A.; Benedetti, S.; Ferrari, G.; Ucuncu, E.; Ragazzi, M.; Constantinou, C.; Louca, I.; Sakai, H.; Ala, P.; et al. Combined Notch and PDGF Signaling Enhances Migration and Expression of Stem Cell Markers While Inducing Perivascular Cell Features in Muscle Satellite Cells. Stem Cell Rep. 2019, 12, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Flintoff-Dye, N.L.; Welser, J.; Rooney, J.; Scowen, P.; Tamowski, S.; Hatton, W.; Burkin, D.J. Role for the A7β1 Integrin in Vascular Development and Integrity. Dev. Dyn. 2005, 234, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.A.; Stahl, H.F.; Becker, C.; Correll, A.; Schneider, F.-J.; Tuettenberg, A.; Jonuleit, H. Soluble GARP Has Potent Antiinflammatory and Immunomodulatory Impact on Human CD4+ T Cells. Blood 2013, 122, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, J.; Dong, X.; Shi, M.; Lu, C.; Springer, T.A. GARP Regulates the Bioavailability and Activation of TGFβ. Mol. Biol. Cell 2012, 23, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Metelli, A.; Wu, B.X.; Riesenberg, B.; Guglietta, S.; Huck, J.D.; Mills, C.; Li, A.; Rachidi, S.; Krieg, C.; Rubinstein, M.P.; et al. Thrombin Contributes to Cancer Immune Evasion via Proteolysis of Platelet-Bound GARP to Activate LTGF-β. Sci. Transl. Med. 2020, 12, eaay4860. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, S.; Devreux, J.; de Streel, G.; van Baren, N.; Havelange, V.; Schröder, D.; Vaherto, N.; Vanhaver, C.; Vanderaa, C.; Dupuis, N.; et al. Therapeutic Activity of GARP:TGF-Β1 Blockade in Murine Primary Myelofibrosis. Blood 2023, 141, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Wallace, C.H.; Wu, B.X.; Salem, M.; Ansa-Addo, E.A.; Metelli, A.; Sun, S.; Gilkeson, G.; Shlomchik, M.J.; Liu, B.; Li, Z. B Lymphocytes Confer Immune Tolerance via Cell Surface GARP-TGF-β Complex. JCI Insight 2018, 3, e99863. [Google Scholar] [CrossRef]
- Carrillo-Gálvez, A.B.; Quintero, J.E.; Rodríguez, R.; Menéndez, S.T.; Victoria González, M.; Blanco-Lorenzo, V.; Allonca, E.; De Araújo Farias, V.; González-Correa, J.E.; Erill-Sagalés, N.; et al. GARP Promotes the Proliferation and Therapeutic Resistance of Bone Sarcoma Cancer Cells through the Activation of TGF-β. Cell Death Dis. 2020, 11, 985. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouaud, L.; Baudin, L.; Gautier-Isola, M.; Van Meerbeeck, P.; Feyereisen, E.; Blacher, S.; van Baren, N.; Kridelka, F.; Lucas, S.; Noel, A. Spatial Distribution of Non-Immune Cells Expressing Glycoprotein A Repetitions Predominant in Human and Murine Metastatic Lymph Nodes. Cancers 2023, 15, 5621. https://doi.org/10.3390/cancers15235621
Rouaud L, Baudin L, Gautier-Isola M, Van Meerbeeck P, Feyereisen E, Blacher S, van Baren N, Kridelka F, Lucas S, Noel A. Spatial Distribution of Non-Immune Cells Expressing Glycoprotein A Repetitions Predominant in Human and Murine Metastatic Lymph Nodes. Cancers. 2023; 15(23):5621. https://doi.org/10.3390/cancers15235621
Chicago/Turabian StyleRouaud, Loïc, Louis Baudin, Marine Gautier-Isola, Pierre Van Meerbeeck, Emilie Feyereisen, Silvia Blacher, Nicolas van Baren, Frédéric Kridelka, Sophie Lucas, and Agnes Noel. 2023. "Spatial Distribution of Non-Immune Cells Expressing Glycoprotein A Repetitions Predominant in Human and Murine Metastatic Lymph Nodes" Cancers 15, no. 23: 5621. https://doi.org/10.3390/cancers15235621
APA StyleRouaud, L., Baudin, L., Gautier-Isola, M., Van Meerbeeck, P., Feyereisen, E., Blacher, S., van Baren, N., Kridelka, F., Lucas, S., & Noel, A. (2023). Spatial Distribution of Non-Immune Cells Expressing Glycoprotein A Repetitions Predominant in Human and Murine Metastatic Lymph Nodes. Cancers, 15(23), 5621. https://doi.org/10.3390/cancers15235621






