The Diversity of Liquid Biopsies and Their Potential in Breast Cancer Management
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Breast Cancer
1.2. Liquid Biopsy (LB)
1.2.1. cfDNA
1.2.2. CTC
1.2.3. EV
1.2.4. Analytical Dimensions
2. Liquid Biopsies for Early Breast Cancer Detection
2.1. cfDNA
2.2. cfDNA and Other Analytes Proposed by Small Non-Interventional Trials
3. Liquid Biopsies for Detailed BC Diagnostic
3.1. cfDNA and Nucleosomes
3.2. Multimodal LB
3.3. Circulating RNA in or Independent of EVs
3.4. CTCs
4. Liquid Biopsies before (Neo)Adjuvant Treatment for Therapy (De)Escalation
4.1. Tissue Analysis
4.2. cfDNA
4.3. CTCs
4.4. EVs and miRNAs
5. Liquid Biopsies under Neoadjuvant Therapy for Therapy Switch/(De)Escalation
6. Liquid Biopsies to Anticipate Minimal Residual Disease
6.1. Persistence of LB Signals under Neoadjuvant Treatment to Anticipate MRD
6.1.1. cfDNA
6.1.2. CTCs
6.2. Liquid Biopsies after Neoadjuvant Treatment to Anticipate MRD
6.2.1. cfDNA
6.2.2. CTCs
6.2.3. Other Analytes
6.2.4. Interventional Trials
6.3. Longitudinal LB Analysis for MRD Detection
6.3.1. CTCs
6.3.2. Multimodal LB
6.3.3. cfDNA
7. Liquid Biopsies for Prognostication in the Metastatic BC Setting
7.1. Other Analytes
7.2. cfDNA
7.3. CTCs
7.4. Multimodal LB
8. Liquid Biopsies for Therapy Guidance in Breast Cancer Management
8.1. Chemotherapy (CTX)
8.2. PARP Inhibition
8.3. Anti-HER2 Therapy
8.4. PIK3CA Inhibition
8.5. Endocrine Therapy (ET)
8.6. AKT Inhibition
8.7. Immune Checkpoint Inhibitors (ICI)
8.8. Tyrosine Receptor Kinase (TRK) Inhibition
8.9. Androgen Receptor Inhibition
8.10. Cylin Dependent Kinase 4/6 (CDK4/6) Inhibition
8.11. Predictive Biomarkers for BC Therapy Guidance
9. Liquid Biopsies for Therapy Monitoring in Breast Cancer
9.1. Circulating Proteins
9.2. CTCs
9.3. CTCs and EVs
9.4. cfDNA
9.5. CTC and cfDNA Results and a Multimodal Approach
10. Challenges for Liquid Biopsy in Breast Cancer Management
11. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Somerfield, M.R.; Dowsett, M.; Hammond, M.E.H.; Hayes, D.F.; McShane, L.M.; Saphner, T.J.; Spears, P.A.; Allison, K.H. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: ASCO-College of American Pathologists Guideline Update. J. Clin. Oncol. 2023, 41, 3867–3872. [Google Scholar] [CrossRef] [PubMed]
- Gennari, A.; André, F.; Barrios, C.H.; Cortés, J.; de Azambuja, E.; DeMichele, A.; Dent, R.; Fenlon, D.; Gligorov, J.; Hurvitz, S.A.; et al. ESMO Clinical Practice Guideline for the diagnosis, staging and treatment of patients with metastatic breast cancer. Ann. Oncol. 2021, 32, 1475–1495. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.-J. Strategies for subtypes—dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Fietz, T.; Tesch, H.; Rauh, J.; Boller, E.; Kruggel, L.; Jänicke, M.; Marschner, N. Palliative systemic therapy and overall survival of 1395 patients with advanced breast cancer—Results from the prospective German TMK cohort study. Breast 2017, 34, 122–130. [Google Scholar] [CrossRef]
- Keup, C.; Suryaprakash, V.; Hauch, S.; Storbeck, M.; Hahn, P.; Sprenger-Haussels, M.; Kolberg, H.-C.; Tewes, M.; Hoffmann, O.; Kimmig, R.; et al. Integrative statistical analyses of multiple liquid biopsy analytes in metastatic breast cancer. Genome Med. 2021, 13, 85. [Google Scholar] [CrossRef]
- Keup, C.; Kimmig, R.; Kasimir-Bauer, S. Combinatorial Power of cfDNA, CTCs and EVs in Oncology. Diagnostics 2022, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Keup, C.; Kimmig, R.; Kasimir-Bauer, S. Multimodality in liquid biopsy: Does a combination uncover insights undetectable in individual blood analytes? J. Lab. Med. 2022, 46, 255–264. [Google Scholar] [CrossRef]
- Koh, W.; Pan, W.; Gawad, C.; Fan, H.C.; Kerchner, G.A.; Wyss-Coray, T.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 7361–7366. [Google Scholar] [CrossRef] [PubMed]
- Lui, Y.Y.N.; Chik, K.-W.; Chiu, R.W.K.; Ho, C.-Y.; Lam, C.W.K.; Lo, Y.M.D. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jiang, P.; Chan, K.C.A.; Wong, J.; Cheng, Y.K.Y.; Liang, R.H.S.; Chan, W.; Ma, E.S.K.; Chan, S.L.; Cheng, S.H.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Les acides nucleiques du plasma sanguin chez l’homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Stroun, M.; Lyautey, J.; Lederrey, C.; Olson-Sand, A.; Anker, P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin. Chim. Acta 2001, 313, 139–142. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Bredno, J.; Lipson, J.; Venn, O.; Aravanis, A.M.; Jamshidi, A. Clinical correlates of circulating cell-free DNA tumor fraction. PLoS ONE 2021, 16, e0256436. [Google Scholar] [CrossRef]
- Yuwono, N.L.; Warton, K.; Ford, C.E. The influence of biological and lifestyle factors on circulating cell-free DNA in blood plasma. Elife 2021, 10, e69679. [Google Scholar] [CrossRef] [PubMed]
- González-Masiá, J.A.; García-Olmo, D.; García-Olmo, D.C. Circulating nucleic acids in plasma and serum (CNAPS): Applications in oncology. Onco. Targets. Ther. 2013, 6, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Elshimali, Y.I.; Khaddour, H.; Sarkissyan, M.; Wu, Y.; Vadgama, J.V. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int. J. Mol. Sci. 2013, 14, 18925–18958. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Piskorz, A.M.; Chandrananda, D.; Moore, E.; Morris, J.; Smith, C.G.; Goranova, T.; Heider, K.; Mair, R.; Supernat, A.; et al. Selecting Short DNA Fragments in Plasma Improves Detection of Circulating Tumour DNA. BioRxiv 2017. [Google Scholar] [CrossRef]
- Karagiannis, G.S.; Pastoriza, J.M.; Wang, Y.; Harney, A.S.; Entenberg, D.; Pignatelli, J.; Sharma, V.P.; Xue, E.A.; Cheng, E.; D’Alfonso, T.M.; et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci. Transl. Med. 2017, 9, eaan0026. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Mohme, M.; Riethdorf, S.; Pantel, K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017, 14, 155–167. [Google Scholar] [CrossRef]
- Theodoropoulos, P.A.; Polioudaki, H.; Agelaki, S.; Kallergi, G.; Saridaki, Z.; Mavroudis, D.; Georgoulias, V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010, 288, 99–106. [Google Scholar] [CrossRef]
- Barrière, G.; Riouallon, A.; Renaudie, J.; Tartary, M.; Rigaud, M. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer 2012, 12, 114. [Google Scholar] [CrossRef]
- Giordano, A.; Gao, H.; Anfossi, S.; Cohen, E.; Mego, M.; Lee, B.-N.; Tin, S.; de Laurentiis, M.; Parker, C.A.; Alvarez, R.H.; et al. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol. Cancer Ther. 2012, 11, 2526–2534. [Google Scholar] [CrossRef]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Welter, L.; Xu, L.; McKinley, D.; Dago, A.E.; Prabakar, R.K.; Restrepo-Vassalli, S.; Xu, K.; Rodriguez-Lee, M.; Kolatkar, A.; Nevarez, R.; et al. Treatment response and tumor evolution: Lessons from an extended series of multianalyte liquid biopsies in a metastatic breast cancer patient. Cold Spring Harb. Mol. Case Stud. 2020, 6, a005819. [Google Scholar] [CrossRef] [PubMed]
- Shishido, S.N.; Masson, R.; Xu, L.; Welter, L.; Prabakar, R.K.; d’Souza, A.; Spicer, D.; Kang, I.; Jayachandran, P.; Hicks, J.; et al. Disease characterization in liquid biopsy from HER2-mutated, non-amplified metastatic breast cancer patients treated with neratinib. NPJ Breast Cancer 2022, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.E.; Vuppalapaty, M.; Wilkerson, C.; Renier, C.; Chiu, M.; Lemaire, C.; Che, J.; Matsumoto, M.; Carroll, J.; Crouse, S.; et al. Detection of EGFR Mutations in cfDNA and CTCs, and Comparison to Tumor Tissue in Non-Small-Cell-Lung-Cancer (NSCLC) Patients. Front. Oncol. 2020, 10, 572895. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.A.; Guttery, D.S.; Hills, A.; Fernandez-Garcia, D.; Page, K.; Rosales, B.M.; Goddard, K.S.; Hastings, R.K.; Luo, J.; Ogle, O.; et al. Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin. Cancer Res. 2017, 23, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Dragovic, R.A.; Southcombe, J.H.; Tannetta, D.S.; Redman, C.W.G.; Sargent, I.L. Multicolor flow cytometry and nanoparticle tracking analysis of extracellular vesicles in the plasma of normal pregnant and pre-eclamptic women. Biol. Reprod. 2013, 89, 151. [Google Scholar] [CrossRef]
- Toth, B.; Nieuwland, R.; Liebhardt, S.; Ditsch, N.; Steinig, K.; Stieber, P.; Rank, A.; Göhring, P.; Thaler, C.J.; Friese, K.; et al. Circulating microparticles in breast cancer patients: A comparative analysis with established biomarkers. Anticancer Res. 2008, 28, 1107–1112. [Google Scholar]
- Mateescu, B.; Kowal, E.J.K.; van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzas, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef]
- Ludwig, A.-K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20389. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023, 23, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, aaw3616. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 2022, 40, 1537–1549.e12. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Cummings, S.R.; Absalan, F.; Alexander, G.; Allen, B.; Amini, H.; et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Bryce, A.H.; Thiel, D.D.; Seiden, M.V.; Richards, D.; Luan, Y.; Coignet, M.; Zhang, Q.; Zhang, N.; Hubbell, E.; Kurtzman, K.N.; et al. Performance of a Cell-Free DNA-Based Multi-cancer Detection Test in Individuals Presenting With Symptoms Suspicious for Cancers. JCO Precis. Oncol. 2023, 7, e2200679. [Google Scholar] [CrossRef]
- Pons-Belda, O.D.; Fernandez-Uriarte, A.; Diamandis, E.P. Multi Cancer Early Detection by Using Circulating Tumor DNA-The Galleri Test. Reply to Klein et al. The Promise of Multicancer Early Detection. Comment on Pons-Belda et al. Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm. Diagnostics 2021, 11, 2171. Diagnostics 2022, 12, 1243. [Google Scholar] [CrossRef]
- Nadauld, L.D.; McDonnell, C.H.; Beer, T.M.; Liu, M.C.; Klein, E.A.; Hudnut, A.; Whittington, R.A.; Taylor, B.; Oxnard, G.R.; Lipson, J.; et al. The PATHFINDER Study: Assessment of the Implementation of an Investigational Multi-Cancer Early Detection Test into Clinical Practice. Cancers 2021, 13, 3501. [Google Scholar] [CrossRef] [PubMed]
- Owens, L.; Gulati, R.; Etzioni, R. Stage Shift as an Endpoint in Cancer Screening Trials: Implications for Evaluating Multicancer Early Detection Tests. Cancer Epidemiol. Biomarkers Prev. 2022, 31, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Li, L.H.; Hua, D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett. 2006, 243, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Mijnes, J.; Tiedemann, J.; Eschenbruch, J.; Gasthaus, J.; Bringezu, S.; Bauerschlag, D.; Maass, N.; Arnold, N.; Weimer, J.; Anzeneder, T.; et al. SNiPER: A novel hypermethylation biomarker panel for liquid biopsy based early breast cancer detection. Oncotarget 2019, 10, 6494–6508. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Ruan, L. APC gene promoter aberrant methylation in serum as a biomarker for breast cancer diagnosis: A meta-analysis. Thorac. Cancer 2018, 9, 284–290. [Google Scholar] [CrossRef]
- Kim, J.-H.; Shin, M.-H.; Kweon, S.-S.; Park, M.H.; Yoon, J.H.; Lee, J.S.; Choi, C.; Fackler, M.J.; Sukumar, S. Evaluation of promoter hypermethylation detection in serum as a diagnostic tool for breast carcinoma in Korean women. Gynecol. Oncol. 2010, 118, 176–181. [Google Scholar] [CrossRef]
- Radpour, R.; Barekati, Z.; Kohler, C.; Lv, Q.; Bürki, N.; Diesch, C.; Bitzer, J.; Zheng, H.; Schmid, S.; Zhong, X.Y. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS ONE 2011, 6, e16080. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nakayama, T.; Kajita, M.; Miyake, T.; Iwamoto, T.; Kim, S.J.; Sakai, A.; Ishihara, H.; Tamaki, Y.; Noguchi, S. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARβ2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res. Treat. 2012, 132, 165–173. [Google Scholar] [CrossRef]
- Li, Z.; Guo, X.; Tang, L.; Peng, L.; Chen, M.; Luo, X.; Wang, S.; Xiao, Z.; Deng, Z.; Dai, L.; et al. Methylation analysis of plasma cell-free DNA for breast cancer early detection using bisulfite next-generation sequencing. Tumour Biol. 2016, 37, 13111–13119. [Google Scholar] [CrossRef] [PubMed]
- Kamel, A.M.; Teama, S.; Fawzy, A.; El Deftar, M. Plasma DNA integrity index as a potential molecular diagnostic marker for breast cancer. Tumour Biol. 2016, 37, 7565–7572. [Google Scholar] [CrossRef] [PubMed]
- Saura, C.; Ortiz, C.; Matito, J.; Arenas, E.J.; Suñol, A.; Martín, Á.; Córdoba, O.; Martínez-Sabadell, A.; García-Ruiz, I.; Miranda, I.; et al. Early-Stage Breast Cancer Detection in Breast Milk. Cancer Discov. 2023, 13, 2180–2191. [Google Scholar] [CrossRef] [PubMed]
- Kruspe, S.; Dickey, D.D.; Urak, K.T.; Blanco, G.N.; Miller, M.J.; Clark, K.C.; Burghardt, E.; Gutierrez, W.R.; Phadke, S.D.; Kamboj, S.; et al. Rapid and Sensitive Detection of Breast Cancer Cells in Patient Blood with Nuclease-Activated Probe Technology. Mol. Ther. Nucleic Acids 2017, 8, 542–557. [Google Scholar] [CrossRef]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, W.; Bu, J.; Li, Y.; Li, R.; Nie, R.; Xiao, C.; Ma, K.; Huang, X.; Li, Y. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol. Carcinog. 2019, 58, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Guzman, N.; Agarwal, K.; Asthagiri, D.; Yu, L.; Saji, M.; Ringel, M.D.; Paulaitis, M.E. Breast Cancer-Specific miR Signature Unique to Extracellular Vesicles Includes “microRNA-like” tRNA Fragments. Mol. Cancer Res. 2015, 13, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef]
- Loke, S.Y.; Munusamy, P.; Koh, G.L.; Chan, C.H.T.; Madhukumar, P.; Thung, J.L.; Tan, K.T.B.; Ong, K.W.; Yong, W.S.; Sim, Y.; et al. A Circulating miRNA Signature for Stratification of Breast Lesions among Women with Abnormal Screening Mammograms. Cancers 2019, 11, 1872. [Google Scholar] [CrossRef]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef]
- Markou, A.; Zavridou, M.; Sourvinou, I.; Yousef, G.; Kounelis, S.; Malamos, N.; Georgoulias, V.; Lianidou, E. Direct Comparison of Metastasis-Related miRNAs Expression Levels in Circulating Tumor Cells, Corresponding Plasma, and Primary Tumors of Breast Cancer Patients. Clin. Chem. 2016, 62, 1002–1011. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhong, G.; Wang, K.; Li, J.; Xiao, S.; Wei, W.; Liu, J. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Breast Cancer Diagnosis. Onco. Targets. Ther. 2020, 13, 2563–2571. [Google Scholar] [CrossRef] [PubMed]
- Opstal-van Winden, A.W.J.; Krop, E.J.M.; Kåredal, M.H.; Gast, M.-C.W.; Lindh, C.H.; Jeppsson, M.C.; Jönsson, B.A.G.; Grobbee, D.E.; Peeters, P.H.M.; Beijnen, J.H.; et al. Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer 2011, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- Fredolini, C.; Pathak, K.V.; Paris, L.; Chapple, K.M.; Tsantilas, K.A.; Rosenow, M.; Tegeler, T.J.; Garcia-Mansfield, K.; Tamburro, D.; Zhou, W.; et al. Shotgun proteomics coupled to nanoparticle-based biomarker enrichment reveals a novel panel of extracellular matrix proteins as candidate serum protein biomarkers for early-stage breast cancer detection. Breast Cancer Res. 2020, 22, 135. [Google Scholar] [CrossRef] [PubMed]
- Bartkowiak, K.; Heidrich, I.; Kwiatkowski, M.; Banys-Paluchowski, M.; Andreas, A.; Wurlitzer, M.; Geffken, M.; Voß, H.; Zeller, T.; Blankenberg, S.; et al. Circulating Cellular Communication Network Factor 1 Protein as a Sensitive Liquid Biopsy Marker for Early Detection of Breast Cancer. Clin. Chem. 2021, 68, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Kure, S.; Satoi, S.; Kitayama, T.; Nagase, Y.; Nakano, N.; Yamada, M.; Uchiyama, N.; Miyashita, S.; Iida, S.; Takei, H.; et al. A prediction model using 2-propanol and 2-butanone in urine distinguishes breast cancer. Sci. Rep. 2021, 11, 19801. [Google Scholar] [CrossRef] [PubMed]
- Tomeva, E.; Switzeny, O.J.; Heitzinger, C.; Hippe, B.; Haslberger, A.G. Comprehensive Approach to Distinguish Patients with Solid Tumors from Healthy Controls by Combining Androgen Receptor Mutation p.H875Y with Cell-Free DNA Methylation and Circulating miRNAs. Cancers 2022, 14, 462. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Wang, W.; Liang, M.; Ma, G.; Li, L.; Zhou, W.; Xia, T.; Xie, H.; Wang, S. Plasma cell-free DNA integrity plus circulating tumor cells: A potential biomarker of no distant metastasis breast cancer. Neoplasma 2017, 64, 611–618. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef]
- S1-Leitlinie: Tumorgenetik—Diagnostik im Kontext maligner Erkrankungen. Med. Genet. 2022, 34, 53–68. [CrossRef]
- Hu, M.; Brown, V.; Jackson, J.M.; Wijerathne, H.; Pathak, H.; Koestler, D.C.; Nissen, E.; Hupert, M.L.; Muller, R.; Godwin, A.K.; et al. Assessing Breast Cancer Molecular Subtypes Using Extracellular Vesicles’ mRNA. Anal. Chem. 2023, 95, 7665–7675. [Google Scholar] [CrossRef] [PubMed]
- Ulz, P.; Thallinger, G.G.; Auer, M.; Graf, R.; Kashofer, K.; Jahn, S.W.; Abete, L.; Pristauz, G.; Petru, E.; Geigl, J.B.; et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 2016, 48, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Brasó-Maristany, F.; Martínez-Sáez, O.; Sanfeliu, E.; Xia, Y.; Bellet, M.; Galván, P.; Martínez, D.; Pascual, T.; Marín-Aguilera, M.; et al. Circulating tumor DNA reveals complex biological features with clinical relevance in metastatic breast cancer. Nat. Commun. 2023, 14, 1157. [Google Scholar] [CrossRef] [PubMed]
- Doebley, A.-L.; Ko, M.; Liao, H.; Cruikshank, A.E.; Santos, K.; Kikawa, C.; Hiatt, J.B.; Patton, R.D.; de Sarkar, N.; Collier, K.A.; et al. A framework for clinical cancer subtyping from nucleosome profiling of cell-free DNA. Nat. Commun. 2022, 13, 7475. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Heitzer, E.; Auer, M.; Hoffmann, E.M.; Pichler, M.; Gasch, C.; Ulz, P.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Mohan, S.; et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int. J. Cancer 2013, 133, 346–356. [Google Scholar] [CrossRef]
- Bortolini Silveira, A.; Bidard, F.-C.; Tanguy, M.-L.; Girard, E.; Trédan, O.; Dubot, C.; Jacot, W.; Goncalves, A.; Debled, M.; Levy, C.; et al. Multimodal liquid biopsy for early monitoring and outcome prediction of chemotherapy in metastatic breast cancer. NPJ Breast Cancer 2021, 7, 115. [Google Scholar] [CrossRef]
- Epic Sciences. Epic Sciences Announces Medicare Coverage For Breast-Cancer Focused ctDNA Gene Panel: Press Release 19.04.2023. Available online: https://www.epicsciences.com/press-releases/epic-sciences-announces-medicare-coverage-for-breast-cancer-focused-ctdna-gene-panel/?goal=0_8c04e2abda-5ca6438936-71670485&mc_cid=5ca6438936 (accessed on 25 July 2023).
- Eichelser, C.; Stückrath, I.; Müller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, A.; de Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Del Garrido-Navas, M.C.; Rolfo, C.; et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Iinuma, H.; Umemoto, Y.; Yanagisawa, T.; Matsumoto, A.; Jinno, H. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol. Lett. 2018, 15, 9584–9592. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.-B.; Yan, M.-G.; Fang, X.; Guo, J.-J.; Xiong, W.; Zhang, R.-P. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin. Chim. Acta 2018, 487, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Narbe, U.; Bendahl, P.-O.; Aaltonen, K.; Fernö, M.; Forsare, C.; Jørgensen, C.L.T.; Larsson, A.-M.; Rydén, L. The Distribution of Circulating Tumor Cells Is Different in Metastatic Lobular Compared to Ductal Carcinoma of the Breast—Long-Term Prognostic Significance. Cells 2020, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Wittner, B.S.; Donaldson, M.C.; O’Keefe, R.; Engstrom, A.; Bersani, F.; Zheng, Y.; Comaills, V.; Niederhoffer, K.; et al. AR Expression in Breast Cancer CTCs Associates with Bone Metastases. Mol. Cancer Res. 2018, 16, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Konstantinidis, G.; Markomanolaki, H.; Papadaki, M.A.; Mavroudis, D.; Stournaras, C.; Georgoulias, V.; Agelaki, S. Apoptotic circulating tumor cells in early and metastatic breast cancer patients. Mol. Cancer Ther. 2013, 12, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Park-Simon, T.-W.; Müller, V.; Jackisch, C.; Albert, U.-S.; Banys-Paluchowski, M.; Bauerfeind, I.; Blohmer, J.-U.; Budach, W.; Dall, P.; Ditsch, N.; et al. AGO Recommendations for the Diagnosis and Treatment of Patients with Early Breast Cancer (EBC): Update 2023. Breast Care 2023, 18, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Lu, X.; Liu, Q.; Zhang, T.; Li, P.; Qiao, W.; Deng, M. Prognostic value of neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio for breast cancer patients: An updated meta-analysis of 17079 individuals. Cancer Med. 2019, 8, 4135–4148. [Google Scholar] [CrossRef]
- Truffi, M.; Piccotti, F.; Albasini, S.; Tibollo, V.; Morasso, C.F.; Sottotetti, F.; Corsi, F. Preoperative Systemic Inflammatory Biomarkers Are Independent Predictors of Disease Recurrence in ER+ HER2- Early Breast Cancer. Front. Oncol. 2021, 11, 773078. [Google Scholar] [CrossRef]
- Moon, G.; Noh, H.; Cho, I.-J.; Lee, J.-I.; Han, A. Prediction of late recurrence in patients with breast cancer: Elevated neutrophil to lymphocyte ratio (NLR) at 5 years after diagnosis and late recurrence. Breast Cancer 2020, 27, 54–61. [Google Scholar] [CrossRef]
- Ferroni, P.; Roselli, M.; Buonomo, O.C.; Spila, A.; Portarena, I.; Laudisi, A.; Valente, M.G.; Pirillo, S.P.; Fortunato, L.; Costarelli, L.; et al. Prognostic Significance of Neutrophil-to-lymphocyte Ratio in the Framework of the 8th TNM Edition for Breast Cancer. Anticancer Res. 2018, 38, 4705–4712. [Google Scholar] [CrossRef] [PubMed]
- Graziano, V.; Grassadonia, A.; Iezzi, L.; Vici, P.; Pizzuti, L.; Barba, M.; Quinzii, A.; Camplese, A.; Di Marino, P.; Peri, M.; et al. Combination of peripheral neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio is predictive of pathological complete response after neoadjuvant chemotherapy in breast cancer patients. Breast 2019, 44, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, A.; Gonzalez, N.S.; Pimentel, I.; Suñol, A.; Zamora, E.; Ortiz, C.; Espinosa-Bravo, M.; Peg, V.; Vivancos, A.; Saura, C.; et al. Prognostic value of ctDNA detection in patients with early breast cancer undergoing neoadjuvant therapy: A systematic review and meta-analysis. Cancer Treat. Rev. 2022, 104, 102362. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Woo, M.; Kim, J.E.; Ahn, J.-H.; Jung, K.H.; Roh, J.; Gong, G.; Kim, S.-B. Efficacy of assessing circulating cell-free DNA using a simple fluorescence assay in patients with triple-negative breast cancer receiving neoadjuvant chemotherapy: A prospective observational study. Oncotarget 2018, 9, 3875–3886. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Silva, M.J.; Venet, D.; Campbell, C.; Bradburry, I.; Rouas, G.; de Azambuja, E.; Maetens, M.; Fumagalli, D.; Rodrik-Outmezguine, V.; et al. Circulating Tumor DNA in HER2-Amplified Breast Cancer: A Translational Research Substudy of the NeoALTTO Phase III Trial. Clin. Cancer Res. 2019, 25, 3581–3588. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Nakayama, T.; Yamamoto, N.; Kim, S.J.; Shimazu, K.; Shimomura, A.; Maruyama, N.; Morimoto, K.; Tamaki, Y.; Noguchi, S. Methylated DNA and total DNA in serum detected by one-step methylation-specific PCR is predictive of poor prognosis for breast cancer patients. Oncology 2012, 83, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Azim, H.A.; Rothé, F.; Aura, C.M.; Bavington, M.; Maetens, M.; Rouas, G.; Gebhart, G.; Gamez, C.; Eidtmann, H.; Baselga, J.; et al. Circulating tumor cells and response to neoadjuvant paclitaxel and HER2-targeted therapy: A sub-study from the NeoALTTO phase III trial. Breast 2013, 22, 1060–1065. [Google Scholar] [CrossRef]
- Riethdorf, S.; Müller, V.; Loibl, S.; Nekljudova, V.; Weber, K.; Huober, J.; Fehm, T.; Schrader, I.; Hilfrich, J.; Holms, F.; et al. Prognostic Impact of Circulating Tumor Cells for Breast Cancer Patients Treated in the Neoadjuvant “Geparquattro” Trial. Clin. Cancer Res. 2017, 23, 5384–5393. [Google Scholar] [CrossRef]
- Pierga, J.-Y.; Bidard, F.-C.; Autret, A.; Petit, T.; Andre, F.; Dalenc, F.; Levy, C.; Ferrero, J.-M.; Romieu, G.; Bonneterre, J.; et al. Circulating tumour cells and pathological complete response: Independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Ann. Oncol. 2017, 28, 103–109. [Google Scholar] [CrossRef]
- Janni, W.J.; Rack, B.; Terstappen, L.W.M.M.; Pierga, J.-Y.; Taran, F.-A.; Fehm, T.; Hall, C.; de Groot, M.R.; Bidard, F.-C.; Friedl, T.W.P.; et al. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin. Cancer Res. 2016, 22, 2583–2593. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Michiels, S.; Riethdorf, S.; Mueller, V.; Esserman, L.J.; Lucci, A.; Naume, B.; Horiguchi, J.; Gisbert-Criado, R.; Sleijfer, S.; et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J. Natl. Cancer Inst. 2018, 110, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl. Cancer Inst. 2014, 106, dju066. [Google Scholar] [CrossRef] [PubMed]
- Todorova, V.K.; Byrum, S.D.; Gies, A.J.; Haynie, C.; Smith, H.; Reyna, N.S.; Makhoul, I. Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Curr. Oncol. 2022, 29, 55. [Google Scholar] [CrossRef] [PubMed]
- Kleivi Sahlberg, K.; Bottai, G.; Naume, B.; Burwinkel, B.; Calin, G.A.; Børresen-Dale, A.-L.; Santarpia, L. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin. Cancer Res. 2015, 21, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- McGuire, A.; Casey, M.-C.; Waldron, R.M.; Heneghan, H.; Kalinina, O.; Holian, E.; McDermott, A.; Lowery, A.J.; Newell, J.; Dwyer, R.M.; et al. Prospective Assessment of Systemic MicroRNAs as Markers of Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancers 2020, 12, 1820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Yu, J.; Xu, L.; Pang, X.; Xiang, Q.; Liu, Q.; Cui, Y. miRNAs as therapeutic predictors and prognostic biomarkers of neoadjuvant chemotherapy in breast cancer: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2022, 194, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Casey, M.C.; McGuire, A.; Waldron, R.M.; Paganga, M.; Holian, E.; Newell, J.; Heneghan, H.M.; McDermott, A.M.; Keane, M.M.; et al. Evaluating the Role of Circulating MicroRNAs to Aid Therapeutic Decision Making for Neoadjuvant Chemotherapy in Breast Cancer: A Prospective, Multicenter Clinical Trial. Ann. Surg. 2022, 276, 905–912. [Google Scholar] [CrossRef]
- Stoetzer, O.J.; Fersching, D.M.I.; Salat, C.; Steinkohl, O.; Gabka, C.J.; Hamann, U.; Braun, M.; Feller, A.-M.; Heinemann, V.; Siegele, B.; et al. Prediction of response to neoadjuvant chemotherapy in breast cancer patients by circulating apoptotic biomarkers nucleosomes, DNAse, cytokeratin-18 fragments and survivin. Cancer Lett. 2013, 336, 140–148. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Silvestri, M.; Baselga, J.; Piccart, M.; Huober, J.; Izquierdo, M.; de la Pena, L.; Hilbers, F.S.; et al. Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. Int. J. Mol. Sci. 2020, 21, 1386. [Google Scholar] [CrossRef]
- Matikas, A.; Wang, K.; Lagoudaki, E.; Acs, B.; Zerdes, I.; Hartman, J.; Azavedo, E.; Bjöhle, J.; Carlsson, L.; Einbeigi, Z.; et al. Prognostic role of serum thymidine kinase 1 kinetics during neoadjuvant chemotherapy for early breast cancer. ESMO Open 2021, 6, 100076. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Swigart, L.B.; Wu, H.-T.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 2021, 32, 229–239. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.-J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.-F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, aax7392. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Gampenrieder, S.P.; Frantal, S.; Rinnerthaler, G.; Singer, C.F.; Egle, D.; Pfeiler, G.; Bartsch, R.; Wette, V.; Pichler, A.; et al. Persistence of ctDNA in Patients with Breast Cancer During Neoadjuvant Treatment Is a Significant Predictor of Poor Tumor Response. Clin. Cancer Res. 2022, 28, 697–707. [Google Scholar] [CrossRef]
- Moss, J.; Zick, A.; Grinshpun, A.; Carmon, E.; Maoz, M.; Ochana, B.L.; Abraham, O.; Arieli, O.; Germansky, L.; Meir, K.; et al. Circulating breast-derived DNA allows universal detection and monitoring of localized breast cancer. Ann. Oncol. 2020, 31, 395–403. [Google Scholar] [CrossRef]
- Hall, C.; Karhade, M.; Laubacher, B.; Anderson, A.; Kuerer, H.; DeSynder, S.; Lucci, A. Circulating Tumor Cells After Neoadjuvant Chemotherapy in Stage I-III Triple-Negative Breast Cancer. Ann. Surg. Oncol. 2015, 22 (Suppl. S3), 552–558. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Chopra, N.; Comino-Méndez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Hancock, B.A.; Solzak, J.P.; Brinza, D.; Scafe, C.; Miller, K.D.; Radovich, M. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 2017, 3, 24. [Google Scholar] [CrossRef]
- Olsson, E.; Winter, C.; George, A.; Chen, Y.; Howlin, J.; Tang, M.-H.E.; Dahlgren, M.; Schulz, R.; Grabau, D.; van Westen, D.; et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 2015, 7, 1034–1047. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Lynce, F.C.; Gallagher, C.; Isaacs, C.; Blaya, M.; et al. Association of Circulating Tumor DNA and Circulating Tumor Cells After Neoadjuvant Chemotherapy With Disease Recurrence in Patients With Triple-Negative Breast Cancer: Preplanned Secondary Analysis of the BRE12-158 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef]
- Barrios, C.H.; Harbeck, N.; Zhang, H.A.; Saji, S.; Jung, K.H.; Hegg, R.; Koehler, A.; Sohn, J.; Iwata, H.; Telli, M.L.; et al. LBA1—Final analysis of the placebo-controlled randomised phase 3 IMpassion031 trial evaluating neoadjuvant atezolizumab (atezo) plus chemotherapy (CT) followed by open-label adjuvant atezo in patients (pts) with early-stage triple-negative breast cancer (eTNBC). Ann. Oncol. 2023, 8, 101220. [Google Scholar] [CrossRef]
- Yoshinami, T.; Kagara, N.; Motooka, D.; Nakamura, S.; Miyake, T.; Tanei, T.; Naoi, Y.; Shimoda, M.; Shimazu, K.; Kim, S.J.; et al. Detection of ctDNA with Personalized Molecular Barcode NGS and Its Clinical Significance in Patients with Early Breast Cancer. Transl. Oncol. 2020, 13, 100787. [Google Scholar] [CrossRef] [PubMed]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic Significance of TWIST1, CD24, CD44, and ALDH1 Transcript Quantification in EpCAM-Positive Circulating Tumor Cells from Early Stage Breast Cancer Patients. Cells 2019, 8, 652. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, J.; Chen, X. Lymphocyte-to-Monocyte Ratio is Associated with the Poor Prognosis of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Cancer Manag. Res. 2021, 13, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, M.A.; Monastirioti, A.; Apostolopoulou, C.A.; Aggouraki, D.; Papadaki, C.; Michaelidou, K.; Vassilakopoulou, M.; Alexakou, K.; Mavroudis, D.; Agelaki, S. TLR4 and pSTAT3 Expression on Circulating Tumor Cells (CTCs) and Immune Cells in the Peripheral Blood of Breast Cancer Patients: Prognostic Implications. Cancers 2022, 14, 1053. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Ciniselli, C.M.; Pizzamiglio, S.; Cappelletti, V.; Silvestri, M.; El-Abed, S.; Izquierdo, M.; Bajji, M.; Nuciforo, P.; Huober, J.; et al. End-of-neoadjuvant treatment circulating microRNAs and HER2-positive breast cancer patient prognosis: An exploratory analysis from NeoALTTO. Front. Oncol. 2022, 12, 1028825. [Google Scholar] [CrossRef]
- Saloustros, E.; Perraki, M.; Apostolaki, S.; Kallergi, G.; Xyrafas, A.; Kalbakis, K.; Agelaki, S.; Kalykaki, A.; Georgoulias, V.; Mavroudis, D. Cytokeratin-19 mRNA-positive circulating tumor cells during follow-up of patients with operable breast cancer: Prognostic relevance for late relapse. Breast Cancer Res. 2011, 13, R60. [Google Scholar] [CrossRef]
- Stergiopoulou, D.; Markou, A.; Strati, A.; Zavridou, M.; Tzanikou, E.; Mastoraki, S.; Kallergi, G.; Georgoulias, V.; Lianidou, E. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci. Rep. 2023, 13, 1258. [Google Scholar] [CrossRef]
- Stewart, C.M.; Kothari, P.D.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.-J.; Arcila, M.E.; et al. The value of cell-free DNA for molecular pathology. J. Pathol. 2018, 244, 616–627. [Google Scholar] [CrossRef]
- Coombes, R.C.; Page, K.; Salari, R.; Hastings, R.K.; Armstrong, A.; Ahmed, S.; Ali, S.; Cleator, S.; Kenny, L.; Stebbing, J.; et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin. Cancer Res. 2019, 25, 4255–4263. [Google Scholar] [CrossRef]
- GenomeWeb Staff Reporter. Medicare to Cover Natera’s Signatera MRD Test in Breast Cancer. Available online: https://www.genomeweb.com/liquid-biopsy/medicare-cover-nateras-signatera-mrd-test-breast-cancer (accessed on 2 August 2023).
- Parsons, H.A.; Rhoades, J.; Reed, S.C.; Gydush, G.; Ram, P.; Exman, P.; Xiong, K.; Lo, C.C.; Li, T.; Fleharty, M.; et al. Sensitive Detection of Minimal Residual Disease in Patients Treated for Early-Stage Breast Cancer. Clin. Cancer Res. 2020, 26, 2556–2564. [Google Scholar] [CrossRef]
- Lipsyc-Sharf, M.; de Bruin, E.C.; Santos, K.; McEwen, R.; Stetson, D.; Patel, A.; Kirkner, G.J.; Hughes, M.E.; Tolaney, S.M.; Partridge, A.H.; et al. Circulating Tumor DNA and Late Recurrence in High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Breast Cancer. J. Clin. Oncol. 2022, 40, 2408–2419. [Google Scholar] [CrossRef]
- GenomeWeb Staff Reporter. NeoGenomics Gets Medicare Coverage for Radar Minimal Residual Disease Assay in Breast Cancer. Available online: https://www.genomeweb.com/business-news/neogenomics-gets-medicare-coverage-radar-minimal-residual-disease-assay-breast-cancer?adobe_mc=MCMID%3D02586013260007224175993110107847566365%7CMCORGID%3D138FFF2554E6E7220A4C98C6%2540AdobeOrg%7CTS%3D1690956320&CSAuthResp=1%3A%3A2385432%3A1911%3A24%3Asuccess%3AE841B8C9EF0F2C2A78147487ED217CFF (accessed on 2 August 2023).
- Turner, N.C.; Swift, C.; Jenkins, B.; Kilburn, L.; Coakley, M.; Beaney, M.; Fox, L.; Goddard, K.; Garcia-Murillas, I.; Proszek, P.; et al. Results of the c-TRAK TN trial: A clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann. Oncol. 2022, 34, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Oncology Center of Excellence. Use of Circulating Tumor Deoxyribonucleic Acid for Early-Stage Solid Tumor Drug Development; Draft Guidance for Industry; Availability: GUIDANCE DOCUMENT. Docket No. FDA-2022-D-0084. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/use-circulating-tumor-deoxyribonucleic-acid-early-stage-solid-tumor-drug-development-draft-guidance (accessed on 10 August 2023).
- Thill, M.; Kolberg-Liedtke, C.; Albert, U.-S.; Banys-Paluchowski, M.; Bauerfeind, I.; Blohmer, J.-U.; Budach, W.; Dall, P.; Ditsch, N.; Fallenberg, E.M.; et al. AGO Recommendations for the Diagnosis and Treatment of Patients with Locally Advanced and Metastatic Breast Cancer: Update 2023. Breast Care 2023, 18, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Duque, G.; Manterola, C.; Otzen, T.; Arias, C.; Galindo, B.; Mora, M.; Guerrero, E.; García, N. Clinical utility of liquid biopsy in breast cancer: A systematic review. Clin. Genet. 2022, 101, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.; Deutsch, T.M.; Feisst, M.; Rippinger, N.; Riedel, F.; Hartkopf, A.D.; Brucker, S.Y.; Domschke, C.; Fremd, C.; Michel, L.; et al. Circulating miR-200 family as predictive markers during systemic therapy of metastatic breast cancer. Arch. Gynecol. Obstet. 2022, 306, 875–885. [Google Scholar] [CrossRef] [PubMed]
- McCartney, A.; Biagioni, C.; Schiavon, G.; Bergqvist, M.; Mattsson, K.; Migliaccio, I.; Benelli, M.; Romagnoli, D.; Bonechi, M.; Boccalini, G.; et al. Prognostic role of serum thymidine kinase 1 activity in patients with hormone receptor-positive metastatic breast cancer: Analysis of the randomised phase III Evaluation of Faslodex versus Exemestane Clinical Trial (EFECT). Eur. J. Cancer 2019, 114, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Veiga, T.; Bravo, S.; Gómez-Tato, A.; Yáñez-Gómez, C.; Abuín, C.; Varela, V.; Cueva, J.; Palacios, P.; Dávila-Ibáñez, A.B.; Piñeiro, R.; et al. Red Blood Cells Protein Profile Is Modified in Breast Cancer Patients. Mol. Cell. Proteomics 2022, 21, 100435. [Google Scholar] [CrossRef] [PubMed]
- Hotz, M.J.; Qing, D.; Shashaty, M.G.S.; Zhang, P.; Faust, H.; Sondheimer, N.; Rivella, S.; Worthen, G.S.; Mangalmurti, N.S. Red Blood Cells Homeostatically Bind Mitochondrial DNA through TLR9 to Maintain Quiescence and to Prevent Lung Injury. Am. J. Respir. Crit. Care Med. 2018, 197, 470–480. [Google Scholar] [CrossRef]
- Rodríguez, M.; Silva, J.; Herrera, A.; Herrera, M.; Peña, C.; Martín, P.; Gil-Calderón, B.; Larriba, M.J.; Coronado, M.J.; Soldevilla, B.; et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget 2015, 6, 40575–40587. [Google Scholar] [CrossRef]
- Visvanathan, K.; Fackler, M.S.; Zhang, Z.; Lopez-Bujanda, Z.A.; Jeter, S.C.; Sokoll, L.J.; Garrett-Mayer, E.; Cope, L.M.; Umbricht, C.B.; Euhus, D.M.; et al. Monitoring of Serum DNA Methylation as an Early Independent Marker of Response and Survival in Metastatic Breast Cancer: TBCRC 005 Prospective Biomarker Study. J. Clin. Oncol. 2017, 35, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Koukaki, T.; Karamitrousis, E.; Nena, E.; Tsamardinos, I.; Kolios, G.; Lianidou, E.; et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2019, 38, 3387–3401. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Mahecha, A.; Lafleur, J.; Brousse, S.; Savichtcheva, O.; Holden, K.A.; Faulkner, N.; McLennan, G.; Jensen, T.J.; Basik, M. Early, On-Treatment Levels and Dynamic Changes of Genomic Instability in Circulating Tumor DNA Predict Response to Treatment and Outcome in Metastatic Breast Cancer Patients. Cancers 2021, 13, 1331. [Google Scholar] [CrossRef] [PubMed]
- Suppan, C.; Brcic, I.; Tiran, V.; Mueller, H.D.; Posch, F.; Auer, M.; Ercan, E.; Ulz, P.; Cote, R.J.; Datar, R.H.; et al. Untargeted Assessment of Tumor Fractions in Plasma for Monitoring and Prognostication from Metastatic Breast Cancer Patients Undergoing Systemic Treatment. Cancers 2019, 11, 1171. [Google Scholar] [CrossRef]
- Hrebien, S.; Citi, V.; Garcia-Murillas, I.; Cutts, R.; Fenwick, K.; Kozarewa, I.; McEwen, R.; Ratnayake, J.; Maudsley, R.; Carr, T.H.; et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019, 30, 945–952. [Google Scholar] [CrossRef]
- Keup, C.; Benyaa, K.; Hauch, S.; Sprenger-Haussels, M.; Tewes, M.; Mach, P.; Bittner, A.-K.; Kimmig, R.; Hahn, P.; Kasimir-Bauer, S. Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell. Mol. Life Sci. 2019, 77, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Garcia, E.; Genta, S.; Chen, X.; Ou, Q.; Araujo, D.V.; Abdul Razak, A.R.; Hansen, A.R.; Spreafico, A.; Bao, H.; Wu, X.; et al. Tumor-Naïve Circulating Tumor DNA as an Early Response Biomarker for Patients Treated With Immunotherapy in Early Phase Clinical Trials. JCO Precis. Oncol. 2023, 7, e2200509. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Tomiguchi, M.; Sueta, A.; Murakami, K.; Omoto, Y.; Iwase, H. Analysis of ESR1 and PIK3CA mutations in plasma cell-free DNA from ER-positive breast cancer patients. Oncotarget 2017, 8, 52142–52155. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Kaklamani, V.G.; Neven, P.; Streich, G.; Montero, A.J.; Forget, F.; Mouret-Reynier, M.-A.; Sohn, J.H.; Taylor, D.; Harnden, K.K.; et al. Elacestrant (oral selective estrogen receptor degrader) versus Standard Endocrine Therapy for Estrogen Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: Results from the Randomized Phase III EMERALD Trial. J. Clin. Oncol. 2022, 40, 3246–3256. [Google Scholar] [CrossRef]
- Baselga, J.; Im, S.-A.; Iwata, H.; Cortés, J.; de Laurentiis, M.; Jiang, Z.; Arteaga, C.L.; Jonat, W.; Clemons, M.; Ito, Y.; et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 904–916. [Google Scholar] [CrossRef]
- Nakauchi, C.; Kagara, N.; Shimazu, K.; Shimomura, A.; Naoi, Y.; Shimoda, M.; Kim, S.J.; Noguchi, S. Detection of TP53/PIK3CA Mutations in Cell-Free Plasma DNA From Metastatic Breast Cancer Patients Using Next Generation Sequencing. Clin. Breast Cancer 2016, 16, 418–423. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Hayes, D.F.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Reuben, J.M.; Doyle, G.V.; Matera, J.; Allard, W.J.; Miller, M.C.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005, 23, 1420–1430. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Magbanua, M.J.M.; Hendrix, L.H.; Hyslop, T.; Barry, W.T.; Winer, E.P.; Hudis, C.; Toppmeyer, D.; Carey, L.A.; Partridge, A.H.; Pierga, J.-Y.; et al. Serial analysis of circulating tumor cells in metastatic breast cancer receiving first-line chemotherapy. J. Natl. Cancer Inst. 2020, 113, 443–452. [Google Scholar] [CrossRef]
- Abreu, M.; Cabezas-Sainz, P.; Pereira-Veiga, T.; Falo, C.; Abalo, A.; Morilla, I.; Curiel, T.; Cueva, J.; Rodríguez, C.; Varela-Pose, V.; et al. Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. J. Clin. Med. 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 2017, 161, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.-M.; Jansson, S.; Bendahl, P.-O.; Levin Tykjaer Jörgensen, C.; Loman, N.; Graffman, C.; Lundgren, L.; Aaltonen, K.; Rydén, L. Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Res. 2018, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, R46. [Google Scholar] [CrossRef]
- Tewes, M.; Aktas, B.; Welt, A.; Mueller, S.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: An option for monitoring response to breast cancer related therapies. Breast Cancer Res. Treat. 2009, 115, 581–590. [Google Scholar] [CrossRef]
- Pereira-Veiga, T.; Martínez-Fernández, M.; Abuin, C.; Piñeiro, R.; Cebey, V.; Cueva, J.; Palacios, P.; Blanco, C.; Muinelo-Romay, L.; Abalo, A.; et al. CTCs Expression Profiling for Advanced Breast Cancer Monitoring. Cancers 2019, 11, 1941. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Horimoto, Y.; Tokuda, E.; Murakami, F.; Uomori, T.; Himuro, T.; Nakai, K.; Orihata, G.; Iijima, K.; Saito, M. Impact of circulating tumour cells on survival of eribulin-treated patients with metastatic breast cancer. Med. Oncol. 2019, 36, 89. [Google Scholar] [CrossRef] [PubMed]
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. RNA-Based CTC Analysis Provides Prognostic Information in Metastatic Breast Cancer. Diagnostics 2021, 11, 513. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Banys-Paluchowski, M.; Friedl, T.W.P.; Fasching, P.A.; Schneeweiss, A.; Hartkopf, A.; Wallwiener, D.; Rack, B.; Meier-Stiegen, F.; Huober, J.; et al. Prognostic relevance of the HER2 status of circulating tumor cells in metastatic breast cancer patients screened for participation in the DETECT study program. ESMO Open 2021, 6, 100299. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.L.; Adams, D.K.; Stefansson, S.; Haudenschild, C.; Martin, S.S.; Charpentier, M.; Chumsri, S.; Cristofanilli, M.; Tang, C.-M.; Alpaugh, R.K. Mitosis in circulating tumor cells stratifies highly aggressive breast carcinomas. Breast Cancer Res. 2016, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Reijm, E.A.; Sieuwerts, A.M.; Smid, M.; Vries, J.B.; Mostert, B.; Onstenk, W.; Peeters, D.; Dirix, L.Y.; Seynaeve, C.M.; Jager, A.; et al. An 8-gene mRNA expression profile in circulating tumor cells predicts response to aromatase inhibitors in metastatic breast cancer patients. BMC Cancer 2016, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, D.; Hills, A.; Page, K.; Hastings, R.K.; Toghill, B.; Goddard, K.S.; Ion, C.; Ogle, O.; Boydell, A.R.; Gleason, K.; et al. Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res. 2019, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wang, C.; Wan, S.; Mu, Z.; Zhang, Z.; Abu-Khalaf, M.M.; Fellin, F.M.; Silver, D.P.; Neupane, M.; Jaslow, R.J.; et al. Association of clinical outcomes in metastatic breast cancer patients with circulating tumour cell and circulating cell-free DNA. Eur. J. Cancer 2019, 106, 133–143. [Google Scholar] [CrossRef]
- Keup, C.; Storbeck, M.; Hauch, S.; Hahn, P.; Sprenger-Haussels, M.; Hoffmann, O.; Kimmig, R.; Kasimir-Bauer, S. Multimodal Targeted Deep Sequencing of Circulating Tumor Cells and Matched Cell-Free DNA Provides a More Comprehensive Tool to Identify Therapeutic Targets in Metastatic Breast Cancer Patients. Cancers 2020, 12, 1084. [Google Scholar] [CrossRef]
- Chimonidou, M.; Strati, A.; Malamos, N.; Kouneli, S.; Georgoulias, V.; Lianidou, E. Direct comparison study of DNA methylation markers in EpCAM-positive circulating tumour cells, corresponding circulating tumour DNA, and paired primary tumours in breast cancer. Oncotarget 2017, 8, 72054–72068. [Google Scholar] [CrossRef]
- Nanou, A.; Zeune, L.L.; Bidard, F.-C.; Pierga, J.-Y.; Terstappen, L.W.M.M. HER2 expression on tumor-derived extracellular vesicles and circulating tumor cells in metastatic breast cancer. Breast Cancer Res. 2020, 22, 86. [Google Scholar] [CrossRef]
- Hartkopf, A.D.; Stefanescu, D.; Wallwiener, M.; Hahn, M.; Becker, S.; Solomayer, E.-F.; Fehm, T.N.; Brucker, S.Y.; Taran, F.-A. Tumor cell dissemination to the bone marrow and blood is associated with poor outcome in patients with metastatic breast cancer. Breast Cancer Res. Treat. 2014, 147, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Keup, C. Comprehensive Molecular Characterization of Circulating Tumor Cells, Extracellular Vesicles and Cell-Free DNA as Matched Multi-Parametric Liquid Biopsy for Therapy Management in Metastatic Breast Cancer Patients; Duisburg-Essen Publications Online; DuEPublico, University of Duisburg-Essen: Duisburg, Germany, 2020. [Google Scholar]
- Zhu, Z.; Li, X.; Dong, H.; Ke, S.; Zheng, W.-H. Let-7f and miRNA-126 correlate with reduced cardiotoxicity risk in triple-negative breast cancer patients who underwent neoadjuvant chemotherapy. Int. J. Clin. Exp. Pathol. 2018, 11, 4987–4995. [Google Scholar] [PubMed]
- Yu, A.F.; Moore, Z.R.; Moskowitz, C.S.; Liu, J.E.; Dang, C.T.; Ramanathan, L.; Oeffinger, K.C.; Steingart, R.M.; Schmitt, A.M. Association of Circulating Cardiomyocyte Cell-Free DNA with Cancer Therapy-Related Cardiac Dysfunction in Patients Undergoing Treatment for ERBB2-Positive Breast Cancer. JAMA Cardiol. 2023, 8, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.A.; Jacob, S.; Gerratana, L.; Shah, A.N.; Wehbe, F.; Katam, N.; Zhang, Q.; Flaum, L.; Siziopikou, K.P.; Platanias, L.C.; et al. Landscape of circulating tumour DNA in metastatic breast cancer. EBioMedicine 2020, 58, 102914. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Davis, A.A.; Carneiro, B.A.; Chandra, S.; Mohindra, N.; Kalyan, A.; Kaplan, J.; Matsangou, M.; Pai, S.; Costa, R.; et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016, 7, 65364–65373. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, C.; Cani, A.K.; Larios, J.M.; Hovelson, D.H.; Aung, K.; Darga, E.P.; Cannell, E.M.; Baratta, P.J.; Liu, C.-J.; Chu, D.; et al. Comprehensive Mutation and Copy Number Profiling in Archived Circulating Breast Cancer Tumor Cells Documents Heterogeneous Resistance Mechanisms. Cancer Res. 2018, 78, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Haselmann, V.; Hedtke, M.; Neumaier, M. Liquid Profiling for Cancer Patient Stratification in Precision Medicine-Current Status and Challenges for Successful Implementation in Standard Care. Diagnostics 2022, 12, 748. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022, 387, 9–20. [Google Scholar] [CrossRef]
- Janiaud, P.; Serghiou, S.; Ioannidis, J.P.A. New clinical trial designs in the era of precision medicine: An overview of definitions, strengths, weaknesses, and current use in oncology. Cancer Treat. Rev. 2019, 73, 20–30. [Google Scholar] [CrossRef]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.D.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, C.; Kernaghan, S.; Moretti, L.; Turner, N.C.; Ring, A.; Wilkinson, K.; Martin, S.; Foster, S.; Kilburn, L.S.; Bliss, J.M. Operational complexity versus design efficiency: Challenges of implementing a phase IIa multiple parallel cohort targeted treatment platform trial in advanced breast cancer. Trials 2022, 23, 372. [Google Scholar] [CrossRef] [PubMed]
- Keup, C.; Storbeck, M.; Hauch, S.; Hahn, P.; Sprenger-Haussels, M.; Tewes, M.; Mach, P.; Hoffmann, O.; Kimmig, R.; Kasimir-Bauer, S. Cell-Free DNA Variant Sequencing Using CTC-Depleted Blood for Comprehensive Liquid Biopsy Testing in Metastatic Breast Cancer. Cancers 2019, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, F.; Ng, C.K.Y.; Patsouris, A.; Droin, N.; Piscuoglio, S.; Carbuccia, N.; Soria, J.C.; Dien, A.T.; Adnani, Y.; Kamal, M.; et al. Genomic characterization of metastatic breast cancers. Nature 2019, 569, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34, 427–438.e6. [Google Scholar] [CrossRef] [PubMed]
- Aftimos, P.; Oliveira, M.; Irrthum, A.; Fumagalli, D.; Sotiriou, C.; Gal-Yam, E.N.; Robson, M.E.; Ndozeng, J.; Di Leo, A.; Ciruelos, E.M.; et al. Genomic and Transcriptomic Analyses of Breast Cancer Primaries and Matched Metastases in AURORA, the Breast International Group (BIG) Molecular Screening Initiative. Cancer Discov. 2021, 11, 2796–2811. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Ma, F.; Li, C.; Chen, R.; Yuan, L.; Sun, X.; Guan, X.; Li, L.; Liu, B.; Guan, Y.; et al. Landscape of somatic mutations in different subtypes of advanced breast cancer with circulating tumor DNA analysis. Sci. Rep. 2017, 7, 5995. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Davis, A.A.; Xie, F.; Gui, X.; Chen, Y.; Zhang, Q.; Gerratana, L.; Zhang, Y.; Shah, A.N.; Behdad, A.; et al. Cell-free DNA comparative analysis of the genomic landscape of first-line hormone receptor-positive metastatic breast cancer from the US and China. Breast Cancer Res. Treat. 2021, 199, 213–226. [Google Scholar] [CrossRef]
- Cheng, S.; Nguyen, E.T.; Pagano, I.; Fukui, J.A. Genomic Landscape of Circulating-Tumor DNA in a Diverse Cohort of Metastatic Breast Cancer Patients. Oncol. Res. Treat. 2023, 46, 26–32. [Google Scholar] [CrossRef]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef]
- Perakis, S.O.; Weber, S.; Zhou, Q.; Graf, R.; Hojas, S.; Riedl, J.M.; Gerger, A.; Dandachi, N.; Balic, M.; Hoefler, G.; et al. Comparison of three commercial decision support platforms for matching of next-generation sequencing results with therapies in patients with cancer. ESMO Open 2020, 5, e000872. [Google Scholar] [CrossRef]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; Ng, C.K.Y.; Bedard, P.L.; Tortora, G.; Douillard, J.-Y.; et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Condorelli, R.; Mosele, F.; Verret, B.; Bachelot, T.; Bedard, P.L.; Cortes, J.; Hyman, D.M.; Juric, D.; Krop, I.; Bieche, I.; et al. Genomic alterations in breast cancer: Level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 2019, 30, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 2017, PO.17.00011. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef] [PubMed]
- MacKay, M.; Manghnani, K.; Hockenberry, A.; Drews, J.; Chen, J.; Ben-Shachar, R.; Guinney, J. Abstract P5-05-08: Dual ctDNA and tissue sequencing improves detection of actionable variants in breast cancer patients. Cancer Res. 2023, 83, P5-05-08. [Google Scholar] [CrossRef]
- Andre, F.; Filleron, T.; Kamal, M.; Mosele, F.; Arnedos, M.; Dalenc, F.; Sablin, M.-P.; Campone, M.; Bonnefoi, H.; Lefeuvre-Plesse, C.; et al. Genomics to select treatment for patients with metastatic breast cancer. Nature 2022, 610, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.A.; Schwaederlé, M.; Scur, M.D.; Boles, S.G.; Helsten, T.; Subramanian, R.; Schwab, R.B.; Kurzrock, R. Breast Cancer Experience of the Molecular Tumor Board at the University of California, San Diego Moores Cancer Center. J. Oncol. Pract. 2015, 11, 442–449. [Google Scholar] [CrossRef] [PubMed]
- van Geelen, C.T.; Savas, P.; Teo, Z.L.; Luen, S.J.; Weng, C.-F.; Ko, Y.-A.; Kuykhoven, K.S.; Caramia, F.; Salgado, R.; Francis, P.A.; et al. Clinical implications of prospective genomic profiling of metastatic breast cancer patients. Breast Cancer Res. 2020, 22, 91. [Google Scholar] [CrossRef]
- Tray, N.; Taff, J.; Singh, B.; Suh, J.; Ngo, N.; Kwa, M.; Troxel, A.B.; Chae, Y.K.; Kurzrock, R.; Patel, S.P.; et al. Metaplastic breast cancers: Genomic profiling, mutational burden and tumor-infiltrating lymphocytes. Breast 2019, 44, 29–32. [Google Scholar] [CrossRef]
- Ross, J.S.; Ali, S.M.; Wang, K.; Khaira, D.; Palma, N.A.; Chmielecki, J.; Palmer, G.A.; Morosini, D.; Elvin, J.A.; Fernandez, S.V.; et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res. Treat. 2015, 154, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Sultova, E.; Westphalen, C.B.; Jung, A.; Kumbrink, J.; Kirchner, T.; Mayr, D.; Rudelius, M.; Ormanns, S.; Heinemann, V.; Metzeler, K.H.; et al. Implementation of Precision Oncology for Patients with Metastatic Breast Cancer in an Interdisciplinary MTB Setting. Diagnostics 2021, 11, 733. [Google Scholar] [CrossRef] [PubMed]
- Bruzas, S.; Kuemmel, S.; Harrach, H.; Breit, E.; Ataseven, B.; Traut, A.; Rüland, A.; Kostara, A.; Chiari, O.; Dittmer-Grabowski, C.; et al. Next-Generation Sequencing-Directed Therapy in Patients with Metastatic Breast Cancer in Routine Clinical Practice. Cancers 2021, 13, 4564. [Google Scholar] [CrossRef] [PubMed]
- Zivanovic Bujak, A.; Weng, C.-F.; Silva, M.J.; Yeung, M.; Lo, L.; Ftouni, S.; Litchfield, C.; Ko, Y.-A.; Kuykhoven, K.; van Geelen, C.; et al. Circulating tumour DNA in metastatic breast cancer to guide clinical trial enrolment and precision oncology: A cohort study. PLoS Med. 2020, 17, e1003363. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Ismaila, N.; Henry, N.L.; Somerfield, M.R.; Bast, R.C.; Barlow, W.; Collyar, D.E.; Hammond, M.E.; Kuderer, N.M.; Liu, M.C.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: ASCO Clinical Practice Guideline Update-Integration of Results from TAILORx. J. Clin. Oncol. 2019, 37, 1956–1964. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Jacot, W.; Kiavue, N.; Dureau, S.; Kadi, A.; Brain, E.; Bachelot, T.; Bourgeois, H.; Gonçalves, A.; Ladoire, S.; et al. Efficacy of Circulating Tumor Cell Count-Driven vs Clinician-Driven First-line Therapy Choice in Hormone Receptor-Positive, ERBB2-Negative Metastatic Breast Cancer: The STIC CTC Randomized Clinical Trial. JAMA Oncol. 2020, 7, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Wang, T.; Sun, X.; Zhang, P.; Chen, Y.; Jin, J.; Hua, D. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J. Surg. Oncol. 2017, 115, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, G.; Dong, L.; Cheng, L.; Li, K.; Wang, Z.; Luo, H. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS ONE 2012, 7, e34210. [Google Scholar] [CrossRef]
- Wang, T.; Ning, K.; Lu, T.-X.; Sun, X.; Jin, L.; Qi, X.; Jin, J.; Hua, D. Increasing circulating exosomes-carrying TRPC5 predicts chemoresistance in metastatic breast cancer patients. Cancer Sci. 2017, 108, 448–454. [Google Scholar] [CrossRef]
- Ma, X.; Chen, Z.; Hua, D.; He, D.; Wang, L.; Zhang, P.; Wang, J.; Cai, Y.; Gao, C.; Zhang, X.; et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 6389–6394. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Singletary, S.E. Rating the risk factors for breast cancer. Ann. Surg. 2003, 237, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Vocka, M.; Zimovjanova, M.; Bielcikova, Z.; Tesarova, P.; Petruzelka, L.; Mateju, M.; Krizova, L.; Kotlas, J.; Soukupova, J.; Janatova, M.; et al. Estrogen Receptor Status Oppositely Modifies Breast Cancer Prognosis in BRCA1/BRCA2 Mutation Carriers Versus Non-Carriers. Cancers 2019, 11, 738. [Google Scholar] [CrossRef]
- Geyer, C.E.; Garber, J.E.; Gelber, R.D.; Yothers, G.; Taboada, M.; Ross, L.; Rastogi, P.; Cui, K.; Arahmani, A.; Aktan, G.; et al. Overall survival in the OlympiA phase III trial of adjuvant olaparib in patients with germline pathogenic variants in BRCA1/2 and high-risk, early breast cancer. Ann. Oncol. 2022, 33, 1250–1268. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Robson, M.E.; Tung, N.; Conte, P.; Im, S.-A.; Senkus, E.; Xu, B.; Masuda, N.; Delaloge, S.; Li, W.; Armstrong, A.; et al. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann. Oncol. 2019, 30, 558–566. [Google Scholar] [CrossRef]
- Litton, J.K.; Hurvitz, S.A.; Mina, L.A.; Rugo, H.S.; Lee, K.-H.; Gonçalves, A.; Diab, S.; Woodward, N.; Goodwin, A.; Yerushalmi, R.; et al. Talazoparib versus chemotherapy in patients with germline BRCA1/2-mutated HER2-negative advanced breast cancer: Final overall survival results from the EMBRACA trial. Ann. Oncol. 2020, 31, 1526–1535. [Google Scholar] [CrossRef]
- Tung, N.M.; Robson, M.E.; Ventz, S.; Santa-Maria, C.A.; Nanda, R.; Marcom, P.K.; Shah, P.D.; Ballinger, T.J.; Yang, E.S.; Vinayak, S.; et al. TBCRC 048: Phase II Study of Olaparib for Metastatic Breast Cancer and Mutations in Homologous Recombination-Related Genes. J. Clin. Oncol. 2020, 38, 4274–4282. [Google Scholar] [CrossRef]
- Loibl, S.; Marmé, F.; Martin, M.; Untch, M.; Bonnefoi, H.; Kim, S.-B.; Bear, H.; McCarthy, N.; Melé Olivé, M.; Gelmon, K.; et al. Palbociclib for Residual High-Risk Invasive HR-Positive and HER2-Negative Early Breast Cancer-The Penelope-B Trial. J. Clin. Oncol. 2021, 39, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- Grüntkemeier, L.; Khurana, A.; Bischoff, F.Z.; Hoffmann, O.; Kimmig, R.; Moore, M.; Cotter, P.; Kasimir-Bauer, S. Single HER2-positive tumor cells are detected in initially HER2-negative breast carcinomas using the DEPArray™-HER2-FISH workflow. Breast Cancer 2022, 29, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Kim, C.; Wolmark, N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N. Engl. J. Med. 2008, 358, 1409–1411. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, P.; Viale, G.; Press, M.F.; Hu, X.; Penault-Llorca, F.; Bardia, A.; Batistatou, A.; Burstein, H.J.; Carey, L.A.; Cortes, J.; et al. ESMO expert consensus statements (ECS) on the definition, diagnosis, and management of HER2-low breast cancer. Ann. Oncol. 2023, 34, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Fehm, T.; Muller, V.; Aktas, B.; Janni, W.; Schneeweiss, A.; Stickeler, E.; Lattrich, C.; Lohberg, C.R.; Solomayer, E.; Rack, B.; et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: A prospective, multicenter trial. Breast Cancer Res. Treat. 2010, 124, 403–412. [Google Scholar] [CrossRef]
- Wallwiener, M.; Hartkopf, A.D.; Riethdorf, S.; Nees, J.; Sprick, M.R.; Schönfisch, B.; Taran, F.-A.; Heil, J.; Sohn, C.; Pantel, K.; et al. The impact of HER2 phenotype of circulating tumor cells in metastatic breast cancer: A retrospective study in 107 patients. BMC Cancer 2015, 15, 403. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Müller, V.; Zhang, L.; Rau, T.; Loibl, S.; Komor, M.; Roller, M.; Huober, J.; Fehm, T.; Schrader, I.; et al. Detection and HER2 expression of circulating tumor cells: Prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin. Cancer Res. 2010, 16, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Georgoulias, V.; Bozionelou, V.; Agelaki, S.; Perraki, M.; Apostolaki, S.; Kallergi, G.; Kalbakis, K.; Xyrafas, A.; Mavroudis, D. Trastuzumab decreases the incidence of clinical relapses in patients with early breast cancer presenting chemotherapy-resistant CK-19mRNA-positive circulating tumor cells: Results of a randomized phase II study. Ann. Oncol. 2012, 23, 1744–1750. [Google Scholar] [CrossRef]
- Krawczyk, N.; Fehm, T.; Banys-Paluchowski, M.; Janni, W.; Schramm, A. Liquid Biopsy in Metastasized Breast Cancer as Basis for Treatment Decisions. Oncol. Res. Treat. 2016, 39, 112–116. [Google Scholar] [CrossRef]
- Pestrin, M.; Bessi, S.; Puglisi, F.; Minisini, A.M.; Masci, G.; Battelli, N.; Ravaioli, A.; Gianni, L.; Di Marsico, R.; Tondini, C.; et al. Final results of a multicenter phase II clinical trial evaluating the activity of single-agent lapatinib in patients with HER2-negative metastatic breast cancer and HER2-positive circulating tumor cells. A proof-of-concept study. Breast Cancer Res. Treat. 2012, 134, 283–289. [Google Scholar] [CrossRef]
- Stebbing, J.; Payne, R.; Reise, J.; Frampton, A.E.; Avery, M.; Woodley, L.; Di Leo, A.; Pestrin, M.; Krell, J.; Coombes, R.C. The efficacy of lapatinib in metastatic breast cancer with HER2 non-amplified primary tumors and EGFR positive circulating tumor cells: A proof-of-concept study. PLoS ONE 2013, 8, e62543. [Google Scholar] [CrossRef] [PubMed]
- Jacot, W.; Cottu, P.; Berger, F.; Dubot, C.; Venat-Bouvet, L.; Lortholary, A.; Bourgeois, H.; Bollet, M.; Servent, V.; Luporsi, E.; et al. Actionability of HER2-amplified circulating tumor cells in HER2-negative metastatic breast cancer: The CirCe T-DM1 trial. Breast Cancer Res. 2019, 21, 121. [Google Scholar] [CrossRef] [PubMed]
- Tellez-Gabriel, M.; Knutsen, E.; Perander, M. Current Status of Circulating Tumor Cells, Circulating Tumor DNA, and Exosomes in Breast Cancer Liquid Biopsies. Int. J. Mol. Sci. 2020, 21, 9457. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.; Kavuri, S.M.; Searleman, A.C.; Shen, W.; Shen, D.; Koboldt, D.C.; Monsey, J.; Goel, N.; Aronson, A.B.; Li, S.; et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013, 3, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Piha-Paul, S.A.; Won, H.; Rodon, J.; Saura, C.; Shapiro, G.I.; Juric, D.; Quinn, D.I.; Moreno, V.; Doger, B.; et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 2018, 554, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.X.; Bose, R.; Gao, F.; Freedman, R.A.; Telli, M.L.; Kimmick, G.; Winer, E.; Naughton, M.; Goetz, M.P.; Russell, C.; et al. Neratinib Efficacy and Circulating Tumor DNA Detection of HER2 Mutations in HER2 Nonamplified Metastatic Breast Cancer. Clin. Cancer Res. 2017, 23, 5687–5695. [Google Scholar] [CrossRef] [PubMed]
- Deniziaut, G.; Tille, J.C.; Bidard, F.-C.; Vacher, S.; Schnitzler, A.; Chemlali, W.; Trémoulet, L.; Fuhrmann, L.; Cottu, P.; Rouzier, R.; et al. ERBB2 mutations associated with solid variant of high-grade invasive lobular breast carcinomas. Oncotarget 2016, 7, 73337–73346. [Google Scholar] [CrossRef]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef]
- Bachman, K.E.; Argani, P.; Samuels, Y.; Silliman, N.; Ptak, J.; Szabo, S.; Konishi, H.; Karakas, B.; Blair, B.G.; Lin, C.; et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol. Ther. 2004, 3, 772–775. [Google Scholar] [CrossRef]
- Saal, L.H.; Holm, K.; Maurer, M.; Memeo, L.; Su, T.; Wang, X.; Yu, J.S.; Malmström, P.-O.; Mansukhani, M.; Enoksson, J.; et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005, 65, 2554–2559. [Google Scholar] [CrossRef]
- Cardoso, F.; Paluch-Shimon, S.; Senkus, E.; Curigliano, G.; Aapro, M.S.; André, F.; Barrios, C.H.; Bergh, J.; Bhattacharyya, G.S.; Biganzoli, L.; et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann. Oncol. 2020, 31, 1623–1649. [Google Scholar] [CrossRef] [PubMed]
- Moynahan, M.E.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Ringeisen, F.; Hortobagyi, G.N.; et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR+, HER2- advanced breast cancer: Results from BOLERO-2. Br. J. Cancer 2017, 116, 726–730. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Cutts, R.J.; Liu, Y.; Hrebien, S.; Huang, X.; Fenwick, K.; André, F.; Loibl, S.; Loi, S.; Garcia-Murillas, I.; et al. The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discov. 2018, 8, 1390–1403. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Hrebien, S.; Morden, J.P.; Beaney, M.; Fribbens, C.; Huang, X.; Liu, Y.; Bartlett, C.H.; Koehler, M.; Cristofanilli, M.; et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018, 9, 896. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, C.; Kagara, N.; Naoi, Y.; Shimoda, M.; Shimomura, A.; Maruyama, N.; Shimazu, K.; Kim, S.J.; Noguchi, S. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res. Treat. 2015, 150, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.J.; Jelovac, D.; Barnathan, E.; Blair, B.; Slater, S.; Powers, P.; Zorzi, J.; Jeter, S.C.; Oliver, G.R.; Fetting, J.; et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res. 2012, 18, 3462–3469. [Google Scholar] [CrossRef] [PubMed]
- Kodahl, A.R.; Ehmsen, S.; Pallisgaard, N.; Jylling, A.M.B.; Jensen, J.D.; Laenkholm, A.-V.; Knoop, A.S.; Ditzel, H.J. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol. Oncol. 2018, 12, 925–935. [Google Scholar] [CrossRef]
- Pestrin, M.; Salvianti, F.; Galardi, F.; de Luca, F.; Turner, N.; Malorni, L.; Pazzagli, M.; Di Leo, A.; Pinzani, P. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 2015, 9, 749–757. [Google Scholar] [CrossRef]
- Schneck, H.; Blassl, C.; Meier-Stiegen, F.; Neves, R.P.; Janni, W.; Fehm, T.; Neubauer, H. Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 2013, 7, 976–986. [Google Scholar] [CrossRef]
- Tzanikou, E.; Markou, A.; Politaki, E.; Koutsopoulos, A.; Psyrri, A.; Mavroudis, D.; Georgoulias, V.; Lianidou, E. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: A direct comparison study. Mol. Oncol. 2019, 13, 2515–2530. [Google Scholar] [CrossRef]
- Campone, M.; Im, S.-A.; Iwata, H.; Clemons, M.; Ito, Y.; Awada, A.; Chia, S.; Jagiełło-Gruszfeld, A.; Pistilli, B.; Tseng, L.-M.; et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant for postmenopausal, hormone receptor-positive, human epidermal growth factor receptor 2-negative, advanced breast cancer: Overall survival results from BELLE-2. Eur. J. Cancer 2018, 103, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Di Leo, A.; Johnston, S.; Lee, K.S.; Ciruelos, E.; Lønning, P.E.; Janni, W.; O’Regan, R.; Mouret-Reynier, M.-A.; Kalev, D.; Egle, D.; et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018, 19, 87–100. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Burstein, H.J.; Somerfield, M.R.; Barton, D.L.; Dorris, A.; Fallowfield, L.J.; Jain, D.; Johnston, S.R.D.; Korde, L.A.; Litton, J.K.; Macrae, E.R.; et al. Endocrine Treatment and Targeted Therapy for Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Metastatic Breast Cancer: ASCO Guideline Update. J. Clin. Oncol. 2021, 39, 3959–3977. [Google Scholar] [CrossRef]
- André, F.; Ciruelos, E.M.; Juric, D.; Loibl, S.; Campone, M.; Mayer, I.A.; Rubovszky, G.; Yamashita, T.; Kaufman, B.; Lu, Y.-S.; et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: Final overall survival results from SOLAR-1. Ann. Oncol. 2021, 32, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.L.; Somerfield, M.R.; Dayao, Z.; Elias, A.; Kalinsky, K.; McShane, L.M.; Moy, B.; Park, B.H.; Shanahan, K.M.; Sharma, P.; et al. Biomarkers for Systemic Therapy in Metastatic Breast Cancer: ASCO Guideline Update. J. Clin. Oncol. 2022, 40, 3205–3221. [Google Scholar] [CrossRef]
- Rugo, H.S.; Lerebours, F.; Ciruelos, E.; Drullinsky, P.; Ruiz-Borrego, M.; Neven, P.; Park, Y.H.; Prat, A.; Bachelot, T.; Juric, D.; et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): One cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol. 2021, 22, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Abramson, V.G.; O’Dea, A.; Nye, L.; Mayer, I.; Pathak, H.B.; Hoffmann, M.; Stecklein, S.R.; Elia, M.; Lewis, S.; et al. Clinical and Biomarker Results from Phase I/II Study of PI3K Inhibitor Alpelisib plus Nab-paclitaxel in HER2-Negative Metastatic Breast Cancer. Clin. Cancer Res. 2021, 27, 3896–3904. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.-M.; Vats, P.; Su, F.; Lonigro, R.J.; Cao, X.; Kalyana-Sundaram, S.; Wang, R.; Ning, Y.; Hodges, L.; et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013, 45, 1446–1451. [Google Scholar] [CrossRef]
- Angus, L.; Beije, N.; Jager, A.; Martens, J.W.M.; Sleijfer, S. ESR1 mutations: Moving towards guiding treatment decision-making in metastatic breast cancer patients. Cancer Treat. Rev. 2017, 52, 33–40. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Hardy-Bessard, A.-C.; Dalenc, F.; Bachelot, T.; Pierga, J.-Y.; de la Motte Rouge, T.; Sabatier, R.; Dubot, C.; Frenel, J.-S.; Ferrero, J.M.; et al. Switch to fulvestrant and palbociclib versus no switch in advanced breast cancer with rising ESR1 mutation during aromatase inhibitor and palbociclib therapy (PADA-1): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2022, 23, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Tomiguchi, M.; Sueta, A.; Murakami, K.; Iwase, H. Clinical significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and ESR1 gene according to treatment lines in ER-positive breast cancer. Mol. Cancer 2018, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016, 2, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Merenbakh-Lamin, K.; Ben-Baruch, N.; Yeheskel, A.; Dvir, A.; Soussan-Gutman, L.; Jeselsohn, R.; Yelensky, R.; Brown, M.; Miller, V.A.; Sarid, D.; et al. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013, 73, 6856–6864. [Google Scholar] [CrossRef] [PubMed]
- Toy, W.; Shen, Y.; Won, H.; Green, B.; Sakr, R.A.; Will, M.; Li, Z.; Gala, K.; Fanning, S.; King, T.A.; et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013, 45, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Jeselsohn, R.; Yelensky, R.; Buchwalter, G.; Frampton, G.; Meric-Bernstam, F.; Gonzalez-Angulo, A.M.; Ferrer-Lozano, J.; Perez-Fidalgo, J.A.; Cristofanilli, M.; Gomez, H.; et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 2014, 20, 1757–1767. [Google Scholar] [CrossRef]
- Schiavon, G.; Hrebien, S.; Garcia-Murillas, I.; Cutts, R.J.; Pearson, A.; Tarazona, N.; Fenwick, K.; Kozarewa, I.; Lopez-Knowles, E.; Ribas, R.; et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 2015, 7, 313ra182. [Google Scholar] [CrossRef]
- Sefrioui, D.; Perdrix, A.; Sarafan-Vasseur, N.; Dolfus, C.; Dujon, A.; Picquenot, J.-M.; Delacour, J.; Cornic, M.; Bohers, E.; Leheurteur, M.; et al. Short report: Monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int. J. Cancer 2015, 137, 2513–2519. [Google Scholar] [CrossRef]
- Paolillo, C.; Mu, Z.; Rossi, G.; Schiewer, M.J.; Nguyen, T.; Austin, L.; Capoluongo, E.; Knudsen, K.E.; Cristofanilli, M.; Fortina, P. Detection of Activating Estrogen Receptor Gene (ESR1) Mutations in Single Circulating Tumor Cells. Clin. Cancer Res. 2017, 23, 6086–6093. [Google Scholar] [CrossRef]
- Beije, N.; Sieuwerts, A.M.; Kraan, J.; Van, N.M.; Onstenk, W.; Vitale, S.R.; van der Vlugt-Daane, M.; Dirix, L.Y.; Brouwer, A.; Hamberg, P.; et al. Estrogen receptor mutations and splice variants determined in liquid biopsies from metastatic breast cancer patients. Mol. Oncol. 2018, 12, 48–57. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Kraan, J.; Bolt-de Vries, J.; van der Spoel, P.; Mostert, B.; Martens, J.W.M.; Gratama, J.-W.; Sleijfer, S.; Foekens, J.A. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res. Treat. 2009, 118, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, K.I. Endocrine therapy: Is the first generation of targeted drugs the last? J. Intern. Med. 2013, 274, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Rondón-Lagos, M.; Villegas, V.E.; Rangel, N.; Sánchez, M.C.; Zaphiropoulos, P.G. Tamoxifen Resistance: Emerging Molecular Targets. Int. J. Mol. Sci. 2016, 17, 1357. [Google Scholar] [CrossRef] [PubMed]
- Elliott, M.J.; Cescon, D.W. Development of novel agents for the treatment of early estrogen receptor positive breast cancer. Breast 2022, 62 (Suppl. S1), S34–S42. [Google Scholar] [CrossRef] [PubMed]
- Fribbens, C.; O’Leary, B.; Kilburn, L.; Hrebien, S.; Garcia-Murillas, I.; Beaney, M.; Cristofanilli, M.; Andre, F.; Loi, S.; Loibl, S.; et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J. Clin. Oncol. 2016, 34, 2961–2968. [Google Scholar] [CrossRef]
- Turner, N.C.; Swift, C.; Kilburn, L.; Fribbens, C.; Beaney, M.; Garcia-Murillas, I.; Budzar, A.U.; Robertson, J.F.R.; Gradishar, W.; Piccart, M.; et al. ESR1 Mutations and Overall Survival on Fulvestrant versus Exemestane in Advanced Hormone Receptor-Positive Breast Cancer: A Combined Analysis of the Phase III SoFEA and EFECT Trials. Clin. Cancer Res. 2020, 26, 5172–5177. [Google Scholar] [CrossRef]
- Burstein, H.J.; DeMichele, A.; Somerfield, M.R.; Henry, N.L. Testing for ESR1 Mutations to Guide Therapy for Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Metastatic Breast Cancer: ASCO Guideline Rapid Recommendation Update. J. Clin. Oncol. 2023, 41, 3423–3425. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Union Register of Medicinal Products for Human Use: ORSERDU. Available online: https://ec.europa.eu/health/documents/community-register/html/h1757.htm (accessed on 31 October 2023).
- Mastoraki, S.; Strati, A.; Tzanikou, E.; Chimonidou, M.; Politaki, E.; Voutsina, A.; Psyrri, A.; Georgoulias, V.; Lianidou, E. ESR1 Methylation: A Liquid Biopsy-Based Epigenetic Assay for the Follow-up of Patients with Metastatic Breast Cancer Receiving Endocrine Treatment. Clin. Cancer Res. 2018, 24, 1500–1510. [Google Scholar] [CrossRef]
- Vitale, S.R.; Ruigrok-Ritstier, K.; Timmermans, A.M.; Foekens, R.; Trapman-Jansen, A.M.A.C.; Beaufort, C.M.; Vigneri, P.; Sleijfer, S.; Martens, J.W.M.; Sieuwerts, A.M.; et al. The prognostic and predictive value of ESR1 fusion gene transcripts in primary breast cancer. BMC Cancer 2022, 22, 165. [Google Scholar] [CrossRef]
- Roßwag, S.; Cotarelo, C.L.; Pantel, K.; Riethdorf, S.; Sleeman, J.P.; Schmidt, M.; Thaler, S. Functional Characterization of Circulating Tumor Cells (CTCs) from Metastatic ER+/HER2- Breast Cancer Reveals Dependence on HER2 and FOXM1 for Endocrine Therapy Resistance and Tumor Cell Survival: Implications for Treatment of ER+/HER2- Breast Cancer. Cancers 2021, 13, 1810. [Google Scholar] [CrossRef]
- Nayar, U.; Cohen, O.; Kapstad, C.; Cuoco, M.S.; Waks, A.G.; Wander, S.A.; Painter, C.; Freeman, S.; Persky, N.S.; Marini, L.; et al. Acquired HER2 mutations in ER+ metastatic breast cancer confer resistance to estrogen receptor-directed therapies. Nat. Genet. 2019, 51, 207–216. [Google Scholar] [CrossRef]
- Formisano, L.; Lu, Y.; Servetto, A.; Hanker, A.B.; Jansen, V.M.; Bauer, J.A.; Sudhan, D.R.; Guerrero-Zotano, A.L.; Croessmann, S.; Guo, Y.; et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 2019, 10, 1373. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Cohen, O.; Kowalski, K.J.; Kusiel, J.G.; Buendia-Buendia, J.E.; Cuoco, M.S.; Exman, P.; Wander, S.A.; Waks, A.G.; Nayar, U.; et al. Acquired FGFR and FGF Alterations Confer Resistance to Estrogen Receptor (ER) Targeted Therapy in ER+ Metastatic Breast Cancer. Clin. Cancer Res. 2020, 26, 5974–5989. [Google Scholar] [CrossRef]
- Rudolph, M.; Anzeneder, T.; Schulz, A.; Beckmann, G.; Byrne, A.T.; Jeffers, M.; Pena, C.; Politz, O.; Köchert, K.; Vonk, R.; et al. AKT1 (E17K) mutation profiling in breast cancer: Prevalence, concurrent oncogenic alterations, and blood-based detection. BMC Cancer 2016, 16, 622. [Google Scholar] [CrossRef] [PubMed]
- Franken, A.; Rivandi, M.; Yang, L.; Jäger, B.; Krawczyk, N.; Honisch, E.; Niederacher, D.; Fehm, T.; Neubauer, H. A Multiplex PCR-Based Next Generation Sequencing-Panel to Identify Mutations for Targeted Therapy in Breast Cancer Circulating Tumor Cells. Appl. Sci. 2020, 10, 3364. [Google Scholar] [CrossRef]
- Franken, A.; Behrens, B.; Reinhardt, F.; Yang, L.; Rivandi, M.; Marass, F.; Jaeger, B.; Krawczyk, N.; Cieslik, J.-P.; Honisch, E.; et al. Multiparametric Circulating Tumor Cell Analysis to Select Targeted Therapies for Breast Cancer Patients. Cancers 2021, 13, 6004. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Oliveira, M.; Howell, S.J.; Dalenc, F.; Cortes, J.; Gomez Moreno, H.L.; Hu, X.; Jhaveri, K.; Krivorotko, P.; Loibl, S.; et al. Capivasertib in Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2023, 388, 2058–2070. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Abraham, J.; Chan, S.; Wheatley, D.; Brunt, A.M.; Nemsadze, G.; Baird, R.D.; Park, Y.H.; Hall, P.S.; Perren, T.; et al. Capivasertib Plus Paclitaxel Versus Placebo Plus Paclitaxel As First-Line Therapy for Metastatic Triple-Negative Breast Cancer: The PAKT Trial. J. Clin. Oncol. 2020, 38, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Keup, C.; Kimmig, R.; Kasimir-Bauer, S. Liquid Biopsies to Evaluate Immunogenicity of Gynecological/Breast Tumors: On the Way to Blood-Based Biomarkers for Immunotherapies. Breast Care 2020, 15, 470–480. [Google Scholar] [CrossRef]
- Stenzinger, A.; van Tilburg, C.M.; Tabatabai, G.; Länger, F.; Graf, N.; Griesinger, F.; Heukamp, L.C.; Hummel, M.; Klingebiel, T.; Hettmer, S.; et al. Diagnostik und Therapie von Tumoren mit NTRK-Genfusionen. Pathologe 2021, 42, 103–115. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Niemeier, L.A.; Dabbs, D.J.; Beriwal, S.; Striebel, J.M.; Bhargava, R. Androgen receptor in breast cancer: Expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod. Pathol. 2010, 23, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.C.; Cole, K.S.; Marotti, J.D.; Hu, R.; Schnitt, S.J.; Tamimi, R.M. Androgen receptor expression in breast cancer in relation to molecular phenotype: Results from the Nurses’ Health Study. Mod. Pathol. 2011, 24, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Vera-Badillo, F.E.; Templeton, A.J.; de Gouveia, P.; Diaz-Padilla, I.; Bedard, P.L.; Al-Mubarak, M.; Seruga, B.; Tannock, I.F.; Ocana, A.; Amir, E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, djt319. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Traina, T.A.; Miller, K.; Yardley, D.A.; Eakle, J.; Schwartzberg, L.S.; O’Shaughnessy, J.; Gradishar, W.; Schmid, P.; Winer, E.; Kelly, C.; et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 884–890. [Google Scholar] [CrossRef] [PubMed]
- de Kruijff, I.E.; Sieuwerts, A.M.; Onstenk, W.; Jager, A.; Hamberg, P.; de Jongh, F.E.; Smid, M.; Kraan, J.; Timmermans, M.A.; Martens, J.W.M.; et al. Androgen receptor expression in circulating tumor cells of patients with metastatic breast cancer. Int. J. Cancer 2019, 145, 1083–1089. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Keup, C.; Hoffmann, O.; Hauch, S.; Kimmig, R.; Bittner, A.-K. Circulating Tumor Cells Expressing the Prostate Specific Membrane Antigen (PSMA) Indicate Worse Outcome in Primary, Non-Metastatic Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 22. [Google Scholar] [CrossRef]
- Galardi, F.; de Luca, F.; Biagioni, C.; Migliaccio, I.; Curigliano, G.; Minisini, A.M.; Bonechi, M.; Moretti, E.; Risi, E.; McCartney, A.; et al. Circulating tumor cells and palbociclib treatment in patients with ER-positive, HER2-negative advanced breast cancer: Results from a translational sub-study of the TREnd trial. Breast Cancer Res. 2021, 23, 38. [Google Scholar] [CrossRef]
- Dandachi, N.; Posch, F.; Graf, R.; Suppan, C.; Klocker, E.V.; Müller, H.D.; Lindenmann, J.; Terbuch, A.; Heitzer, E.; Balic, M. Longitudinal tumor fraction trajectories predict risk of progression in metastatic HR+ breast cancer patients undergoing CDK4/6 treatment. Mol. Oncol. 2020, 15, 2390–2400. [Google Scholar] [CrossRef] [PubMed]
- Gyanchandani, R.; Kota, K.J.; Jonnalagadda, A.R.; Minteer, T.; Knapick, B.A.; Oesterreich, S.; Brufsky, A.M.; Lee, A.V.; Puhalla, S.L. Detection of ESR1 mutations in circulating cell-free DNA from patients with metastatic breast cancer treated with palbociclib and letrozole. Oncotarget 2017, 8, 66901–66911. [Google Scholar] [CrossRef] [PubMed]
- Tolaney, S.M.; Toi, M.; Neven, P.; Sohn, J.; Grischke, E.-M.; Llombart-Cussac, A.; Soliman, H.; Wang, H.; Wijayawardana, S.; Jansen, V.M.; et al. Clinical Significance of PIK3CA and ESR1 Mutations in Circulating Tumor DNA: Analysis from the MONARCH 2 Study of Abemaciclib plus Fulvestrant. Clin. Cancer Res. 2022, 28, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Turner, N.C.; Bondarenko, I.; Ro, J.; Im, S.-A.; Masuda, N.; Colleoni, M.; DeMichele, A.; Loi, S.; Verma, S.; et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016, 17, 425–439. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Cutts, R.; Huang, X.; Hrebien, S.; Liu, Y.; Garcia-Murillas, I.; Andre, F.; Loi, S.; Loibl, S.; Cristofanilli, M.; et al. Genomic markers of early progression on fulvestrant with or without palbociclib for ER+ advanced breast cancer in the PALOMA-3 trial. J. Clin. Oncol. 2019, 37, 1010. [Google Scholar] [CrossRef]
- O’Leary, B.; Cutts, R.J.; Huang, X.; Hrebien, S.; Liu, Y.; André, F.; Loibl, S.; Loi, S.; Garcia-Murillas, I.; Cristofanilli, M.; et al. Circulating Tumor DNA Markers for Early Progression on Fulvestrant With or Without Palbociclib in ER+ Advanced Breast Cancer. J. Natl. Cancer Inst. 2021, 113, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Crucitta, S.; Lorenzini, G.; de Angelis, C.; Diodati, L.; Cavallero, D.; Bargagna, I.; Cinacchi, P.; Fratini, B.; Salvadori, B.; et al. PI3K mutations detected in liquid biopsy are associated to reduced sensitivity to CDK4/6 inhibitors in metastatic breast cancer patients. Pharmacol. Res. 2021, 163, 105241. [Google Scholar] [CrossRef]
- Bardia, A.; Su, F.; Solovieff, N.; Im, S.-A.; Sohn, J.; Lee, K.S.; Campos-Gomez, S.; Jung, K.H.; Colleoni, M.; Vázquez, R.V.; et al. Genomic Profiling of Premenopausal HR+ and HER2- Metastatic Breast Cancer by Circulating Tumor DNA and Association of Genetic Alterations With Therapeutic Response to Endocrine Therapy and Ribociclib. JCO Precis. Oncol. 2021, 5, 1408–1420. [Google Scholar] [CrossRef]
- Finn, R.S.; Crown, J.P.; Lang, I.; Boer, K.; Bondarenko, I.M.; Kulyk, S.O.; Ettl, J.; Patel, R.; Pinter, T.; Schmidt, M.; et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015, 16, 25–35. [Google Scholar] [CrossRef]
- Bourrier, C.; Pierga, J.-Y.; Xuereb, L.; Salaun, H.; Proudhon, C.; Speicher, M.R.; Belic, J.; Heitzer, E.; Lockhart, B.P.; Guigal-Stephan, N. Shallow Whole-Genome Sequencing from Plasma Identifies FGFR1 Amplified Breast Cancers and Predicts Overall Survival. Cancers 2020, 12, 1481. [Google Scholar] [CrossRef]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Petrakova, K.; Blackwell, K.L.; Winer, E.P.; et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann. Oncol. 2018, 29, 1541–1547. [Google Scholar] [CrossRef]
- Slamon, D.J.; Neven, P.; Chia, S.; Fasching, P.A.; de Laurentiis, M.; Im, S.-A.; Petrakova, K.; Bianchi, G.V.; Esteva, F.J.; Martín, M.; et al. Phase III Randomized Study of Ribociclib and Fulvestrant in Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: MONALEESA-3. J. Clin. Oncol. 2018, 36, 2465–2472. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Neven, P.; Chia, S.; Jerusalem, G.; de Laurentiis, M.; Im, S.; Petrakova, K.; Valeria Bianchi, G.; Martín, M.; Nusch, A.; et al. Ribociclib plus fulvestrant for postmenopausal women with hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer in the phase III randomized MONALEESA-3 trial: Updated overall survival. Ann. Oncol. 2021, 32, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Bertolini, I.; Crucitta, S.; Fontanelli, L.; Rofi, E.; de Angelis, C.; Diodati, L.; Cavallero, D.; Gianfilippo, G.; Salvadori, B.; et al. Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res. Treat. 2019, 178, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.S.; Cobain, E.F.; Dayao, Z.; Somerfield, M.R.; DeMichele, A.; Henry, N.L. Biomarkers for Systemic Therapy in Metastatic Breast Cancer: ASCO Guideline Update Q and A. JCO Oncol. Pract. 2022, 18, 830–832. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Attard, G.; Bidard, F.-C.; Curigliano, G.; de Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- Jongbloed, E.M.; Deger, T.; Sleijfer, S.; Martens, J.W.M.; Jager, A.; Wilting, S.M. A Systematic Review of the Use of Circulating Cell-Free DNA Dynamics to Monitor Response to Treatment in Metastatic Breast Cancer Patients. Cancers 2021, 13, 1811. [Google Scholar] [CrossRef] [PubMed]
- van Poznak, C.; Somerfield, M.R.; Bast, R.C.; Cristofanilli, M.; Goetz, M.P.; Gonzalez-Angulo, A.M.; Hicks, D.G.; Hill, E.G.; Liu, M.C.; Lucas, W.; et al. Use of Biomarkers to Guide Decisions on Systemic Therapy for Women With Metastatic Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2015, 33, 2695–2704. [Google Scholar] [CrossRef]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W.W.M. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef]
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W.M.M.; et al. Circulating tumor cells versus imaging—Predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef] [PubMed]
- Cabel, L.; Berger, F.; Cottu, P.; Loirat, D.; Rampanou, A.; Brain, E.; Cyrille, S.; Bourgeois, H.; Kiavue, N.; Deluche, E.; et al. Clinical utility of circulating tumour cell-based monitoring of late-line chemotherapy for metastatic breast cancer: The randomised CirCe01 trial. Br. J. Cancer 2021, 124, 1207–1213. [Google Scholar] [CrossRef]
- Helissey, C.; Berger, F.; Cottu, P.; Diéras, V.; Mignot, L.; Servois, V.; Bouleuc, C.; Asselain, B.; Pelissier, S.; Vaucher, I.; et al. Circulating tumor cell thresholds and survival scores in advanced metastatic breast cancer: The observational step of the CirCe01 phase III trial. Cancer Lett. 2015, 360, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, T.M.; Stefanovic, S.; Feisst, M.; Fischer, C.; Riedel, F.; Fremd, C.; Domschke, C.; Pantel, K.; Hartkopf, A.D.; Sutterlin, M.; et al. Cut-Off Analysis of CTC Change under Systemic Therapy for Defining Early Therapy Response in Metastatic Breast Cancer. Cancers 2020, 12, 1055. [Google Scholar] [CrossRef] [PubMed]
- Agelaki, S.; Kalykaki, A.; Markomanolaki, H.; Papadaki, M.A.; Kallergi, G.; Hatzidaki, D.; Kalbakis, K.; Mavroudis, D.; Georgoulias, V. Efficacy of Lapatinib in Therapy-Resistant HER2-Positive Circulating Tumor Cells in Metastatic Breast Cancer. PLoS ONE 2015, 10, e0123683. [Google Scholar] [CrossRef] [PubMed]
- Pantano, F.; Rossi, E.; Iuliani, M.; Facchinetti, A.; Simonetti, S.; Ribelli, G.; Zoccoli, A.; Vincenzi, B.; Tonini, G.; Zamarchi, R.; et al. Dynamic changes of Receptor activator of nuclear factor-κB expression in Circulating Tumor Cells during Denosumab predict treatment effectiveness in Metastatic Breast Cancer. Sci. Rep. 2020, 10, 1288. [Google Scholar] [CrossRef] [PubMed]
- Bredemeier, M.; Edimiris, P.; Tewes, M.; Mach, P.; Aktas, B.; Schellbach, D.; Wagner, J.; Kimmig, R.; Kasimir-Bauer, S. Establishment of a multimarker qPCR panel for the molecular characterization of circulating tumor cells in blood samples of metastatic breast cancer patients during the course of palliative treatment. Oncotarget 2016, 7, 41677–41690. [Google Scholar] [CrossRef]
- Bredemeier, M.; Edimiris, P.; Mach, P.; Kubista, M.; Sjöback, R.; Rohlova, E.; Kolostova, K.; Hauch, S.; Aktas, B.; Tewes, M.; et al. Gene Expression Signatures in Circulating Tumor Cells Correlate with Response to Therapy in Metastatic Breast Cancer. Clin. Chem. 2017, 63, 1585–1593. [Google Scholar] [CrossRef]
- Keup, C.; Mach, P.; Aktas, B.; Tewes, M.; Kolberg, H.-C.; Hauch, S.; Sprenger-Haussels, M.; Kimmig, R.; Kasimir-Bauer, S. RNA Profiles of Circulating Tumor Cells and Extracellular Vesicles for Therapy Stratification of Metastatic Breast Cancer Patients. Clin. Chem. 2018, 64, 1054–1062. [Google Scholar] [CrossRef]
- Visvanathan, K.; Cope, L.; Fackler, M.J.; Considine, M.; Sokoll, L.; Carey, L.A.; Forero-Torres, A.; Ingle, J.N.; Lin, N.U.; Nanda, R.; et al. Evaluation of a Liquid Biopsy-Breast Cancer Methylation (LBx-BCM) Cartridge Assay for Predicting Early Disease Progression and Survival: TBCRC 005 Prospective Trial. Clin. Cancer Res. 2023, 29, 784–790. [Google Scholar] [CrossRef]
- Gerratana, L.; Davis, A.A.; Zhang, Q.; Basile, D.; Rossi, G.; Strickland, K.; Franzoni, A.; Allegri, L.; Mu, Z.; Zhang, Y.; et al. Longitudinal Dynamics of Circulating Tumor Cells and Circulating Tumor DNA for Treatment Monitoring in Metastatic Breast Cancer. JCO Precis. Oncol. 2021, 5, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.Y.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S.C.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, T.; Yang, Z.; Zheng, Y.; Yu, R.; Wu, X.; Yan, J.; Shao, Y.W.; Shao, X.; Cao, W.; et al. Monitoring treatment efficacy and resistance in breast cancer patients via circulating tumor DNA genomic profiling. Mol. Genet. Genomic Med. 2020, 8, e1079. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sáez, O.; Pascual, T.; Brasó-Maristany, F.; Chic, N.; González-Farré, B.; Sanfeliu, E.; Rodríguez, A.; Martínez, D.; Galván, P.; Rodríguez, A.B.; et al. Circulating tumor DNA dynamics in advanced breast cancer treated with CDK4/6 inhibition and endocrine therapy. NPJ Breast Cancer 2021, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-B.; Dent, R.; Im, S.-A.; Espié, M.; Blau, S.; Tan, A.R.; Isakoff, S.J.; Oliveira, M.; Saura, C.; Wongchenko, M.J.; et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2017, 18, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Clouthier, D.L.; Lien, S.C.; Yang, S.Y.C.; Nguyen, L.T.; Manem, V.S.K.; Gray, D.; Ryczko, M.; Razak, A.R.A.; Lewin, J.; Lheureux, S.; et al. An interim report on the investigator-initiated phase 2 study of pembrolizumab immunological response evaluation (INSPIRE). J. Immunother. Cancer 2019, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Baird, R.D.; van Rossum, A.G.J.; Oliveira, M.; Beelen, K.; Gao, M.; Schrier, M.; Mandjes, I.A.M.; Garcia-Corbacho, J.; Vallier, A.-L.; Dougall, G.; et al. POSEIDON Trial Phase 1b Results: Safety, Efficacy and Circulating Tumor DNA Response of the Beta Isoform-Sparing PI3K Inhibitor Taselisib (GDC-0032) Combined with Tamoxifen in Hormone Receptor Positive Metastatic Breast Cancer Patients. Clin. Cancer Res. 2019, 25, 6598–6605. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Geuna, E.; Mussolin, B.; Crisafulli, G.; Bartolini, A.; Galizia, D.; Casorzo, L.; Sarotto, I.; Scaltriti, M.; Sapino, A.; et al. Genotyping tumour DNA in cerebrospinal fluid and plasma of a HER2-positive breast cancer patient with brain metastases. ESMO Open 2017, 2, e000253. [Google Scholar] [CrossRef]
- Bratman, S.V.; Yang, S.Y.C.; Iafolla, M.A.J.; Liu, Z.; Hansen, A.R.; Bedard, P.L.; Lheureux, S.; Spreafico, A.; Razak, A.A.; Shchegrova, S.; et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer 2020, 7, 131. [Google Scholar] [CrossRef]
- Darrigues, L.; Pierga, J.-Y.; Bernard-Tessier, A.; Bièche, I.; Silveira, A.B.; Michel, M.; Loirat, D.; Cottu, P.; Cabel, L.; Dubot, C.; et al. Circulating tumor DNA as a dynamic biomarker of response to palbociclib and fulvestrant in metastatic breast cancer patients. Breast Cancer Res. 2021, 23, 31. [Google Scholar] [CrossRef]
- Jeannot, E.; Darrigues, L.; Michel, M.; Stern, M.-H.; Pierga, J.-Y.; Rampanou, A.; Melaabi, S.; Benoist, C.; Bièche, I.; Vincent-Salomon, A.; et al. A single droplet digital PCR for ESR1 activating mutations detection in plasma. Oncogene 2020, 39, 2987–2995. [Google Scholar] [CrossRef] [PubMed]
- Clatot, F.; Perdrix, A.; Beaussire, L.; Lequesne, J.; Lévy, C.; Emile, G.; Bubenheim, M.; Lacaille, S.; Calbrix, C.; Augusto, L.; et al. Risk of early progression according to circulating ESR1 mutation, CA-15.3 and cfDNA increases under first-line anti-aromatase treatment in metastatic breast cancer. Breast Cancer Res. 2020, 22, 56. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.-J.; Rosenfeld, N.; Caldas, C. Circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 369, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Keup, C.; Suryaprakash, V.; Storbeck, M.; Hoffmann, O.; Kimmig, R.; Kasimir-Bauer, S. Longitudinal Multi-Parametric Liquid Biopsy Approach Identifies Unique Features of Circulating Tumor Cell, Extracellular Vesicle, and Cell-Free DNA Characterization for Disease Monitoring in Metastatic Breast Cancer Patients. Cells 2021, 10, 212. [Google Scholar] [CrossRef]
- Venesio, T.; Siravegna, G.; Bardelli, A.; Sapino, A. Liquid Biopsies for Monitoring Temporal Genomic Heterogeneity in Breast and Colon Cancers. Pathobiology 2017, 85, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Aucamp, J.; Pretorius, P.J. Cell-free DNA: Preanalytical variables. Clin. Chim. Acta 2015, 450, 243–253. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Lampignano, R.; Neumann, M.H.D.; Weber, S.; Kloten, V.; Herdean, A.; Voss, T.; Groelz, D.; Babayan, A.; Tibbesma, M.; Schlumpberger, M.; et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2019, 66, 149–160. [Google Scholar] [CrossRef]
- Godsey, J.H.; Silvestro, A.; Barrett, J.C.; Bramlett, K.; Chudova, D.; Deras, I.; Dickey, J.; Hicks, J.; Johann, D.J.; Leary, R.; et al. Generic Protocols for the Analytical Validation of Next-Generation Sequencing-Based ctDNA Assays: A Joint Consensus Recommendation of the BloodPAC’s Analytical Variables Working Group. Clin. Chem. 2020, 66, 1156–1166. [Google Scholar] [CrossRef]
- Bossuyt, P.M.M. Clinical validity: Defining biomarker performance. Scand. J. Clin. Lab. Investig. Suppl. 2010, 242, 46–52. [Google Scholar] [CrossRef]
- Pletcher, M.J.; Pignone, M. Evaluating the clinical utility of a biomarker: A review of methods for estimating health impact. Circulation 2011, 123, 1116–1124. [Google Scholar] [CrossRef]
- IJzerman, M.J.; de Boer, J.; Azad, A.; Degeling, K.; Geoghegan, J.; Hewitt, C.; Hollande, F.; Lee, B.; To, Y.H.; Tothill, R.W.; et al. Towards Routine Implementation of Liquid Biopsies in Cancer Management: It Is Always Too Early, until Suddenly It Is Too Late. Diagnostics 2021, 11, 103. [Google Scholar] [CrossRef]
- Douglas, M.P.; Gray, S.W.; Phillips, K.A. Private Payer and Medicare Coverage for Circulating Tumor DNA Testing: A Historical Analysis of Coverage Policies from 2015 to 2019. J. Natl. Compr. Canc. Netw. 2020, 18, 866–872. [Google Scholar] [CrossRef] [PubMed]
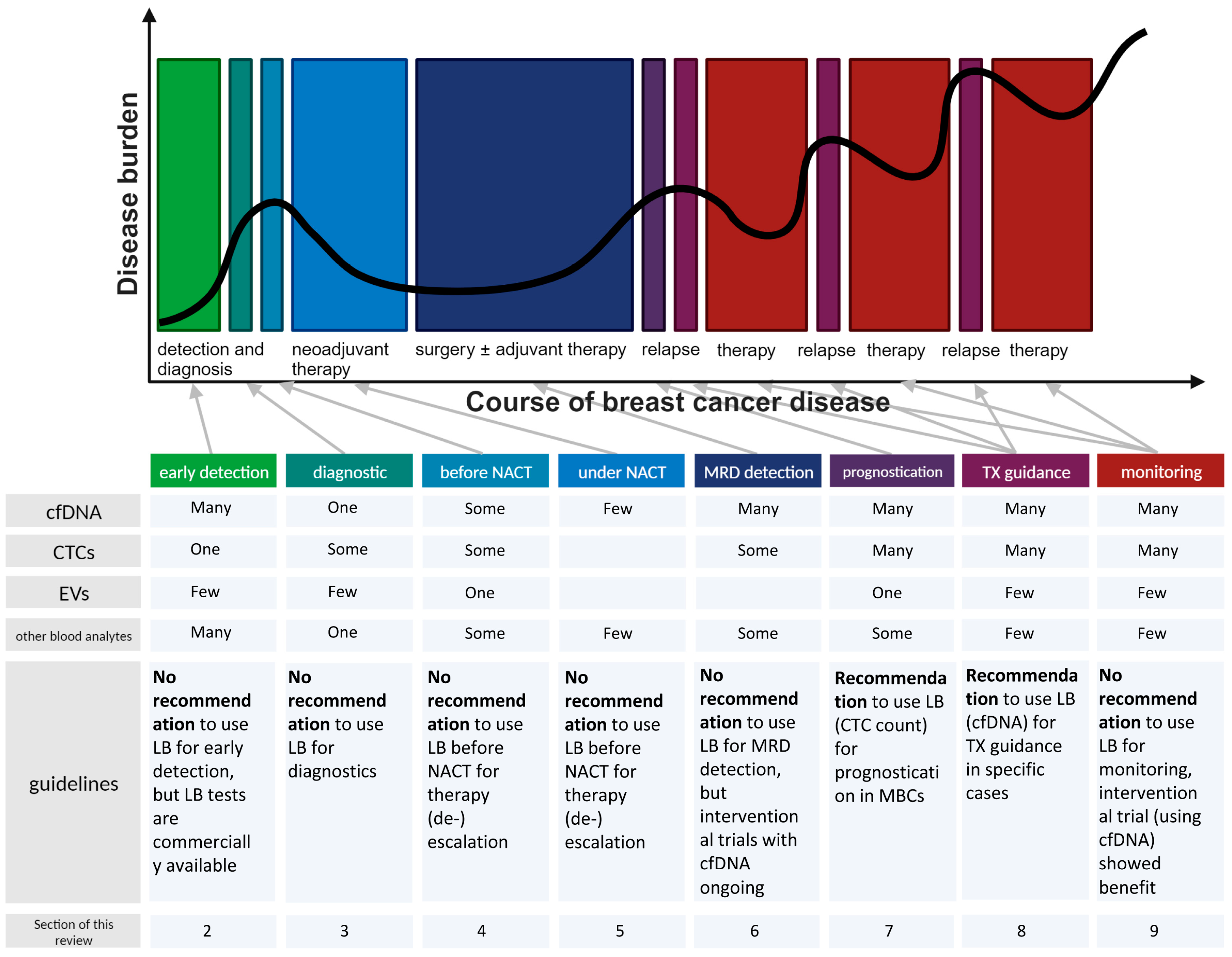

| Early Detection | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| targeted cfDNA mutation analysis combined with the evaluation of circulating proteins | DETECT-A study, n = 10,006 | specificity of 99% and a sensitivity of 33% to detect eight tumor types, including BC | 10.1126/science.aar3247 |
| different dimensions for cfDNA analysis | CCGA, NCT02889978 | Whole genome bisulphite sequencing for methylation analysis and targeted sequencing single nucleotide variants with paired white blood cell background removal showed the lowest limit of detection | 10.1016/j.ccell.2022.10.022 |
| targeted methylation cfDNA sequencing | CCGA sub-study 2 and the STRIVE study (NCT03085888) | The specificity within all covered cancer entities was 99.3% and the sensitivity to detect BCs with stage I disease was <10% (stage I), 50% (stage II) and >80% (stage III or IV). | 10.1016/j.annonc.2020.02.011 |
| targeted methylation cfDNA sequencing panel of >100,000 regions | CCGA sub-study 3 | specificity was 99.5% (low false-positive rate of 0.5%). Overall sensitivity across cancer classes and stages was 51.5%, but for BC only 30.5% across all stages | 10.1016/j.annonc.2021.05.806 |
| targeted methylation cfDNA sequencing panel of >100,000 regions | CCGA sub-study 3 in patients with symptoms only | increased overall sensitivity of 64.3% (52.8% for BC) and the overall accuracy of the cancer site of origin prediction in true positives was 90.3% | 10.1200/PO.22.00679 |
| targeted cfDNA methylation-based MCED test, here referred to as Galleri (MCED-Scr; 30,000 CpG fragments covered) | PATHFINDER study (NCT04241796), in adults with elevated cancer risk | Adding Galleri test to standard of care screening more than doubled the number of cancers detected. Half of the cancers detected with the blood test, were stage I or II. Accuracy of tissue of origin was 97.1%. In total, 71% of participants with a Galleri detected cancer had cancer types with no routine screening test available. PPV was 43.1% and the false-positive rate was less than 1% | 10.3390/diagnostics12051244 and ESMO Congress 2022 |
| cfDNA concentration | 61 patients with breast cancer, 33 control patients with benign breast diseases and 27 healthy control individuals | cfDNA concentration in the BC patients was significantly higher than that in the control patients or healthy control. | 10.1016/j.canlet.2005.11.027 |
| cfDNA PIK3CA mutations | sensitivity of 93.3% and a specificity of 100% for detecting early-stage BC | 10.1158/1078-0432.CCR-13-2933 | |
| cfDNA methylation (pyroseq of 3 genes: SPAG6, PER1 and ITIH5; SNiPER) | plasma cohort (n = 125) | 64% sensitivity for breast cancer detection using SPAG6, PER1 and ITIH5 | 10.18632/oncotarget.27303 |
| cfDNA methylation (one gene: APC) | meta-analysis of 12 studies | low sensitivity (20%) but high specificity (96%) for detecting breast cancer | 10.1111/1759-7714.12580 |
| cfDNA methylation (2 genes) | 94.1% sensitivity | 10.1016/j.ygyno.2010.04.016 | |
| cfDNA methylation (8 genes) | 90% sensitivity | 10.1371/journal.pone.0016080 | |
| cfDNA methylation (3 genes) | greater sensitivity than the serum markers CEA and/or CA15-3 | 10.1007/s10549-011-1575-2 | |
| cfDNA methylation (EGFR and PPM1E promoter) | patients with BC had significantly higher methylation levels than healthy controls | 10.1007/s13277-016-5190-z | |
| cfDNA integrity index | patients with confirmed malignancy had significantly greater DNA damage than those with benign breast lesions and healthy controls | 10.1007/s13277-015-4624-3 | |
| cfDNA mutations in breast milk | 10 women diagnosed with BC during pregnancy and 9 diagnosed during breastfeeding: 12 healthy donors | Variants in cfDNA from breast milk detected in 87% of the cases, while undetected in 92% of matched plasma. Overall clinical sensitivity of 71.4% and specificity of 100%. In two cases, ctDNA was detectable in BM collected 18 and 6 months prior to standard diagnosis. | 10.1158/2159-8290.CD-22-1340 |
| (2) CTCs | |||
| CTCs by nuclease-activated probe technology | discrimination between BC patients and healthy controls | 10.1016/j.omtn.2017.08.004 | |
| (3) EVs | |||
| EVs with specific proteomic profiles, including immunoglobulins | 426 human samples | 95% sensitivity/90% specificity in detecting cancer | 10.1016/j.cell.2020.07.009 |
| EVs with CD82 | quantification for early diagnosis of BC | 10.1002/mc.22960 | |
| EVs including unique tRNAs and miR-10b and miR-21 | EVs BC patients contain miR-10b and miR21 and unique tRNA (in contrast to healthy controls that do not convert pre-miRNA of these two types) | 10.1158/1541-7786.MCR-14-0533 | |
| EVs including miR-21 and miR-1246 | significant higher concentration in BC patients than in healthy controls | 10.1186/s13058-016-0753-x | |
| EVs with long non-coding RNA (lncRNA H19) | H19 expression in EVs was significantly upregulated in the serum of patients with BC as compared to patients without malignancy | 10.2147/OTT.S243601 | |
| (4) other blood analytes | |||
| circulating miRNAs (8 miRNAs) | Serum samples from 116 malignant breast lesions and 64 benign breast lesions | area under the curve (AUC) of 0.9542 with eight-miRNA signature | 10.3390/cancers11121872 |
| circulating miRNAs (5 miRNAs) | Combination of miR-1246, miR-1307-3p, miR-4634, miR-6861-5p, and miR-6875-5p was shown to detect early-stage BC with sensitivity of 97.3%, specificity of 82.9% and accuracy of 89.7%. | 10.1111/cas.12880 | |
| circulating miRNAs | 55 patients with metastatic breast cancer and 20 healthy donors | miR-21, miR-146a, and miR-210 could discriminate patients from healthy individuals | 10.1373/clinchem.2015.253716 |
| proteins (afamin, apolipoprotein E, alpha-2-macroglobulin and ceruloplasmin) | 68 women diagnosed with BC within three years after enrollment, with 68 matched controls | Afamin, apolipoprotein E and ITIH4 were found in higher concentration in pre-diagnostic breast cancer (p < 0.05), while alpha-2-macroglobulin and ceruloplasmin were lower (p < 0.05). | 10.1186/1471-2407-11-381 |
| proteins (integrin subunit alpha, Filamin A, Ras-associated protein-1A and Talin-1) | 20 patients with BC and 20 female control individuals with positive mammograms and benign pathology at biopsy | 4 proteins classified breast cancer patients with 100% sensitivity and 85% specificity (AUC of 0.93) | 10.1186/s13058-020-01373-9 |
| proteins (Cyr 61) in plasma | 544 patients BC and 427 healthy controls | specificity of 99.0% and sensitivity of 80.0% for cancer detection | 10.1093/clinchem/hvab153 |
| volatile organic compounds in the urine | sensitivity of 93.3% and specificity of 83.3% | 10.1038/s41598-021-99396-5 | |
| (5) multiple blood analytes | |||
| cfDNA mutation and cfDNA methylation as well as circulating miRNA information | 205 patients with stage I, II, or III cancer prior to cancer therapy and 15 healthy controls | combination of three different analytes could improve the sensitivity for cancer detection | 10.3390/cancers14020462 |
| cfDNA fragmentation combined with cfDNA mutation analysis | sensitivity 91% and specificity 98% with combined workflow | 10.1038/s41586-019-1272-6 | |
| cfDNA integrity, in combination with the detection of CTCs | 84 patients with no-distant metastatic BC and 30 patients with benign breast tumors | Combination of CTCs with cfDI: false positive rate 10.71% andarea under the curve value 0.68. | 10.4149/neo_2017_417 |
| combination of circulating mRNAs and a protein | Eight mRNAs (S100A8, GRIK1, GRM1, H6PD, IGF2BP1, CSTA, MDM4,and TPT1) and the CA6 protein were able to distinguish BC patients and healthy controls. Diagnostic accuracy: 92% (sensitivity of 83% and specificity of 97%). | 10.1371/journal.pone.0015573 | |
| Diagnostic | ||
|---|---|---|
| Specific Analyte | Conclusion | Reference |
| (1) cfDNA | ||
| nucleosome position and accessibility of cfDNA | differentiate ER-positive from ER-negative MBCs | 10.1038/s41467-022-35076-w |
| ctDNA fraction | High ctDNA fraction itself has already been shown to correlate with TNBC status, and also high tumor grade and metastatic status | 10.1038/s41523-021-00319-4 |
| (2) CTCs | ||
| CTC number | A significantly increased number of CTCs, determined by CellSearch, before therapy was reported for MBCs with lobular compared to ductal histology | 10.3390/cells9071718 |
| AR on CTCs | androgen receptor (AR) expression on CTCs was correlated with bone metastasis | 10.1158/1541-7786.MCR-17-0480 |
| number of apoptotic CTCs | higher number of apoptotic CTCs was detected in early in contrast to metastatic BC patients | 10.1158/1535-7163.MCT-12-1167 |
| (3) EVs | ||
| EV miRNA-373 | EV miRNA-373 was increased in the blood of TNBCs patients compared to patients with other BC subtypes | 10.18632/oncotarget.2520 |
| EV mRNA | Profiling of PAM50 transcripts in EVs showed good concordance with tissue results. | 10.1021/acs.analchem.3c00624 |
| 45 miRNAs in EVs | panel of 45 miRNAs detected in plasma EVs of BC patients differentiated HER2-positive from TNBC patients | 10.1186/s12916-018-1163-y |
| EV miR-21 | MBCs were shown to have significantly higher EV miR-21 levels compared to BC patients with no metastases | 10.1186/s13058-019-1109-0 |
| EV miR-223-3p | EV miR-223-3p was significantly increased in invasive ductal carcinoma patients compared to subjects with ductal carcinoma in situ (DCIS) | 10.3892/ol.2018.8457 |
| (4) other blood analytes | ||
| Circular RNA, circ_0001785 | circ_0001785 is proposed to be correlated with distant metastasis and histology | 10.1016/j.cca.2017.10.011 |
| (5) multiple blood analytes | ||
| 56-gene cfDNA Panel (including SNV, CNV, MSI, TMB analysis) and ER and HER2 protein expression and ERBB2 amp in/on CTCs | to subtype MBC | https://www.epicsciences.com/press-releases/epic-sciences-announces-medicare-coverage-for-breast-cancer-focused-ctdna-gene-panel/?goal=0_8c04e2abda-5ca6438936-71670485&mc_cid=5ca6438936 |
| Therapy (De)Escalation | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| ctDNA quantity | before neoadjuvant therapy | ctDNA quantity predicted the risk of relapse and OS | 10.1016/j.ctrv.2022.102362 |
| ctDNA quantity | before neoadjuvant chemotherapy in TNBC patients | predicting the risk for recurrence | 10.18632/oncotarget.23520 |
| PIK3CA and/or TP53 mutation detection in cfDNA | NeoALTTO trial (before neoadjuvant anti-HER2 treatment in HER2-positive BC patients) | PIK3CA and/or TP53 mutations in cfDNA correlated with lower pCR rates | 10.1158/1078-0432.CCR-18-2521 |
| cfDNA methylation of GASTP1, RASSF1A and RARB2 | before neoadjuvant treatment in early BC patients | cfDNA methylation of GASTP1, RASSF1A and RARB2 was associated with OS independent of pCR | 10.1159/000342083 |
| ctDNA clearance | I-SPY 2 study (neoadjuvant therapy with an AKT inhibitor); 84 high risk early BC patients; from baseline to three weeks after therapy initiation | ctDNA clearance from baseline to three weeks was related to an increased pCR rate and even in patients with no pCR, the ctDNA clearance within the first three weeks of therapy was correlated with improved survival compared to patients achieving no pCR and no ctDNA clearance | 10.1016/j.annonc.2020.11.007 |
| mutations in cfDNA | under neoadjuvant therapy | Detection of mutations in cfDNA based on tumor-informed personalized assays under neoadjuvant therapy were correlated with a lower chance of pCR | 10.1126/scitranslmed.aax7392 |
| (2) CTCs | |||
| CTC detection by CellSearch | NeoALTTO trial (before neoadjuvant anti-HER2 treatment in HER2-positive BC patients) | CTC detection resulted in numerically lower pCR rates (pCR in 27.3% patients with detectable CTCs and pCR in 42.5% with no detectable CTCs). | 10.1016/j.breast.2013.08.014 |
| CTC detection | GeparQuattro trial (before neoadjuvant chemotherapy) | CTC detection correlated significantly with disease-free (DSF) and OS | 10.1158/1078-0432.CCR-17-0255 |
| CTC quantity by CellSearch | BEVERLY-1 and -2 trials (before neoadjuvant chemotherapy) | CTC quantity did not show any correlation to pCR rates, but CTC detection was associated with significantly decreased DFS and OS | 10.1093/annonc/mdw535 |
| CTC detection | 2000 early BC patients (before neoadjuvant therapy) | presence of CTCs was an independent predictor of poor DSF, distant disease free (DDSF) and OS. | 10.1158/1078-0432.CCR-15-1603 and 10.1093/jnci/djy018 |
| (3) EVs | |||
| EV miRNA (miR-30b, miR-328 and miR-423) | before neoadjuvant therapy in BC patients | levels of specific EV miRNA (miR-30b, miR-328 and miR-423) forecast pCR | 10.3390/curroncol29020055 |
| miR-141, miR-34a, miR-182 and miR-183 in EVs | after the first dose of neoadjuvant therapy | miR-141, miR-34a, miR-182 and miR-183 in EVs after the first dose of neoadjuvant therapy predicted pCR/non-pCR | 10.3390/curroncol29020055 |
| (4) other blood analytes | |||
| miRNAs in plasma | TNBC patients before neoadjuvant therapy | miRNAs in plasma correlate with relapse and OS | 10.1158/1078-0432.CCR-14-2011 |
| miRNAs in plasma | before neoadjuvant therapy | miRNAs in plasma correlate with pCR | 10.3390/cancers12071820 and 10.1007/s10549-022-06642-z |
| miR-145 in plasma | HER2-positive BC, before neoadjuvant therapy | Reduced miR-145 levels were related to pCR in HER2-positive BC | 10.1097/SLA.0000000000005613 |
| let7a in plasma | luminal BC, before neoadjuvant therapy | let7a correlated with pCR | 10.1097/SLA.0000000000005613 |
| level of circulating nucleosomes | before neoadjuvant therapy in early BC patients | level of circulating nucleosomes had prognostic value | 10.1016/j.canlet.2013.04.013 |
| circulating miR-148a-3p and miR-374a-5p | from baseline to two weeks of trastuzumab-based neoadjuvant chemotherapy in the NeoALTTO trial | increased levels of circulating miR-148a-3p and miR-374a-5p from baseline to two weeks were related to pCR | 10.3390/ijms21041386 |
| thymidine kinase activity in the plasma | early under neoadjuvant treatment | prognostic value | 10.1016/j.esmoop.2021.100076 |
| Minimal Residual Disease Detection | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| ctDNA dynamics | before and after neoadjuvant treatment | ctDNA clearance during neoadjuvant therapy was informative regarding the existence of MRD | 10.1158/1078-0432.CCR-21-3231 |
| ctDNA dynamics by personalized mutation assays | I-SPY 2 study (neoadjuvant therapy with an AKT inhibitor); 84 high risk early BC patients; from baseline to end of neoadjuvant treatment | cfDNA clearance from baseline to the end of treatment correlated with a pCR. Patients with ctDNA detection after therapy showed a significantly increased risk for metastatic recurrence. In particular, patients not achieving a pCR but with no ctDNA detection after therapy had an excellent outcome, similar to the patients who achieved a pCR. | 10.1016/j.annonc.2020.11.007 |
| ctDNA dynamics by targeted digital sequencing | before and after neoadjuvant treatment | patients with a pCR showed a larger decrease in ctDNA during neoadjuvant therapy compared to the patients with no pCR | 10.1126/scitranslmed.aax7392 |
| ctDNA dynamics by BC-specific methylation pattern | before and after neoadjuvant treatment | ctDNA persistence even after neoadjuvant therapy indicated the existence of MRD | 10.1016/j.annonc.2019.11.014 |
| cfDNA integrity | before and after neoadjuvant treatment | longitudinal cfDNA integrity analysis indicated tumor shrinkage | 10.4149/neo_2017_417 |
| cfDNA integrity index | after neoadjuvant treatment | Patients who achieved a pCR, but showed an reduced cfDNA integrity index after neoadjuvant therapy had a higher risk for distant metastases | 10.4149/neo_2017_417 |
| ctDNA detection by mutation analysis | after neoadjuvant treatment | prognostic value of ctDNA detection by mutation analysis in all BC subgroups after neo-adjuvant therapy | 10.1016/j.ctrv.2022.102362 and 10.1001/jamaoncol.2019.1838 and 10.1038/s41523-017-0028-4 and 10.15252/emmm.201404913 and 10.1126/scitranslmed.aab0021 and 10.1001/jamaoncol.2020.2295 |
| ctDNA concentration and presence | after neoadjuvant treatment | ctDNA presence after neoadjuvant therapy was detected in 12/13 patients with no pCR, but also in 5/9 patients achieving a pCR. ctDNA concentration but not ctDNA presence after neoadjuvant therapy was significantly correlated with a pCR | 10.1126/scitranslmed.aax7392 |
| ctDNA detection | IMPASSION031 trial, TNBC patients, after neoadjuvant treatment | Patients achieving a pCR and who had no detectable ctDNA showed the best DSF and OS while the non-pCR cohort could be differentiated by ctDNA presence in patients with increased DSF and OS (ctDNA negative) and patients with worse DSF and OS (ctDNA positive) | 10.1016/esmoop/esmoop101220 |
| ctDNA detection by personalized mutation sequencing panels | in the follow-up | sensitivity of 89% for MRD detection with a lead time of up to 24 months (median 8.9 months) with a specificity of 100% with none of the non-relapsing patients being ctDNA-positive | 10.1158/1078-0432.CCR-18-3663 |
| ctDNA detection by patient-specific digital droplet PCR (ddPCR) panels | within one year after surgery | MRD was detected with 19% sensitivity and median lead time from first positive test to recurrence was 18.9 months | 10.1158/1078-0432.CCR-19-3005 |
| ctDNA detection by RaDaR assays | high-risk HR-positive, HER2-negative BC patients with no evidence of recurrence five years after diagnosis, serial blood analysis | RaDaR assays identified all patients with distant metastatic recurrences (7.2%) with a median ctDNA lead time of 12.4 months. However, 2/8 patients with ctDNA-positive results had not had clinical recurrence. In total, 1.2% of patients had no MRD but local recurrence. | 10.1200/JCO.22.00908 |
| (2) CTCs | |||
| CTC detection | before and after neoadjuvant treatment | presence of persisting CTCs correlated with shorter DSF and OS | 10.1093/jnci/dju066 |
| CTC detection | before and after neoadjuvant treatment | presence of persisting CTCs correlated with an increased risk of relapse | 10.1245/s10434-015-4600-6 |
| CTC number | in TNBC patients after neoadjuvant chemotherapy | one or more CTCs present after neoadjuvant chemotherapy predicted relapse and survival in TNBC patients | 10.1245/s10434-015-4600-6 |
| TWIST transcripts in CTCs | early BC patients after surgery and before adjuvant therapy | prognostication of DSF in early BC patients after surgery and before adjuvant therapy | 10.3390/cells8070652 |
| CK19 mRNA positive CTCs | during the first five years of BC follow-up | persistent detection of CK19 mRNA positive CTCs during the first five years of BC follow-up increased the risk of late relapse | 10.1186/bcr2897 |
| (3) EVs | |||
| EV miR-21 | before and after neoadjuvant treatment | Persisting high levels of circulating miR-21 after neoadjuvant treatment were associated with poor prognosis | 10.1186/s13058-019-1109-0 |
| (4) other blood analytes | |||
| circulating miR-21 | before and after neoadjuvant treatment | Persisting high levels of circulating miR-21 after neoadjuvant treatment were associated with poor prognosis | 10.1007/s10549-022-06642-z |
| circulating miRNAs | NeoALTTO trial, after completion of neoadjuvant therapy | miR-185-5p, miR-146a-5p and miR-22-3p are prognostic marker independent of pCR | 10.3389/fonc.2022.1028825 |
| lymphocyte-to-monocyte ratio | after surgery and neoadjuvant therapy | lymphocyte-to-monocyte ratio was shown to be significantly associated with worse prognosis | 10.2147/CMAR.S292048 |
| expression of TLR4 on peripheral blood mononuclear cells | at the time point of surgery in early BC patients | the expression of TLR4 on peripheral blood mononuclear cells predicted high risk of relapse | 10.3390/cancers14041053 |
| (5) multiple blood analytes | |||
| cfDNA and CTC analysis | TNBC patients after neoadjuvant treatment | MRD sensitivity was 79% with ctDNA analysis alone, 62% with CTC analysis alone and 90% with the combination of both analytes | 10.1001/jamaoncol.2020.2295 |
| CTC quantification, phenotypic, transcriptomic, and genomic profiling of CTCs as well as mutation and methylation profiling of cfDNA | early BC patients in the follow-up | Multimodal approach identified a relapse at least four years earlier than metastases could clinically be detected | 10.1038/s41598-022-25400-1 |
| Prognostification in MBCs | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| genome-wide cfDNA methylation | MBCs, at baseline or week four after therapy initiation and dynamics | Methylation pattern on genome-wide scale in cfDNA was shown to correlate with OS—even prognostic in case evaluated at week four after therapy initiation and dynamics from baseline to four weeks were informative about OS as well. | 10.1200/JCO.2015.66.2080 |
| cfDNA methylation (5 genes) | at baseline in MBCs | correlate with OS | 10.1038/s41388-018-0660-y |
| copy number changes and genomic instability score from cfDNA | MBCs, at baseline, after one week and two weeks after treatment initiation | The genomic instability score at baseline, after one week and two weeks after treatment initiation were significantly associated with poor OS. | 10.3390/cancers13061331 |
| Tumor-derived cfDNA fractions | MBC | associated with clinical outcome (PFS and OS) | 10.3390/cancers11081171 |
| ctDNA abundance by mutation-specific ddPCR | BEECH trial, ER-positive advanced BCs, after four weeks of therapy | ctDNA abundance after four weeks of therapy revealed significant correlation with PFS | 10.1093/annonc/mdz085 |
| pathogenic or likely pathogenic variants in the cfDNA | MBCs | Higher number of pathogenic or likely pathogenic variants in the cfDNA associated with worse OS | 10.1007/s00018-019-03189-z |
| mean variant allele frequency of cfDNA mutations | from baseline to cycle two in advanced BC patients treated with ICI | It was also described that a decrease in the mean variant allele frequency of cfDNA mutations in any of the 425 genes sequenced from baseline to cycle two was related to a longer PFS | 10.1200/PO.22.00509 |
| cfDNA ESR1 mutations | 42 pre-treated MBCs | cfDNA ESR1 mutations were found to indicate worse OS and were associated with shorter duration of endocrine treatment effectiveness | 10.18632/oncotarget.18479 |
| cfDNA TP53 and/or PIK3CA | MBCs | TP53 and/or PIK3CA mutations detected in cfDNA of MBCs were shown to indicate worse OS | 10.1016/S1470-2045(17)30376-5 and 10.1016/j.clbc.2016.05.004 |
| cfDNA specific BRCA1 mutation | 44 HR-positive/HER2-negative MBC | Specific BRCA1 mutation detected in the cfDNA was associated worse OS | 10.1007/s00018-019-03189-z |
| (2) CTCs | |||
| CTCs number by the CellSearch system | 177 MBC patients before therapy initiation of any therapy in any therapy line | A cut-off of five CTCs in 7.5ml blood differentiated patients with good (mean 7.0 months) versus worse (mean 2.7 months) PFS and correlated significantly with OS | 10.1056/NEJMoa040766 |
| CTCs number by the CellSearch system | 83 newly diagnosed, measurable MBC who were about to start their first line of systemic therapy | cut-off of five CTCs per 7.5ml blood was applied: 52% of patients had ≥ five CTCs and these patients had a decreased PFS and OS compared to the patients with no or less than five CTCs | 10.1200/JCO.2005.08.140 |
| CTCs number by the CellSearch system | meta-analysis including 1944 MBC pa-tients | significant association of CTC quantity (cut-off: 5 CTCs) regarding PFS and OS | 10.1016/S1470-2045(14)70069-5. |
| CTC clusters | MBCs (first line treated) | presence of CTC clusters has additional prognostic value when compared to the single CTC quantification alone. number of CTCs within the clusters might also be related to OS | 10.1007/s10549-016-4026-2 and 10.1186/s13058-018-0976-0 |
| CTC detection by positive immunomagnetic selection and molecular characterization | MBCs | CTC presence was significant associated with PFS; patients with CTCs showing high PALB2 or MYC transcript expression had a shorter PFS and OS; patients with CTCs showing epithelial-stem cell like features also showed shorter PFS and OS | 10.1007/s10549-008-0143-x and 10.3390/cancers11121941 |
| CTC isolation by a microfluidic chip and molecular characterization | MBC patients before eribulin treatment | entirety of epithelial and mesenchymal CTCs, as well as only the epithelial or mesenchymal CTCs was related to OS | 10.1007/s12032-019-1314-9 |
| CTC isolation by density and EpCAM expression and molecular characterization | MBCs | univariate Cox regression model showed prognostic value for the presence of CTCs with either CK-19 overexpression, HER2 overexpression or CTCs with CD44high/CD24low or ALDH1high/CD24low features | 10.3390/diagnostics11030513 |
| HER2+ CTCs | HER2-negative MBC | reduced OS in case CTCs with strong HER2 staining were detectable | 10.1016/j.esmoop.2021.100299 |
| mitotic activity of CTCs | MBCs | characterization of CTCs with regard to their mitotic activity increased the hazard ratio for association with OS dramatically compared to CTC quantification itself | 10.1186/s13058-016-0706-4 |
| CTC mRNA profile | MBCs (first-line aromatase inhibitor (AI) treated patients vs. treated with other therapy regimens | a-8-gene predictor (EEF1A, PTRF, CXCL14, ERBB3, EGFR, PTPRK, KRT81, TWIST1) was published to be related to PFS in first-line aromatase inhibitor (AI) treated patients, while the same predictor was not related to PFS in MBCs treated with other therapy regimens | 10.1186/s12885-016-2155-y |
| (3) EVs | |||
| metastasis- and stemness-related mRNAs in EVs | A set of metastasis- and stemness-related mRNAs were higher expressed in EVs from BC patients with poor OS than in those patients with increased OS | 10.18632/oncotarget.5818 | |
| (4) other blood analytes | |||
| circulating miR-200 family members | at baseline of new line of systemic therapy in MBCs | miR-200a, miR-200b, miR-141, and miR-429 were shown to significantly correlate with progression-free survival (PFS) | 10.1007/s00404-022-06442-2 |
| thymidine kinase 1 (sTK1) in plasma | EFECT trial in MBCs, at baseline | prognostic value | 10.1016/j.ejca.2019.04.002 |
| LAMP2 protein levels in red blood cells | MBCs | related to OS | 10.1016/j.mcpro.2022.100435 |
| (5) multiple blood analytes | |||
| CTC counts and total cfDNA level | MBCs | CTC counts and total cfDNA level were associated with OS in MBCs and thus, concluded CTCs and cfDNA to be equally valuable OS markers. the combined analysis of CTCs and cfDNA was more informative regarding OS than the sole analysis of one of the analytes | 10.1158/1078-0432.CCR-16-0825 and 10.1186/s13058-019-1235-8 and 10.1016/j.ejca.2018.10.012 |
| CTC counts by CellSearch and ctDNA identified by targeted NGS | UCBG COMET study (NCT01745757), first-line paclitaxel and bevacizumab | CTC counts and ctDNA had non-overlapping profiles and correlated in sole and also in combined analysis with OS | 10.1038/s41523-021-00319-4 |
| ESR1 variants in CTCs and cfDNA | MBCs | ESR1 variants in CTCs and cfDNA to indicate worse OS | 10.3390/cancers12051084 |
| SOX17 promotor methylation in cfDNA and CTCs | MBCs | SOX17 promotor methylation in cfDNA and CTCs to be of prognostic relevance | 10.18632/oncotarget.18679 |
| HER2+ CTCs and HER2+ EVs | MBCs | the heterogeneity of CTCs or EVs within one blood sample was shown to be inversely associated with OS | 10.1186/s13058-020-01323-5 |
| CTCs and disseminated tumor cells (DTCs) in the bone marrow | MBCs | DTCs as well as CTCs were significantly associated with worse OS, no significant association of DTC and CTC results | 10.1007/s10549-014-3113-5 |
| cfDNA, CTC genomic DNA, CTC mRNA and EV mRNA | ELIMA project, MBCs | additive value for prognostication: ‘ELIMA.score’ showed a significant correlation with OS with a decreased p-value when compared to each single analyte | 10.1186/s13073-021-00902-1 |
| Decision for/against CTX | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (2) CTCs | |||
| CTC quantity by CellSearch | STIC CTC trial, first-line therapy selection in HER2-negative MBCs, before therapy initiation | Therapy selection was conducted either based on the CTC quantity or clinicians’ choice. In general, PFS and OS were equally distributed in all groups, however, in patients with no concordant stratification status (high risk by clinicians/low CTC number or low risk by clinicians/high CTC number), chemotherapy prolonged PFS and OS compared to endocrine therapy | 10.1001/jamaoncol.2020.5660 |
| (3) EVs | |||
| Ubiquitin carboxyl-terminal hydrolase-L1 protein levels in EVs | before therapy initiation | Ubiquitin carboxyl-terminal hydrolase-L1 protein levels in EVs were shown to predict response to CTX | 10.1002/jso.24614 |
| (4) other blood analytes | |||
| Circulating miR-125b | before therapy initiation | Circulating miR-125b was shown to predict response to CTX | 10.1371/journal.pone.0034210 |
| Decision for/against PARP Inhibition | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| Somatic BRCA1/2 mutations (from cfDNA) | Olaparib Expanded trial, MBCs | The Olaparib Expanded trial also showed the effectiveness of Olaparib in MBC patients with somatic BRCA1/2 mutations. Olaparib therapy is rated as an option for MBC patients with somatic BRCA1/2 mutations (ESCAT scale IIA) by the ESMO guideline | 10.1200/JCO.20.02151 and 10.1016/j.annonc.2021.09.019 |
| (4) other blood analytes | |||
| Germline BRCA1/2 mutations (from blood cells) | OLYMPIA trial, HER2-negative early BC patients | In early BC, gBRCA1/2 mutations are of prognostic value to achieve a pCR under chemotherapy and forecast DFS under PARP inhibition. Olaparib is recommended for early TNBC patients showing no pCR and harboring gBRCA1/2 mutations as well as for high risk gBRCA1/2 mutant HR-positive/HER2-negative early BC patients as proven in the OLYMPIA trial. Standard to test HER2-negative BC patients for gBRCA1/2 mutations (ESCAT scale IA | 10.1159/000531578 and 10.1016/j.annonc.2022.09.159 and 10.1093/annonc/mdz036 |
| Germline PALB2 mutations (from blood cells) | Olaparib Expanded trial, MBCs | The Olaparib Expanded trial also showed the effectiveness of Olaparib in MBC patients with germline PALB2 mutations. Olaparib therapy is rated as an option for MBC patients with germline PALB2 mutations (ESCAT scale IIA) by the ESMO guideline | 10.1200/JCO.20.02151 and 10.1016/j.annonc.2021.09.019 |
| Decision for/against Anti-HER2 Therapy | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| cfDNA ERBB2 mutations | MBCs | MBC patients with ERBB2 mutations were resistant to lapatinib, but sensitive to neratinib | 10.1158/2159-8290.CD-12-0349 and 10.1038/nature25475 and 10.1158/1078-0432.CCR-17-0900 |
| cfDNA ERBB2 mutations | plasmaMATCH trial, cohort B, MBC patients | Cohort B in the plasmaMATCH trial also showed a benefit of neratinib treatment in ERBB2 mutant MBC patients | 10.1016/S1470-2045(20)30444-7 |
| (2) CTCs | |||
| CK19-positive CTCs | HER2-negative early BC patients | Treatment with trastuzumab prolonged the DFS in HER2-negative patients with CK19-positive CTCs present before and after adjuvant chemotherapy compared to observation. The fraction of patients with CK19-positive CTCs after trastuzumab treatment was reduced down to 14%, while observation led to 17.9% of patients with CK19-positive CTCs. | 10.1093/annonc/mds020 |
| Decision for/against PIK3CA Inhibition | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| cfDNA PIK3CA mutation | BELLE-2 trial, HR-positive/HER2-negative MBC patients progressing under aromatase inhibitor (AI) therapy | addition of buparlisib improved the PFS in PIK3CA mutant patients | 10.1016/S1470-2045(17)30376-5 and 10.1016/j.ejca.2018.08.002 |
| cfDNA PIK3CA mutation | SOLAR-1 study, MBCs | Worse PFS of PIK3CA mutant MBC patients was improved by the application of alpesilib to an extend of a PFS achieved in PIK3CA wildtype MBC patients, that did not benefit from alpesilib treatment. | 10.1056/NEJMoa1813904 |
| cfDNA PIK3CA mutation | HR-positive/HER2-negative MBC patients after progression under AI | Alpelisib in combination with fulvestrant for PIK3CA mutant HR-positive/HER2-negative MBC patients after progression under AI is recommended by the ESMO stating that ctDNA assessment for PIK3CA mutation analysis is an option besides mutational profiling in tissue samples. In patients with no available archival tumor tissue, ctDNA assessment is recommended. PIK3CA mutations are classified as tier IA by the ESMOs’ ESCAT scale. Recommendation for PIK3CA mutation profiling in primary tumor tissue, metastasis or plasma was confirmed in 2023. | 10.1016/j.annonc.2021.09.019 and 10.1016/j.annonc.2020.09.010 and 10.1159/000531579 |
| cfDNA PIK3CA mutation | NCT02379247, HER2-negative heavily pre-treated patients | Alpesilib might be applied in more HER2-negative patients because its application demonstrated that in combination with nab-paclitaxel a prolonged PFS could be achieved in heavily pre-treated patients with PIK3CA mutation in tumor or plasma compared to PIK3CA wildtype patients | 10.1158/1078-0432.CCR-20-4879 |
| (2) CTCs | |||
| CTC PIK3CA mutations | MBCs | 16% to 33% of all MBCs were reported to harbor PIK3CA mutant CTCs | 10.1016/j.molonc.2014.12.001 and 10.1016/j.molonc.2013.07.007 |
| (5) multiple blood analytes | |||
| PIK3CA mutation in cfDNA and CTCs | MBCs | PIK3CA mutational status was found concordant in cfDNA and CTCs isolated from the same sample from MBC patients | 10.1002/1878-0261.12540 |
| Decision for/against Endocrine Therapy | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| cfDNA ESR1 mutations | SoFEA trial, HR-positive MBC patients | Direct comparison of fulvestrant (SERD) with exemestane (AI), showed a significantly prolonged PFS using fulvestrant compared to exemestane in ESR1 mutant HR-positive MBC patients | 10.1200/JCO.2016.67.3061 |
| cfDNA ESR1 mutations | SoFEA and EFECT trial | OS benefit for ESR1 mutant MBC patients treated with fulvestrant compared to exemestane | 10.1158/1078-0432.CCR-20-0224 |
| cfDNA ESR1 mutations | PADA-1 trial | Longitudinal monitoring via ESR1 mutation detection in the plasma under AI treatment and switch to fulvestrant plus CDK4/6i compared to continuation of AI after emergence of ESR1 mutations without radiographic evidence for progression increased the PFS from 5.7 months to 11.9 months. | 10.1016/S1470-2045(22)00555-1. |
| cfDNA ESR1 mutations | EMERALD trial, ER-positive/HER2-negative MBC in the second or more therapy line after progression under CDK4/6i and one previous chemotherapy line at maximum | Elacestrant was recently shown to significantly increase the PFS compared to standard endocrine monotherapy. This effect was shown for both, ESR1 mutant and ESR1 wild-type patients. The hazard ratio however, showed a greater effect of PFS prolongation from elacestrant compared to fulvestrant in ESR1 mutant patients compared to all patients, independent of their ESR1 status. | 10.1200/JCO.22.00338 |
| cfDNA ESR1 mutations | ER-positive/HER2-negative MBCs at the time of recurrence or progression on endocrine therapy | Testing for the emergence of ESR1 mutations is now recommended by the ASCO. Blood-based ESR1 mutation detection is preferred over tumor tissue testing due to the higher sensitivity. In HR-positive/HER2-negative MBC patients with prior CDK4/6i therapy and presence of ESR1 mutation in blood or tissue, elacestrant is recommended by the ASCO. | 10.1016/S1470-2045(20)30444-7 and 10.1200/JCO.23.00638 |
| cfDNA ESR1 promotor methylation | Methylation of the ESR1 promotor in cfDNA might become relevant for selection of an endocrine therapy | 10.1158/1078-0432.CCR-17-1181 | |
| (2) CTCs | |||
| CTC ESR1 promotor methylation | Methylation of the ESR1 promotor in CTCs might become relevant for selection of an endocrine therapy | 10.1158/1078-0432.CCR-17-1181 | |
| Decision for/against AKT Inhibition | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| cfDNA AKT1 mutation | plasmaMATCH trial, cohort C, ER-positive/HER2-negative MBC patients | Patients with AKT1 mutation in the cfDNA received capivasertib plus fulvestrant and this cohort met or exceeded the target number of responses with 4/18 patients | 10.1016/S1470-2045(20)30444-7 |
| cfDNA PIK3CA, AKT1 or PTEN alterations | PAKT trial, metastatic TNBC | Addition of capivasertib to paclitaxel compared to paclitaxel alone correlated with a prolonged PFS and OS, especially in patients with PIK3CA, AKT1 or PTEN alterations. | 10.1200/JCO.19.00368 |
| Decision for/against Tyrosine Receptor Kinase (TRK) Inhibition | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| cfDNA NTRK1/2/3 fusion | BC patients | Trk inhibitors for BC patients with NTRK fusions are recommended. FDA approved blood-based evaluation of NTRK1/2/3 fusions in cfDNA available. | 10.1159/000531579 and 10.1016/j.annonc.2020.09.010 and 10.1200/JCO.22.01063 |
| Decision for/against Androgen Receptor Inhibition | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (2) CTCs | |||
| AR + CTCs | metastatic TNBC patients | AR protein expression analysis on CTCs in the blood might be usable as predictive marker for anti-AR therapy. | 10.1002/ijc.32209 |
| CTC AR_v7 mRNA | early TNBC patients | CTC mRNA analysis showed a minority of early TNBC patients to potentially benefit from anti-AR therapy based on AR_v7 transcript expression. | 10.3389/fonc.2020.01658 |
| Decision for/against CDK4/6i | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| Tumor fraction in cfDNA | MBCs, before CDK4/6i initiation | Correlation of tumor fraction in cfDNA with PFS but not OS | 10.1038/s41467-023-36801-9 |
| cfDNA PIK3CA mutation | MBCs, before CDK4/6i initiation, n = 30 or in the MONALEESA-7 trial | potential of plasma PIK3CA mutations before CDK4/6i as predictive markers | 10.1016/j.phrs.2020.105241 and 10.1200/PO.20.00445 |
| cfDNA KRAS mutation | MONALEESA-7 trial, MBCs, before CDK4/6i initiation | Patients treated with palbociclib and fulvestrant with baseline KRAS mutations had a worse median PFS compared to patients with KRAS wild-type | 10.1200/PO.20.00445 |
| cfDNA RB1 mutation | PALOMA-3 trial, MBCs, before CDK4/6i initiation | patients with RB loss (17.3% prevalence) at baseline had a significantly worse PFS under palbociclib plus fulvestant compared to RB wild-type patients | 10.1093/jnci/djaa087 |
| cfDNA RB-LOH signature | 245 patients treated with ET + CDK4/6i from two independent cohorts | RB-LOH signature, consisting of 224 copy number features in the entire cfDNA genome showed a strong correlation with poor response and poor survival following CDK4/6i plus endocrine therapy | 10.1038/s41467-023-36801-9 |
| (2) CTCs | |||
| Single CTC RB1 transcript expression | within the TREnd trial, small cohort of MBC before Palbociclib | Gene expression regarding RB1 in single CTCs revealed a prolonged PFS | 10.1186/s13058-021-01415-w |
| (3) EVs | |||
| EV CDK4 mRNA expression | 40 HR-positive/HER2-negative advanced BC patients receiving palbociclib plus endocrine therapy, at baseline | High mRNA expression levels of CDK4 in EVs correlated significantly with a longer PFS | 10.1007/s10549-019-05365-y |
| Gene | Alteration | Cancer Type | Drugs | ESCAT Scale 2019/2020 | ASCO 2022 | AGO 2023 |
|---|---|---|---|---|---|---|
| PIK3CA | C420R, E542K, E545A, E545D, E545G, E545K, Q546E, Q546R H1047L, H1047R, H1047Y and other oncogenic mutations | Breast Cancer | Alpelisib + Fulvestrant | IA | recommended | ++ |
| ERBB2 | Amplification | Breast Cancer | Trastuzumab, Trastuzumab + Chemotherapy | IA | ||
| ERBB2 | Amplification | Breast Cancer | Trastuzumab Deruxtecan | IA | ||
| ERBB2 | Amplification | Breast Cancer | Trastuzumab + Pertuzumab + Chemotherapy | IA | ||
| ERBB2 | Amplification | Breast Cancer | Trastuzumab + Tucatinib + Capecitabine | IA | ||
| NTRK1 | Fusion | All Solid Tumors | Larotrectinib | IC | recommended | + |
| NTRK2 | Fusion | All Solid Tumors | Larotrectinib | IC | recommended | + |
| NTRK3 | Fusion | All Solid Tumors | Larotrectinib | IC | recommended | + |
| NTRK1 | Fusion | All Solid Tumors | Entrectinib | IC | recommended | + |
| NTRK2 | Fusion | All Solid Tumors | Entrectinib | IC | recommended | + |
| NTRK3 | Fusion | All Solid Tumors | Entrectinib | IC | recommended | + |
| Microsatellite Instability-High | All Solid Tumors | Pembrolizumab | IC | recommended | + | |
| Tumor Mutational Burden-High | All Solid Tumors | Pembrolizumab | IC | recommended | ||
| ESR1 | D538, E380, L469V, L536, S436P, Y537, V422del | Breast Cancer | Elacestrant | IIA | not recommended in 2022, but recommended in 2023 | + |
| ERBB2 | Amplification | Breast Cancer | Ado-Trastuzumab Emtansine | |||
| ERBB2 | Amplification | Breast Cancer | Lapatinib + Capecitabine, Lapatinib + Letrozole | |||
| ERBB2 | Amplification | Breast Cancer | Margetuximab + Chemotherapy | |||
| ERBB2 | Amplification | Breast Cancer | Neratinib, Neratinib + Capecitabine | |||
| BRAF | V600E | All Solid Tumors (excluding Colorectal Cancer) | Dabrafenib + Trametinib | |||
| NTRK1 | G595R | All Solid Tumors | Resistance to Larotrectinib | |||
| NTRK3 | F617L | All Solid Tumors | Resistance to Larotrectinib | |||
| NTRK3 | G623R | All Solid Tumors | Resistance to Larotrectinib | |||
| NTRK3 | G696A | All Solid Tumors | Resistance to Larotrectinib | |||
| RET | Fusion | All Solid Tumors (excluding Thyroid Cancer, Non-Small Cell Lung Cancer) | Selpercatinib |
| Therapy Monitoring | |||
|---|---|---|---|
| Specific Analyte | Clinical Setting | Conclusion | Reference |
| (1) cfDNA | |||
| cfDNA methylation (9 marker) | TBCRC 005 study, MBCs, under therapy | 9-marker cfDNA methylation assay was shown to forecast disease progression three months earlier than radiographic staging in MBC patients | 10.1158/1078-0432.CCR-22-2128 |
| genomic instability of cfDNA | 25 MBC patients, at baseline, one week under therapy, three months after therapy initiation | More than 50% reduction in genomic instability number (GIN) from low-pass WGS of cfDNA at baseline to one week under therapy was shown to associate with the stable disease proven by staging after 3 months and also with OS. A rise in GIN from baseline to two weeks under therapy associated with poor response, evaluated three months after therapy initiation by staging. | 10.3390/cancers13061331 |
| cfDNA CNVs | HR-positive/HER2-negative MBC patients treated with CDK4/6i, at baseline and under therapy | comparison of z-scores at baseline and under therapy (z-score trajectories) has monitoring value | 10.1002/1878-0261.12870 |
| mean allele frequency dynamics in cfDNA | LOTUS and INSPIRE trials, MBCs treated with different therapy regimens, baseline to a time point under therapy | Mean allele frequency dynamics from baseline to a time point under therapy related to therapy response at the time of blood draw or to PFS and OS | 10.1200/PO.20.00345 and 10.1200/PO.20.00345 and 10.1002/mgg3.1079 and 10.1038/s41523-021-00218-8 and 10.1016/S1470-2045(17)30450-3 and 10.1186/s40425-019-0541-0 |
| ctDNA mutations | POSEIDON and SUMMIT trials, under therapy | In the POSEIDON and SUMMIT trials, early evaluation of ctDNA changes forecasted the radiologic treatment response and the emergence of specific mutations correlated with clinical drug resistance. Allele frequency of HER2 mutations in cfDNA decreased under pan-HER inhibitor neratinib, but increased upon radiographically proven progression. | 10.1158/1078-0432.CCR-19-0508 and 10.1158/1078-0432.CCR-17-0900. |
| ctDNA in CSF and plasma | HER2-positive MBCs with brain metastases | dynamic changes in ctDNA in CSF and plasma under therapy revealed decreased allele frequencies in the plasma to be consistent with extra-CNS disease control and increased allele frequencies in the CSF to be related to poor treatment benefit in CNS | 10.1136/esmoopen-2017-000253 |
| ctDNA level | INSPIRE trial, TNBC patients and patients with other tumor entities, from baseline to six weeks under treatment with pembrolizumab | ctDNA level changes from baseline to six weeks under treatment forecasted the therapy benefit. In all patients who responded to therapy, ctDNA clearance was detected before visible radiological response. | 10.1038/s43018-020-0096-5 |
| cfDNA PIK3CA mutations | PALOMA-3 trial, palbociclib treated patients from baseline to two weeks | cfDNA PIK3CA mutation dynamics had significant monitoring value. Decrease in PIK3CA mutations in the cfDNA correlated significantly with increased PFS and long-term clinical benefit. | 10.1038/s41467-018-03215-x |
| cfDNA level and mutations | ALCINA trial, at day 15/30 under palbociclib plus fulvestrant | cfDNA evaluation showed a decrease in all patients independent of their PFS. On day 30, undetectable cfDNA mutations (PIK3CA, TP53 and AKT1 studied) associated with improved PFS. | 10.1186/s13058-021-01411-0 and 10.1038/s41388-020-1174-y |
| cfDNA ESR1 mutations | MBCs under first-line AI treatment | ESR1 mutation detection in the plasma revealed a direct association with progressive disease with a 100% specificity. ESR1 mutations were detectable prior to progression with median lead time of 110 days. | 10.1186/s13058-020-01290-x |
| cfDNA ESR1 mutations | PADA-1 trial, under palbociclib and AI | Rising allele frequencies of cfDNA ESR1 mutations were used to identify patients with no radiographically proven progressive disease suitable for therapy switch of endocrine therapy. Significant clinical benefit with regard to PFS in case the therapy switch was conducted in patients with rising ESR1 mutations detectable under therapy | 10.1016/S1470-2045(22)00555-1 |
| (2) CTCs | |||
| CTC count by CellSearch | 3–5 or 6–8 weeks after initiation of therapy | It was shown that the CTC count itself by CellSearch evaluated 3–5 or 6–8 weeks after initiation of therapy was significantly associated with PFS and OS | 10.1016/S1470-2045(14)70069-5. |
| CTC count | from baseline to a time point under therapy | A decrease in CTC counts from baseline to a time point under therapy was related to an increased PFS and OS. Persistently high CTC counts from baseline to under therapy, despite radiologically proven therapy response, associated with worse outcome | 10.1158/1078-0432.CCR-05-2821 and 10.1158/1078-0432.CCR-05-1769 |
| apoptotic CTCs | baseline to under therapy | number of apoptotic CTCs from baseline to under therapy revealed a 50% apoptotic CTC reduction to differentiate between patients showing stable versus progressive disease and in case the apoptotic CTC number decreased from baseline to under therapy by less than 10%, progressive disease was identified with 74% specificity | 10.3390/cancers12041055 |
| HER2+ CTCs | MBC patients treated with anti-HER2 treatment lapatinib | significant decrease in HER2-positive CTCs was only detected in MBC patients responding to anti-HER2 treatment with lapatinib, but not in patients progressing under lapatinib | 10.1371/journal.pone.0123683 |
| RANK-positive CTCs | MBCs treated with Denosumab, baseline to day 2 | increase in RANK-positive CTCs from baseline to day 2 and persistence of RANK-positive CTCs was related to a longer time to progress of the bone metastasis | 10.1038/s41598-020-58339-2 |
| CTCs overexpressing EpCAM, MUC1 or HER2 | under therapy in MBCs | The persistence of CTCs overexpressing EpCAM, MUC1 or HER2 transcripts under therapy in MBC patients correlated with shorter OS | 10.1007/s10549-008-0143-x |
| CTCs overexpressing either EMT markers or the stem cell marker ALDH1 | MBCs, at the staging time point | 74% of all patients with progressive disease have CTCs overexpressing either EMT markers or the stem cell marker ALDH1 in contrast to only 10% of patients with stable disease | 10.1186/bcr2333 |
| CTC mRNA profile | MBCs, at the staging time point | overexpression of ERBB2, ERBB3, ERCC1 alone or in combination with AURKA in CTCs of MBCs was significantly more prevalent in patients showing progressive disease at the time of blood draw compared to patients with stable disease. Identification of CTCs with overexpression of ERBB2, ERBB3, ERCC1 alone or in combination with AURKA during therapy in MBCs was furthermore related to a shorter OS. ERBB2 overexpression in CTCs was related to therapy failure at the time of blood draw and to a reduced OS | 10.18632/oncotarget.9528 |
| CTC mRNA profile | MBCs, at the staging time point | Patients with progressive disease at the time of blood draw were more likely to have CTC overexpression signals than patients with stable disease. Two different gene expression patterns in CTCs were shown for patients with progressive disease, but a homogeneous expression pattern in patients with stable disease | 10.1373/clinchem.2016.269605 |
| ERBB2 and/or ERBB3 overexpression in CTCs | MBCs, at the staging time point | ERBB2 and/or ERBB3 overexpression in CTCs was significantly correlated with progressive disease at the time of blood draw | 10.1373/clinchem.2017.283531 |
| (4) other blood analytes | |||
| CEA, CA 15-3, and CA 27-29 | BC | The circulating proteins CEA, CA 15-3, and CA 27-29 were recommended for therapy monitoring in 2015 by the ASCO | 10.1200/JCO.2015.61.1459 |
| (5) multiple blood analytes | |||
| CTC and EV mRNA profiles | MBCs, at the staging time point | Stronger correlation of ERBB2 and ERBB3 signals in CTCs and EVs with disease progression was identified compared to ERBB2 and ERBB3 signals in CTCs alone, revealing a synergistic value of CTCs and EVs for therapy monitoring. mTOR overexpression signals in EVs of MBCs under therapy was related to consecutive therapy failure while mTOR overexpression in CTCs was related to patients showing therapy response over at least six months | 10.1373/clinchem.2017.283531 |
| ctDNA, CTC, CA 15-3 | MBCs | ctDNA evaluation was shown to have a higher sensitivity and higher correlation with tumor burden compared to CA 15-3 and CTC evaluations. | 10.1056/NEJMc1306040 |
| cfDNA, CTC genomic DNA, CTC mRNA and EV mRNA | MBCs, at the staging time point | Additive value of these analytes in treatment monitoring. Presence of either ERBB3 overexpression signals or ERBB2 overexpression signals in CTCs were related significantly to the staging result. Combined evaluation of ERBB3 in all three analytes associated with therapy response. Dynamics from one time point to the next time point were more informative than single time point evaluations. Overexpression signals in EVs were the most dynamic ones during therapy and newly occurring ERCC1 overexpression signals in EVs from one time point to the next had a specificity of 97% but sensitivity of 18% to determine therapy response. The accuracy for detecting disease progression was 70% and 66% for PIK3CA and ESR1 variant appearances and the combined evaluation of ESR1 or PIK3CA allele frequency development was significantly correlated with disease progression. | 10.3390/cells10020212 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keup, C.; Kimmig, R.; Kasimir-Bauer, S. The Diversity of Liquid Biopsies and Their Potential in Breast Cancer Management. Cancers 2023, 15, 5463. https://doi.org/10.3390/cancers15225463
Keup C, Kimmig R, Kasimir-Bauer S. The Diversity of Liquid Biopsies and Their Potential in Breast Cancer Management. Cancers. 2023; 15(22):5463. https://doi.org/10.3390/cancers15225463
Chicago/Turabian StyleKeup, Corinna, Rainer Kimmig, and Sabine Kasimir-Bauer. 2023. "The Diversity of Liquid Biopsies and Their Potential in Breast Cancer Management" Cancers 15, no. 22: 5463. https://doi.org/10.3390/cancers15225463
APA StyleKeup, C., Kimmig, R., & Kasimir-Bauer, S. (2023). The Diversity of Liquid Biopsies and Their Potential in Breast Cancer Management. Cancers, 15(22), 5463. https://doi.org/10.3390/cancers15225463






