Current Knowledge about the Peritumoral Microenvironment in Glioblastoma
Abstract
Simple Summary
Abstract
1. Introduction
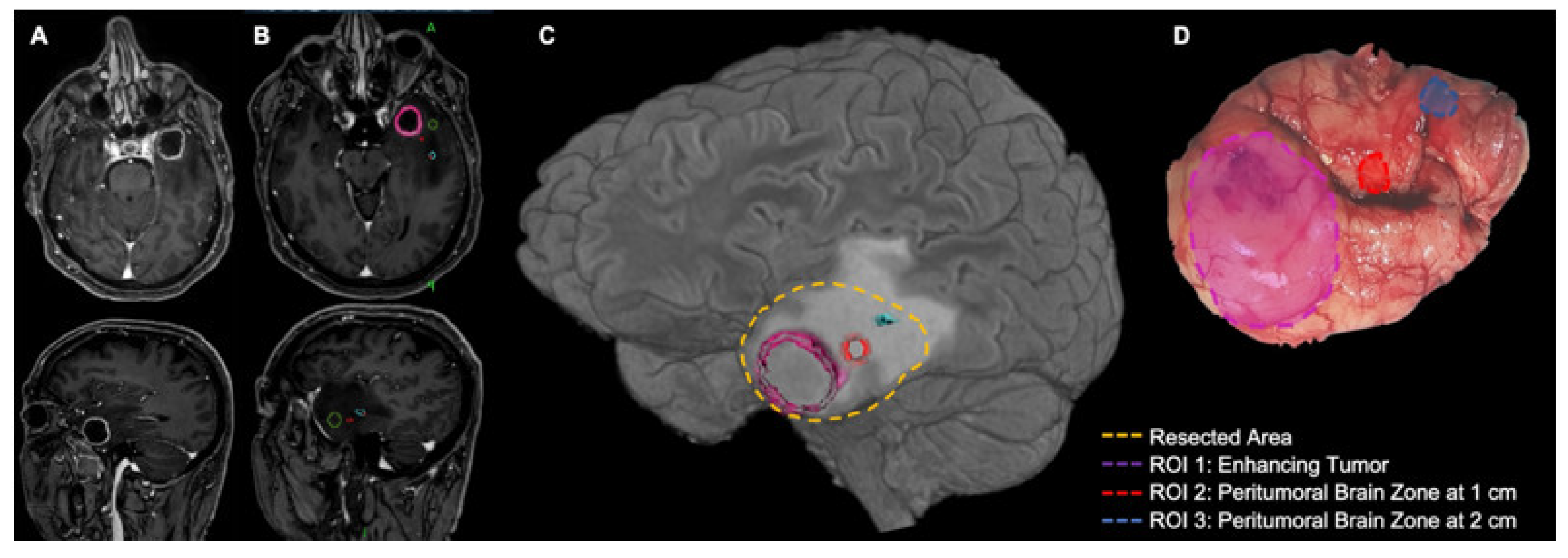
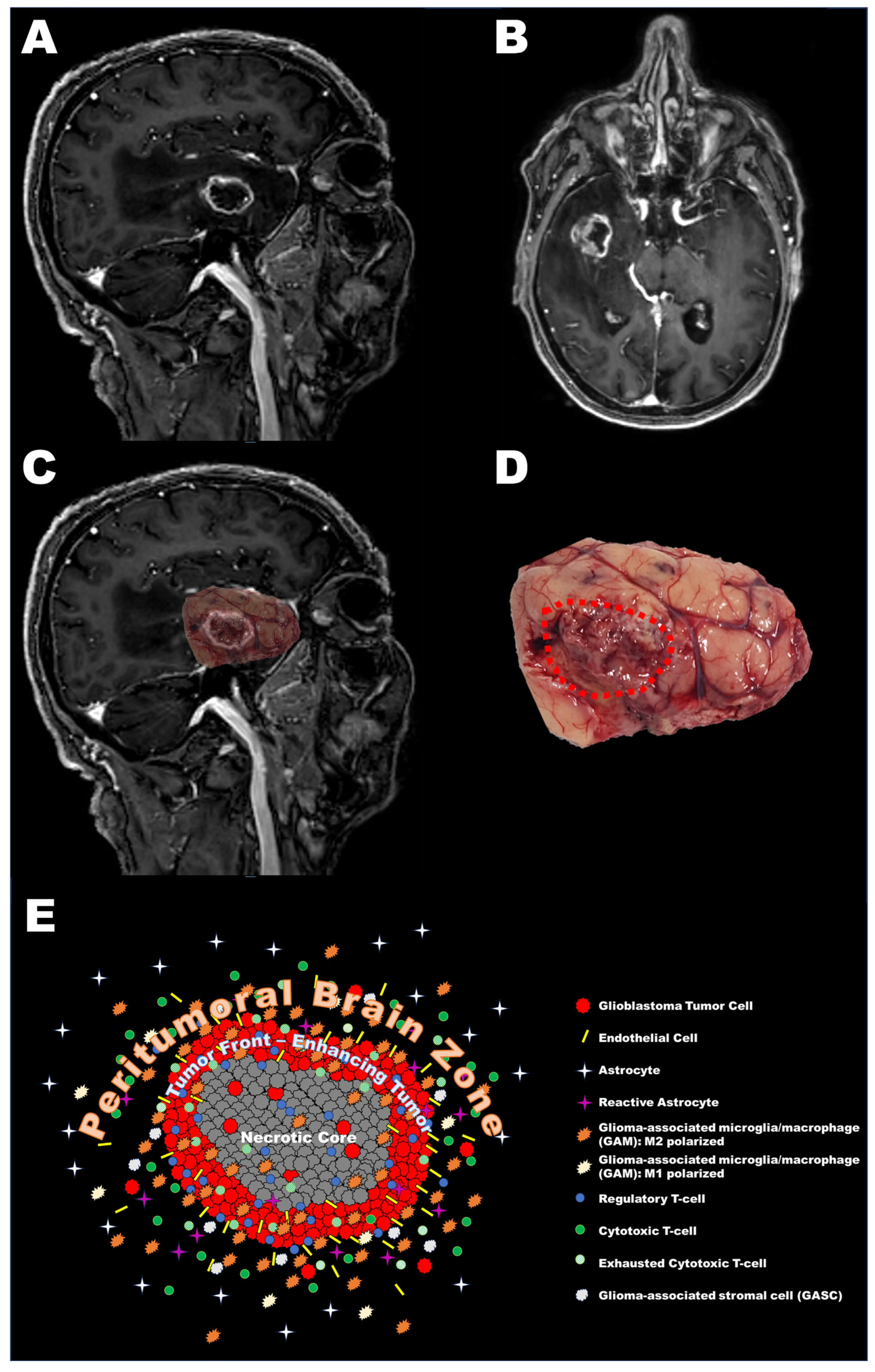
2. Definition of the Peritumoral Brain Zone (PBZ)
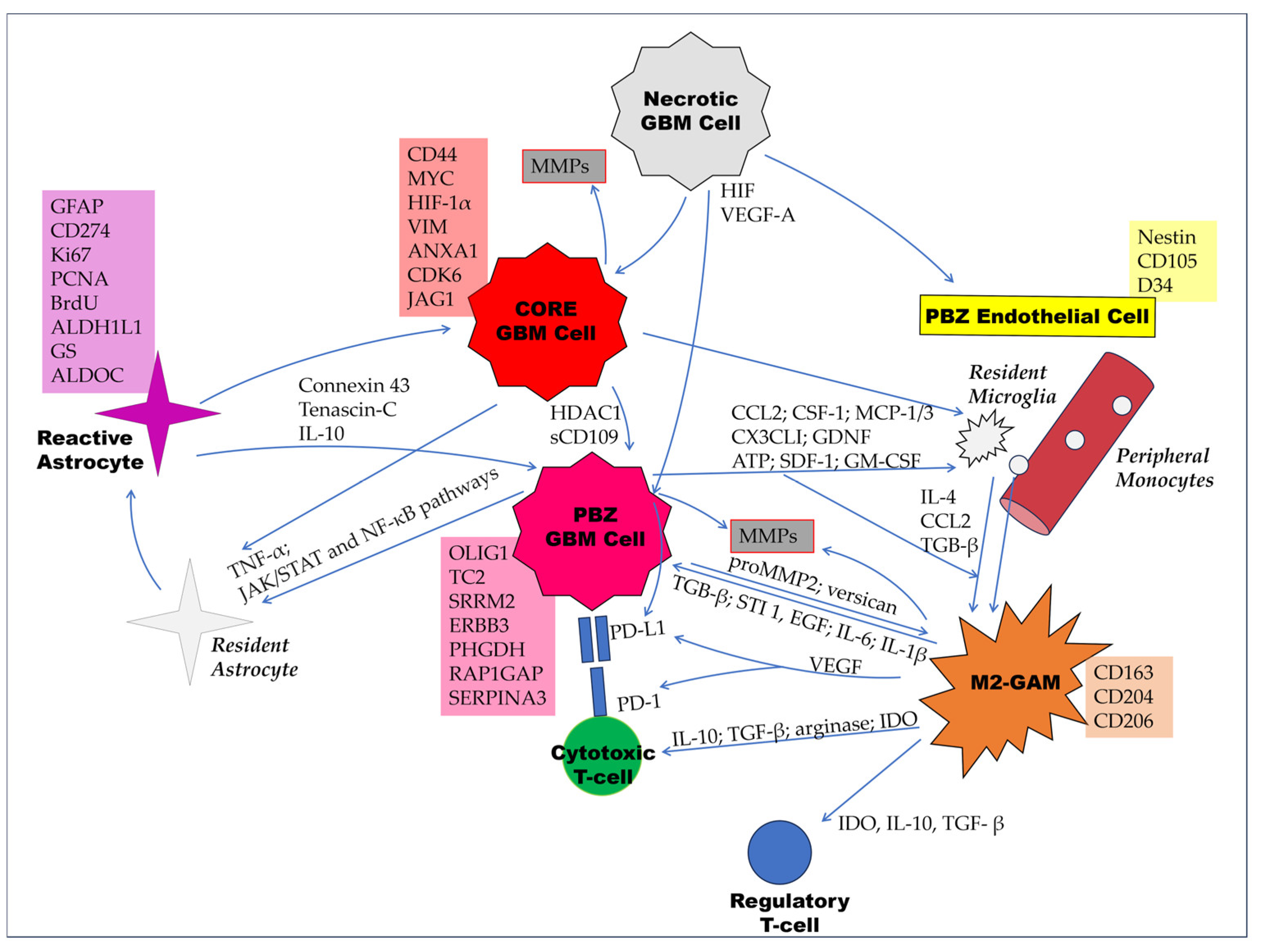
3. Cellular Characteristics of the PBZ
3.1. Tumor Cells
3.2. Non-Neoplastic Cells
3.2.1. Endothelial Cells
3.2.2. Reactive Astrocytes
3.2.3. Glioma-Associated Microglia/Macrophages (GAMs)
3.2.4. Tumor-Infiltrating Lymphocytes (TILs)
3.2.5. Glioma-Associated Stromal Cells (GASCs)
3.3. Summary of Cellular Characteristics of the PBZ
4. Molecular Characteristics of the PBZ
4.1. Immunohistochemistry
4.2. Genomic and Transcriptomic Characteristics
5. Discussion
- -
- The PBZ is the real target of post-surgical therapies; therefore, understanding it is of paramount translational significance.
- -
- Only in about one third of cases is it possible to detect tumor cells via microscopic analysis of the PBZ; nonetheless, we can observe distinct abnormal features also in cases of no infiltration.
- -
- If some data on the PBZ can suggest the influence on the features of this area of tumor cells migrated well beyond the tumor border, diluted in brain parenchyma, and possibly having a “dormant” feature and therefore liable to be missed by pathological examination, others suggest the recruitment of surrounding cells reprogrammed towards a neoplastic phenotype.
- -
- Angiogenesis and hypoxia play a pivotal role in influencing the tumor and PBZ microenvironment.
- -
- An increasing immunocompromised gradient is seen from the PBZ to the tumor core.
- -
- The role of inflammatory cells has been illuminated in recent years, and several alterations in the PBZ can be related to an activation of microglia and astrocytes that could promote tumor growth and invasion.
- -
- The transcriptomic profile of the PBZ is extremely complex, since this tissue differs from normal white matter, shares some tumor core characteristics, and possibly shows some peculiar alterations. Overall, the PBZ seems to be “half-way” on the road towards malignancy. However, the data are highly heterogeneous due to possible infiltration of glioma cells/glioma stem cells. Single-cell analysis could improve our understanding.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncology 2021, 23, iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Ziu, M.; Kim, B.Y.S.; Jiang, W.; Ryken, T.; Olson, J.J. The role of radiation therapy in treatment of adults with newly diagnosed glioblastoma multiforme: A systematic review and evidence-based clinical practice guideline update. J. Neuro-Oncol. 2020, 150, 215–267. [Google Scholar] [CrossRef]
- Redjal, N.; Nahed, B.V.; Dietrich, J.; Kalkanis, S.N.; Olson, J.J. Congress of neurological surgeons systematic review and evidence-based guidelines update on the role of chemotherapeutic management and antiangiogenic treatment of newly diagnosed glioblastoma in adults. J. Neuro-Oncol. 2020, 150, 165–213. [Google Scholar] [CrossRef]
- Domino, J.S.; Ormond, D.R.; Germano, I.M.; Sami, M.; Ryken, T.C.; Olson, J.J. Cytoreductive surgery in the management of newly diagnosed glioblastoma in adults: A systematic review and evidence-based clinical practice guideline update. J. Neuro-Oncol. 2020, 150, 121–142. [Google Scholar] [CrossRef]
- Ius, T.; Sabatino, G.; Panciani, P.P.; Fontanella, M.M.; Rudà, R.; Castellano, A.; Barbagallo, G.M.V.; Belotti, F.; Boccaletti, R.; Catapano, G.; et al. Surgical management of Glioma Grade 4: Technical update from the neuro-oncology section of the Italian Society of Neurosurgery (SINch®): A systematic review. J. Neuro-Oncol. 2023, 162, 267–293. [Google Scholar] [CrossRef]
- Mier-García, J.F.; Ospina-Santa, S.; Orozco-Mera, J.; Ma, R.; Plaha, P. Supramaximal versus gross total resection in Glioblastoma, IDH wild-type and Astrocytoma, IDH-mutant, grade 4, effect on overall and progression free survival: Systematic review and meta-analysis. J. Neuro-Oncol. 2023, 164, 31–41. [Google Scholar] [CrossRef]
- Petrecca, K.; Guiot, M.-C.; Panet-Raymond, V.; Souhami, L. Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J. Neuro-Oncol. 2013, 111, 19–23. [Google Scholar] [CrossRef]
- Dandy, W.E. Removal of Right Cerebral Hemisphere for Certain Tumors with Hemiplegia. J. Am. Med. Assoc. 1928, 90, 823–825. [Google Scholar] [CrossRef]
- Capper, D.; Sahm, F.; Jeibmann, A.; Habel, A.; Paulus, W.; Troost, D.; von Deimling, A. Addressing Diffuse Glioma as a Systemic Brain Disease with Single-Cell Analysis. Arch. Neurol. 2012, 69, 523–526. [Google Scholar] [CrossRef]
- Silbergeld, D.L.; Chicoine, M.R. Isolation and characterization of human malignant glioma cells from histologically normal brain. J. Neurosurg. 1997, 86, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Mangiola, A.; de Bonis, P.; Maira, G.; Balducci, M.; Sica, G.; Lama, G.; Lauriola, L.; Anile, C. Invasive tumor cells and prognosis in a selected population of patients with glioblastoma multiforme. Cancer 2008, 113, 841–846. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Fidoamore, A.; Cristiano, L.; Antonosante, A.; D’angelo, M.; Di Giacomo, E.; Astarita, C.; Giordano, A.; Ippoliti, R.; Benedetti, E.; Cimini, A. Glioblastoma Stem Cells Microenvironment: The Paracrine Roles of the Niche in Drug and Radioresistance. Stem Cells Int. 2016, 2016, 6809105. [Google Scholar] [CrossRef] [PubMed]
- Mangiola, A.; Saulnier, N.; De Bonis, P.; Orteschi, D.; Sica, G.; Lama, G.; Pettorini, B.L.; Sabatino, G.; Zollino, M.; Lauriola, L.; et al. Gene Expression Profile of Glioblastoma Peritumoral Tissue: An Ex Vivo Study. PLoS ONE 2013, 8, e57145. [Google Scholar] [CrossRef]
- Dapash, M.; Hou, D.; Castro, B.; Lee-Chang, C.; Lesniak, M.S. The Interplay between Glioblastoma and Its Microenvironment. Cells 2021, 10, 2257. [Google Scholar] [CrossRef] [PubMed]
- DeCordova, S.; Shastri, A.; Tsolaki, A.G.; Yasmin, H.; Klein, L.; Singh, S.K.; Kishore, U. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front. Immunol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Simon, T.; Jackson, E.; Giamas, G. Breaking through the glioblastoma micro-environment via extracellular vesicles. Oncogene 2020, 39, 4477–4490. [Google Scholar] [CrossRef]
- Ye, Z.; Ai, X.; Zhao, L.; Fei, F.; Wang, P.; Zhou, S. Phenotypic plasticity of myeloid cells in glioblastoma development, progression, and therapeutics. Oncogene 2021, 40, 6059–6070. [Google Scholar] [CrossRef]
- Faisal, S.M.; Comba, A.; Varela, M.L.; Argento, A.E.; Brumley, E.; Abel, C.; Castro, M.G.; Lowenstein, P.R. The complex interactions between the cellular and non-cellular components of the brain tumor microenvironmental landscape and their therapeutic implications. Front. Oncol. 2022, 12, 1005069. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. The Definition of Primary and Secondary Glioblastoma. Clin. Cancer Res. 2013, 19, 764–772. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Genetic Pathways to Primary and Secondary Glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Lemée, J.-M.; Clavreul, A.; Aubry, M.; Com, E.; De Tayrac, M.; Eliat, P.-A.; Henry, C.; Rousseau, A.; Mosser, J.; Menei, P. Characterizing the peritumoral brain zone in glioblastoma: A multidisciplinary analysis. J. Neuro-Oncol. 2015, 122, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Broggi, G.; Altieri, R.; Barresi, V.; Certo, F.; Barbagallo, G.M.V.; Zanelli, M.; Palicelli, A.; Magro, G.; Caltabiano, R. Histologic Definition of Enhancing Core and FLAIR Hyperintensity Region of Glioblastoma, IDH-Wild Type: A Clinico-Pathologic Study on a Single-Institution Series. Brain Sci. 2023, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Altieri, R.; Barbagallo, D.; Certo, F.; Broggi, G.; Ragusa, M.; Di Pietro, C.; Caltabiano, R.; Magro, G.; Peschillo, S.; Purrello, M.; et al. Peritumoral Microenvironment in High-Grade Gliomas: From FLAIRectomy to Microglia–Glioma Cross-Talk. Brain Sci. 2021, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Certo, F.; Altieri, R.; Maione, M.; Schonauer, C.; Sortino, G.; Fiumanò, G.; Tirrò, E.; Massimino, M.; Broggi, G.; Vigneri, P.; et al. FLAIRectomy in Supramarginal Resection of Glioblastoma Correlates with Clinical Outcome and Survival Analysis: A Prospective, Single Institution, Case Series. Oper. Neurosurg. 2021, 20, 151–163. [Google Scholar] [CrossRef]
- Deng, Z.; Yan, Y.; Zhong, D.; Yang, G.; Tang, W.; Lü, F.; Xie, B.; Liu, B. Quantitative analysis of glioma cell invasion by diffusion tensor imaging. J. Clin. Neurosci. 2010, 17, 1530–1536. [Google Scholar] [CrossRef]
- Lu, S.; Ahn, D.; Johnson, G.; Law, M.; Zagzag, D.; Grossman, R.I.; Bette, S.; Huber, T.; Gempt, J.; Boeckh-Behrens, T.; et al. Diffusion-Tensor MR Imaging of Intracranial Neoplasia and Associated Peritumoral Edema: Introduction of the Tumor Infiltration Index. Radiology 2004, 232, 221–228. [Google Scholar] [CrossRef]
- Prasanna, P.; Patel, J.; Partovi, S.; Madabhushi, A.; Tiwari, P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur. Radiol. 2017, 27, 4188–4197. [Google Scholar] [CrossRef]
- Dasgupta, A.; Geraghty, B.; Maralani, P.J.; Malik, N.; Sandhu, M.; Detsky, J.; Tseng, C.-L.; Soliman, H.; Myrehaug, S.; Husain, Z.; et al. Quantitative mapping of individual voxels in the peritumoral region of IDH-wildtype glioblastoma to distinguish between tumor infiltration and edema. J. Neuro-Oncol. 2021, 153, 251–261. [Google Scholar] [CrossRef]
- Malik, N.; Geraghty, B.; Dasgupta, A.; Maralani, P.J.; Sandhu, M.; Detsky, J.; Tseng, C.-L.; Soliman, H.; Myrehaug, S.; Husain, Z.; et al. MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J. Neuro-Oncol. 2021, 155, 181–191. [Google Scholar] [CrossRef]
- Guerrini, F.; Roca, E.; Spena, G. Supramarginal Resection for Glioblastoma: It Is Time to Set Boundaries! A Critical Review on a Hot Topic. Brain Sci. 2022, 12, 652. [Google Scholar] [CrossRef]
- Mangiola, A.; Lama, G.; Giannitelli, C.; De Bonis, P.; Anile, C.; Lauriola, L.; La Torre, G.; Sabatino, G.; Maira, G.; Jhanwar-Uniyal, M.; et al. Stem Cell Marker Nestin and c-Jun NH2-Terminal Kinases in Tumor and Peritumor Areas of Glioblastoma Multiforme: Possible Prognostic Implications. Clin. Cancer Res. 2007, 13, 6970–6977. [Google Scholar] [CrossRef] [PubMed]
- Aubry, M.; de Tayrac, M.; Etcheverry, A.; Clavreul, A.; Saikali, S.; Menei, P.; Mosser, J. From the core to beyond the margin: A genomic picture of glioblastoma intratumor heterogeneity. Oncotarget 2015, 6, 12094–12109. [Google Scholar] [CrossRef] [PubMed]
- Idoate, M.A.; Díez Valle, R.; Echeveste, J.; Tejada, S. Pathological characterization of the glioblastoma border as shown during surgery using 5-aminolevulinic acid-induced fluorescence. Neuropathology 2011, 31, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Clavreul, A.; Etcheverry, A.; Tétaud, C.; Rousseau, A.; Avril, T.; Henry, C.; Mosser, J.; Menei, P. Identification of two glioblastoma-associated stromal cell subtypes with different carcinogenic properties in histologically normal surgical margins. J. Neuro-Oncol. 2015, 122, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Glas, M.; Rath, B.H.; Simon, M.; Reinartz, R.; Schramme, A.; Trageser, D.; Eisenreich, R.; Leinhaas, A.; Keller, M.; Schildhaus, H.; et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann. Neurol. 2010, 68, 264–269. [Google Scholar] [CrossRef]
- Piccirillo, S.G.M.; Dietz, S.; Madhu, B.; Griffiths, J.; Price, S.J.; Collins, V.P.; Watts, C. Fluorescence-guided surgical sampling of glioblastoma identifies phenotypically distinct tumour-initiating cell populations in the tumour mass and margin. Br. J. Cancer 2012, 107, 462–468. [Google Scholar] [CrossRef]
- Piccirillo, S.G.M.; Combi, R.; Cajola, L.; Patrizi, A.; Redaelli, S.; Bentivegna, A.; Baronchelli, S.; Maira, G.; Pollo, B.; Mangiola, A.; et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene 2009, 28, 1807–1811. [Google Scholar] [CrossRef]
- Bastola, S.; Pavlyukov, M.S.; Yamashita, D.; Ghosh, S.; Cho, H.; Kagaya, N.; Zhang, Z.; Minata, M.; Lee, Y.; Sadahiro, H.; et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun. 2020, 11, 4660. [Google Scholar] [CrossRef]
- Sica; Sica, G.; Lama, G.; Anile, C.; Geloso, M.C.; La Torre, G.; De Bonis, P.; Maira, G.; Lauriola, L.; Jhanwar-Uniyal, M.; et al. Assessment of angiogenesis by CD105 and nestin expression in peritumor tissue of glioblastoma. Int. J. Oncol. 2011, 38, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Lemée, J.-M.; Clavreul, A.; Menei, P. Intratumoral heterogeneity in glioblastoma: Don’t forget the peritumoral brain zone. Neuro-Oncology 2015, 17, 1322–1332. [Google Scholar] [CrossRef]
- Nimbalkar, V.P.; Kruthika, B.S.; Sravya, P.; Rao, S.; Sugur, H.S.; Verma, B.K.; Chickabasaviah, Y.T.; Arivazhagan, A.; Kondaiah, P.; Santosh, V. Differential gene expression in peritumoral brain zone of glioblastoma: Role of SERPINA3 in promoting invasion, stemness and radioresistance of glioma cells and association with poor patient prognosis and recurrence. J. Neuro-Oncol. 2021, 152, 55–65. [Google Scholar] [CrossRef]
- Gill, B.J.; Pisapia, D.J.; Malone, H.R.; Goldstein, H.; Lei, L.; Sonabend, A.; Yun, J.; Samanamud, J.; Sims, J.S.; Banu, M.; et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 12550–12555. [Google Scholar] [CrossRef]
- Charles, N.A.; Holland, E.C.; Gilbertson, R.; Glass, R.; Kettenmann, H. The brain tumor microenvironment. Glia 2011, 59, 1169–1180. [Google Scholar] [CrossRef]
- Ahir, B.K.; Engelhard, H.H.; Lakka, S.S. Tumor Development and Angiogenesis in Adult Brain Tumor: Glioblastoma. Mol. Neurobiol. 2020, 57, 2461–2478. [Google Scholar] [CrossRef] [PubMed]
- Domènech, M.; Hernández, A.; Plaja, A.; Martínez-Balibrea, E.; Balañà, C. Hypoxia: The Cornerstone of Glioblastoma. Int. J. Mol. Sci. 2021, 22, 12608. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Zhu, J.; Zhang, X.; Mao, X. The Role of Hypoxia and Cancer Stem Cells in Development of Glioblastoma. Cancers 2023, 15, 2613. [Google Scholar] [CrossRef]
- D’alessio, A.; Proietti, G.; Lama, G.; Biamonte, F.; Lauriola, L.; Moscato, U.; Vescovi, A.; Mangiola, A.; Angelucci, C.; Sica, G. Analysis of angiogenesis related factors in glioblastoma, peritumoral tissue and their derived cancer stem cells. Oncotarget 2016, 7, 78541–78556. [Google Scholar] [CrossRef]
- Tamura, R.; Ohara, K.; Sasaki, H.; Morimoto, Y.; Yoshida, K.; Toda, M. Histopathological vascular investigation of the peritumoral brain zone of glioblastomas. J. Neuro-Oncol. 2018, 136, 233–241. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Chen, H.L.; Girgis, K.R.; Cunningham, H.T.; Meny, G.M.; Nadaf, S.; Kavanaugh, D.; Carbone, D.P. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat. Med. 1996, 2, 1096–1103. [Google Scholar] [CrossRef]
- Ohm, J.E.; Gabrilovich, D.I.; Sempowski, G.D.; Kisseleva, E.; Parman, K.S.; Nadaf, S.; Carbone, D.P. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood 2003, 101, 4878–4886. [Google Scholar] [CrossRef] [PubMed]
- Escartin, C.; Galea, E.; Lakatos, A.; O’callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Steinhäuser, C.; Volterra, A.; Carmignoto, G.; Agarwal, A.; et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Heiland, D.H.; Ravi, V.M.; Behringer, S.P.; Frenking, J.H.; Wurm, J.; Joseph, K.; Garrelfs, N.W.C.; Strähle, J.; Heynckes, S.; Grauvogel, J.; et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat. Commun. 2019, 10, 2541. [Google Scholar] [CrossRef] [PubMed]
- Placone, A.L.; Quiñones-Hinojosa, A.; Searson, P.C. The role of astrocytes in the progression of brain cancer: Complicating the picture of the tumor microenvironment. Tumor Biol. 2016, 37, 61–69. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef]
- Lin, Q.; Liu, Z.; Ling, F.; Xu, G. Astrocytes protect glioma cells from chemotherapy and upregulate survival genes via gap junctional communication. Mol. Med. Rep. 2016, 13, 1329–1335. [Google Scholar] [CrossRef]
- Munoz, J.L.; Rodriguez-Cruz, V.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Temozolomide resistance in glioblastoma cells occurs partly through epidermal growth factor receptor-mediated induction of connexin. Cell Death Dis. 2014, 5, e1145. [Google Scholar] [CrossRef]
- Huang, J.-Y.; Cheng, Y.-J.; Lin, Y.-P.; Lin, H.-C.; Su, C.-C.; Juliano, R.; Yang, B.-C. Extracellular Matrix of Glioblastoma Inhibits Polarization and Transmigration of T Cells: The Role of Tenascin-C in Immune Suppression. J. Immunol. 2010, 185, 1450–1459. [Google Scholar] [CrossRef]
- Fujita, M.; Zhu, X.; Sasaki, K.; Ueda, R.; Low, K.L.; Pollack, I.F.; Okada, H. Inhibition of STAT3 Promotes the Efficacy of Adoptive Transfer Therapy Using Type-1 CTLs by Modulation of the Immunological Microenvironment in a Murine Intracranial Glioma. J. Immunol. 2008, 180, 2089–2098. [Google Scholar] [CrossRef]
- Fazi, B.; Felsani, A.; Grassi, L.; Moles, A.; D’andrea, D.; Toschi, N.; Sicari, D.; De Bonis, P.; Anile, C.; Guerrisi, M.G.; et al. The transcriptome and miRNome profiling of glioblastoma tissues and peritumoral regions highlights molecular pathways shared by tumors and surrounding areas and reveals differences between short-term and long-term survivors. Oncotarget 2015, 6, 22526–22552. [Google Scholar] [CrossRef] [PubMed]
- Roggendorf, W.; Strupp, S.; Paulus, W. Distribution and characterization of microglia/macrophages in human brain tumors. Acta Neuropathol. 1996, 92, 288–293. [Google Scholar] [CrossRef] [PubMed]
- da Fonseca, A.C.C.; Badie, B. Microglia and Macrophages in Malignant Gliomas: Recent Discoveries and Implications for Promising Therapies. J. Immunol. Res. 2013, 2013, 264124. [Google Scholar] [CrossRef]
- Hussain, S.F.; Yang, D.; Suki, D.; Aldape, K.; Grimm, E.; Heimberger, A.B. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro-Oncology 2006, 8, 261–279. [Google Scholar] [CrossRef]
- Watters, J.J.; Schartner, J.M.; Badie, B. Microglia function in brain tumors. J. Neurosci. Res. 2005, 81, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; E Doykan, C.; et al. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef]
- Michelucci, A.; Heurtaux, T.; Grandbarbe, L.; Morga, E.; Heuschling, P. Characterization of the microglial phenotype under specific pro-inflammatory and anti-inflammatory conditions: Effects of oligomeric and fibrillar amyloid-β. J. Neuroimmunol. 2009, 210, 3–12. [Google Scholar] [CrossRef]
- Tamura, R.; Tamura, R.; Tanaka, T.; Tanaka, T.; Yamamoto, Y.; Yamamoto, Y.; Akasaki, Y.; Akasaki, Y.; Sasaki, H.; Sasaki, H.; et al. Dual role of macrophage in tumor immunity. Immunotherapy 2018, 10, 899–909. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.-E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Lisi, L.; Stigliano, E.; Lauriola, L.; Navarra, P.; Russo, C.D. Proinflammatory-Activated Glioma Cells Induce a Switch in Microglial Polarization and Activation Status, from a Predominant M2b Phenotype to a Mixture of M1 and M2a/B Polarized Cells. ASN Neuro 2014, 6, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef]
- Lisi, L.; Ciotti, G.M.P.; Braun, D.; Kalinin, S.; Currò, D.; Dello Russo, C.; Coli, A.; Mangiola, A.; Anile, C.; Feinstein, D.L.; et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci. Lett. 2017, 645, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Rahimi Koshkaki, H.; Minasi, S.; Ugolini, A.; Trevisi, G.; Napoletano, C.; Zizzari, I.G.; Gessi, M.; Giangaspero, F.; Mangiola, A.; Nuti, M.; et al. Immunohistochemical Characterization of Immune Infiltrate in Tumor Microenvironment of Glioblastoma. J. Pers. Med. 2020, 10, 112. [Google Scholar] [CrossRef]
- Annovazzi, L.; Mellai, M.; Bovio, E.; Mazzetti, S.; Pollo, B.; Schiffer, D. Microglia immunophenotyping in gliomas. Oncol. Lett. 2018, 15, 998–1006. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Sampson, J.H.; Gunn, M.D.; Fecci, P.E.; Ashley, D.M. Brain immunology and immunotherapy in brain tumours. Nat. Rev. Cancer 2020, 20, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Ding, S. The Crosstalk between Tumor-Associated Macrophages (TAMs) and Tumor Cells and the Corresponding Targeted Therapy. Front. Oncol. 2020, 10, 590941. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Jung, T.-Y.; Jung, S.; Jang, W.-Y.; Moon, K.-S.; Kim, I.-Y.; Lee, M.-C.; Lee, J.-J. Tumour-infiltrating T-cell subpopulations in glioblastomas. Br. J. Neurosurg. 2012, 26, 21–27. [Google Scholar] [CrossRef]
- Woroniecka, K.; Chongsathidkiet, P.; Rhodin, K.; Kemeny, H.; Dechant, C.; Farber, S.H.; Elsamadicy, A.A.; Cui, X.; Koyama, S.; Jackson, C.; et al. T-Cell Exhaustion Signatures Vary with Tumor Type and Are Severe in Glioblastoma. Clin. Cancer Res. 2018, 24, 4175–4186. [Google Scholar] [CrossRef]
- Gong, D.; Shi, W.; Yi, S.-J.; Chen, H.; Groffen, J.; Heisterkamp, N. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 2012, 13, 31. [Google Scholar] [CrossRef]
- Vitkovic, L.; Maeda, S.; Sternberg, E. Anti-Inflammatory Cytokines: Expression and Action in the Brain. Neuroimmunomodulation 2001, 9, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, I.; Alizadeh, D.; Liang, J.; Zhang, L.; Gao, H.; Song, Y.; Ren, H.; Ouyang, M.; Wu, X.; D’apuzzo, M.; et al. Characterization of Arginase Expression in Glioma-Associated Microglia and Macrophages. PLoS ONE 2016, 11, e0165118. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, D.A.; Balyasnikova, I.V.; Chang, A.L.; Ahmed, A.U.; Moon, K.-S.; Auffinger, B.; Tobias, A.L.; Han, Y.; Lesniak, M.S. IDO Expression in Brain Tumors Increases the Recruitment of Regulatory T Cells and Negatively Impacts Survival. Clin. Cancer Res. 2012, 18, 6110–6121. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Théate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef]
- Tamura, R.; Ohara, K.; Sasaki, H.; Morimoto, Y.; Kosugi, K.; Yoshida, K.; Toda, M. Difference in Immunosuppressive Cells Between Peritumoral Area and Tumor Core in Glioblastoma. World Neurosurg. 2018, 120, e601–e610. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2017, 18, 153–167. [Google Scholar] [CrossRef]
- Voron, T.; Colussi, O.; Marcheteau, E.; Pernot, S.; Nizard, M.; Pointet, A.-L.; Latreche, S.; Bergaya, S.; Benhamouda, N.; Tanchot, C.; et al. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J. Exp. Med. 2015, 212, 139–148. [Google Scholar] [CrossRef]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Polanska, U.M.; Orimo, A. Carcinoma-associated fibroblasts: Non-neoplastic tumour-promoting mesenchymal cells. J. Cell. Physiol. 2013, 228, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Clavreul, A.; Guette, C.; Faguer, R.; Tétaud, C.; Boissard, A.; Lemaire, L.; Rousseau, A.; Avril, T.; Henry, C.; Coqueret, O.; et al. Glioblastoma-associated stromal cells (GASCs) from histologically normal surgical margins have a myofibroblast phenotype and angiogenic properties. J. Pathol. 2014, 233, 74–88. [Google Scholar] [CrossRef]
- Clavreul, A.; Menei, P. Mesenchymal Stromal-Like Cells in the Glioma Microenvironment: What Are These Cells? Cancers 2020, 12, 2628. [Google Scholar] [CrossRef] [PubMed]
- Clavreul, A.; Network, T.G.O.G.P.; Etcheverry, A.; Chassevent, A.; Quillien, V.; Avril, T.; Jourdan, M.-L.; Michalak, S.; François, P.; Carré, J.-L.; et al. Isolation of a new cell population in the glioblastoma microenvironment. J. Neuro-Oncol. 2012, 106, 493–504. [Google Scholar] [CrossRef]
- Hossain, A.; Gumin, J.; Gao, F.; Figueroa, J.; Shinojima, N.; Takezaki, T.; Priebe, W.; Villarreal, D.; Kang, S.-G.; Joyce, C.; et al. Mesenchymal Stem Cells Isolated from Human Gliomas Increase Proliferation and Maintain Stemness of Glioma Stem Cells Through the IL-6/gp130/STAT3 Pathway. Stem Cells 2015, 33, 2400–2415. [Google Scholar] [CrossRef]
- Ho, I.A.; Toh, H.C.; Ng, W.H.; Teo, Y.L.; Guo, C.M.; Hui, K.M.; Lam, P.Y. Human Bone Marrow-Derived Mesenchymal Stem Cells Suppress Human Glioma Growth through Inhibition of Angiogenesis. Stem Cells 2013, 31, 146–155. [Google Scholar] [CrossRef]
- Hmadcha, A.; Martin-Montalvo, A.; Gauthier, B.R.; Soria, B.; Capilla-Gonzalez, V. Therapeutic Potential of Mesen-chymal Stem Cells for Cancer Therapy. Front. Bioeng. Biotechnol. 2020, 8, 43. [Google Scholar] [CrossRef]
- Lim, E.-J.; Kim, S.; Oh, Y.; Suh, Y.; Kaushik, N.; Lee, J.-H.; Lee, H.-J.; Kim, M.-J.; Park, M.-J.; Kim, R.-K.; et al. Crosstalk between GBM cells and mesenchymal stemlike cells promotes the invasiveness of GBM through the C5a/p38/ZEB1 axis. Neuro-Oncology 2020, 22, 1452–1462. [Google Scholar] [CrossRef]
- D’alessandris, Q.G.; Della Pepa, G.M.; Noya, C.; Olivi, A.; Pallini, R. Mesenchymal stem cells: Are they the good or the bad? Neuro-Oncology 2021, 23, 1203–1204. [Google Scholar] [CrossRef]
- Kang, S.-G.; Lee, S.-J. Reply to D’Alessandris et al.: Clear evidence of differences between tumor-resident mesenchymal stemlike cells and bone marrow-derived mesenchymal stem cells. Neuro-Oncology 2021, 23, 1205–1206. [Google Scholar] [CrossRef]
- Bajetto, A.; Thellung, S.; Dellacasagrande, I.; Pagano, A.; Barbieri, F.; Florio, T. Cross talk between mesenchymal and glioblastoma stem cells: Communication beyond controversies. Stem Cells Transl. Med. 2020, 9, 1310–1330. [Google Scholar] [CrossRef]
- Schichor, C.; Birnbaum, T.; Etminan, N.; Schnell, O.; Grau, S.; Miebach, S.; Aboody, K.; Padovan, C.; Straube, A.; Tonn, J.-C.; et al. Vascular endothelial growth factor A contributes to glioma-induced migration of human marrow stromal cells (hMSC). Exp. Neurol. 2006, 199, 301–310. [Google Scholar] [CrossRef]
- Ho, I.A.W.; Chan, K.Y.W.; Ng, W.-H.; Guo, C.M.; Hui, K.M.; Cheang, P.; Lam, P.Y.P. Matrix Metalloproteinase 1 Is Necessary for the Migration of Human Bone Marrow-Derived Mesenchymal Stem Cells Toward Human Glioma. Stem Cells 2009, 27, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, T.; Hildebrandt, J.; Nuebling, G.; Sostak, P.; Straube, A. Glioblastoma-dependent differentiation and angiogenic potential of human mesenchymal stem cells in vitro. J. Neuro-Oncol. 2011, 105, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiang, W.; Xue, B.-Z.; Yi, D.-Y.; Zhao, H.-Y.; Fu, P. Growth factors contribute to the mediation of angiogenic capacity of glioma-associated mesenchymal stem cells. Oncol. Lett. 2021, 21, 215. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.-J.; Suh, Y.; Kim, S.; Kang, S.-G.; Lee, S.-J. Force-mediated proinvasive matrix remodeling driven by tumor-associated mesenchymal stem-like cells in glioblastoma. BMB Rep. 2018, 51, 182–187. [Google Scholar] [CrossRef]
- Oliveira, M.N.; Pillat, M.M.; Motaln, H.; Ulrich, H.; Lah, T.T. Kinin-B1 Receptor Stimulation Promotes Invasion and is Involved in Cell-Cell Interaction of Co-Cultured Glioblastoma and Mesenchymal Stem Cells. Sci. Rep. 2018, 8, 1299. [Google Scholar] [CrossRef]
- Sun, C.; Dai, X.; Zhao, D.; Wang, H.; Rong, X.; Huang, Q.; Lan, Q. Mesenchymal stem cells promote glioma neovascularization in vivo by fusing with cancer stem cells. BMC Cancer 2019, 19, 1240. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Yuan, F.; Zhu, J.; Yang, J.; Tang, C.; Cong, Z.; Ma, C. Glioma-Associated Stromal Cells Stimulate Glioma Malignancy by Regulating the Tumor Immune Microenvironment. Front. Oncol. 2021, 11, 672928. [Google Scholar] [CrossRef]
- Lama, G.; Mangiola, A.; Anile, C.; Sabatino, G.; De Bonis, P.; Lauriola, L.; Giannitelli, C.; La Torre, G.; Jhanwar-Uniyal, M.; Sica, G.; et al. Activated ERK1/2 Expression in Glioblastoma Multiforme and in Peritumor Tissue. Int. J. Oncol. 2007, 30, 1333–1342. [Google Scholar] [CrossRef]
- Antonyak, A.M.; Kenyon, L.C.; Godwin, A.K.; James, D.C.; Emlet, D.R.; Okamoto, I.; Tnani, M.; Holgado-Madruga, M.; Moscatello, D.K.; Wong, A.J. Elevated JNK activation contributes to the pathogenesis of human brain tumors. Oncogene 2002, 21, 5038–5046. [Google Scholar] [CrossRef]
- Cui, J.; Han, S.-Y.; Wang, C.; Su, W.; Harshyne, L.; Holgado-Madruga, M.; Wong, A.J. c-Jun NH2-Terminal Kinase 2α2 Promotes the Tumorigenicity of Human Glioblastoma Cells. Cancer Res. 2006, 66, 10024–10031. [Google Scholar] [CrossRef]
- Sluss, H.K.; Barrett, T.; Dérijard, B.; Davis, R.J. Signal Transduction by Tumor Necrosis Factor Mediated by JNK Protein Kinases. Mol. Cell Biol. 1994, 14, 8376–8384. [Google Scholar] [PubMed]
- Jensen, R.L.; Mumert, M.L.; Gillespie, D.L.; Kinney, A.Y.; Schabel, M.C.; Salzman, K.L. Preoperative dynamic contrast-enhanced MRI correlates with molecular markers of hypoxia and vascularity in specific areas of intratumoral microenvironment and is predictive of patient outcome. Neuro-Oncology 2014, 16, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Haybaeck, J.; Obrist, P.; Schindler, C.U.; Spizzo, G.; Doppler, W. STAT-1 Expression in Human Glioblastoma and Peritumoral Tissue. Anticancer Res. 2007, 27, 3829–3835. [Google Scholar]
- Thota, B.; Arimappamagan, A.; Kandavel, T.; Shastry, A.H.; Pandey, P.; Chandramouli, B.A.; Hegde, A.S.; Kondaiah, P.; Santosh, V. STAT-1 expression is regulated by IGFBP-3 in malignant glioma cells and is a strong predictor of poor survival in patients with glioblastoma. J. Neurosurg. 2014, 121, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Synowitz, M.; Glass, R.; Fäber, K.; Markovic, D.; Kronenberg, G.; Herrmann, K.; Schnermann, J.; Nolte, C.; van Rooijen, N.; Kiwit, J.; et al. A1 Adenosine Receptors in Microglia Control Glioblastoma-Host Interaction. Cancer Res. 2006, 66, 8550–8557. [Google Scholar] [CrossRef] [PubMed]
- Dufrusine, B.; Capone, E.; Ponziani, S.; Lattanzio, R.; Lanuti, P.; Giansanti, F.; De Laurenzi, V.; Iacobelli, S.; Ippoliti, R.; Mangiola, A.; et al. Extracellular LGALS3BP: A potential disease marker and actionable target for antibody–drug conjugate therapy in glioblastoma. Mol. Oncol. 2023, 17, 1460–1473. [Google Scholar] [CrossRef]
- Capone, E.; Iacobelli, S.; Sala, G. Role of galectin 3 binding protein in cancer progression: A potential novel therapeutic target. J. Transl. Med. 2021, 19, 405. [Google Scholar] [CrossRef]
- Rana, R.; Chauhan, K.; Gautam, P.; Kulkarni, M.; Banarjee, R.; Chugh, P.; Chhabra, S.S.; Acharya, R.; Kalra, S.K.; Gupta, A.; et al. Plasma-Derived Extracellular Vesicles Reveal Galectin-3 Binding Protein as Potential Biomarker for Early Detection of Glioma. Front. Oncol. 2021, 11, 778754. [Google Scholar] [CrossRef]
- Giansanti, F.; Capone, E.; Ponziani, S.; Piccolo, E.; Gentile, R.; Lamolinara, A.; Di Campli, A.; Sallese, M.; Iacobelli, V.; Cimini, A.; et al. Secreted Gal-3BP is a novel promising target for non-internalizing Antibody–Drug Conjugates. J. Control. Release 2019, 294, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, S.; Di Vittorio, G.; Pitari, G.; Cimini, A.M.; Ardini, M.; Gentile, R.; Iacobelli, S.; Sala, G.; Capone, E.; Flavell, D.J.; et al. Antibody-Drug Conjugates: The New Frontier of Chemotherapy. Int. J. Mol. Sci. 2020, 21, 5510. [Google Scholar] [CrossRef]
- Giambra, M.; Messuti, E.; Di Cristofori, A.; Cavandoli, C.; Bruno, R.; Buonanno, R.; Marzorati, M.; Zambuto, M.; Rodriguez-Menendez, V.; Redaelli, S.; et al. Characterizing the Genomic Profile in High-Grade Gliomas: From Tumor Core to Peritumoral Brain Zone, Passing through Glioma-Derived Tumorspheres. Biology 2021, 10, 1157. [Google Scholar] [CrossRef] [PubMed]
- Giambra, M.; Di Cristofori, A.; Conconi, D.; Marzorati, M.; Redaelli, S.; Zambuto, M.; Rocca, A.; Roumy, L.; Carrabba, G.; Lavitrano, M.; et al. Insights into the Peritumoural Brain Zone of Glioblastoma: CDK4 and EXT2 May Be Potential Drivers of Malignancy. Int. J. Mol. Sci. 2023, 24, 2835. [Google Scholar] [CrossRef]
- Cen, L.; Carlson, B.L.; Schroeder, M.A.; Ostrem, J.L.; Kitange, G.J.; Mladek, A.C.; Fink, S.R.; Decker, P.A.; Wu, W.; Kim, J.-S.; et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro-Oncology 2012, 14, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Giambra, M.; Di Cristofori, A.; Valtorta, S.; Manfrellotti, R.; Bigiogera, V.; Basso, G.; Moresco, R.M.; Giussani, C.; Bentivegna, A. The peritumoral brain zone in glioblastoma: Where we are and where we are going. J. Neurosci. Res. 2023, 101, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Jill, P.; Alexe, G.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chatterjee, M.; Schmid, H.; Beck, S.; Gawaz, M. CXCL14 as an emerging immune and inflammatory modulator. J. Inflamm. 2016, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Ellert-Miklaszewska, A.; Dabrowski, M.; Lipko, M.; Sliwa, M.; Maleszewska, M.; Kaminska, B. Molecular definition of the pro-tumorigenic phenotype of glioma-activated microglia. Glia 2013, 61, 1178–1190. [Google Scholar] [CrossRef]
- Lin, B.; Madan, A.; Yoon, J.-G.; Fang, X.; Yan, X.; Kim, T.-K.; Hwang, D.; Hood, L.; Foltz, G. Massively Parallel Signature Sequencing and Bioinformatics Analysis Identifies Up-Regulation of TGFBI and SOX4 in Human Glioblastoma. PLoS ONE 2010, 5, e10210. [Google Scholar] [CrossRef]
- Peng, P.; Zhu, H.; Liu, D.; Chen, Z.; Zhang, X.; Guo, Z.; Dong, M.; Wan, L.; Zhang, P.; Liu, G.; et al. TGFBI secreted by tumor-associated macrophages promotes glioblastoma stem cell-driven tumor growth via integrin αvβ5-Src-Stat3 signaling. Theranostics 2022, 12, 4221–4236. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Truty, M.A.; Kang, Y.; Chopin-Laly, X.; Zhang, R.; Roife, D.; Chatterjee, D.; Lin, E.; Thomas, R.M.; Wang, H.; et al. Extracellular Lumican Inhibits Pancreatic Cancer Cell Growth and Is Associated with Prolonged Survival after Surgery. Clin. Cancer Res. 2014, 20, 6529–6540. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, L.; Wu, Z.; Li, Y.; Ying, Z.; Lin, C.; Wu, J.; Hu, B.; Cheng, S.-Y.; Li, M.; et al. TGF-β induces miR-182 to sustain NF-κB activation in glioma subsets. J. Clin. Investig. 2012, 122, 3563–3578. [Google Scholar] [CrossRef] [PubMed]
- Ciafrè, S.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.-G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.; Farace, M. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Rolle, K.; Belter, A.; Barciszewska, A.M.; Żywicki, M.; Michalak, M.; Nowak, S.; Naskręt-Barciszewska, M.Z.; Barciszewski, J. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol. Oncol. 2015, 9, 1324–1340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, J.; Zhang, J.; Qiu, W.; Xu, S.; Yu, Q.; Liu, C.; Wang, Y.; Lu, A.; Zhang, J.; et al. MicroRNA-625 inhibits the proliferation and increases the chemosensitivity of glioma by directly targeting AKTAm. J. Cancer Res. 2017, 7, 1835–1849. [Google Scholar]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef]
- Shi, J.; Wang, Y.-J.; Sun, C.-R.; Qin, B.; Zhang, Y.; Chen, G. Long noncoding RNA lncHERG promotes cell proliferation, migration and invasion in glioblastoma. Oncotarget 2017, 8, 108031–108041. [Google Scholar] [CrossRef]
- Lemée, J.-M.; Com, E.; Clavreul, A.; Avril, T.; Quillien, V.; de Tayrac, M.; Pineau, C.; Menei, P. Proteomic analysis of glioblastomas: What is the best brain control sample? J. Proteom. 2013, 85, 165–173. [Google Scholar] [CrossRef][Green Version]
- Zhang, N.; Wei, L.; Ye, M.; Kang, C.; You, H. Treatment Progress of Immune Checkpoint Blockade Therapy for Glioblastoma. Front. Immunol. 2020, 11, 592612. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Li, X.; Li, Y.; Zhang, J.; Zong, Z.; Zhang, H. Current Immunotherapies for Glioblastoma Multiforme. Front. Immunol. 2020, 11, 603911. [Google Scholar] [CrossRef] [PubMed]

| Peritumoral Brain Zone Cellular Characteristics | ||
|---|---|---|
| Cell Type | Details | Main References |
| Tumor-infiltrating cells |
|
|
| Endothelial cells |
| |
| Reactive astrocytes |
|
|
| Glioma-associated microglia and macrophages |
| |
| Tumor-infiltrating lymphocytes |
| |
| Glioma-associated stromal cells |
| |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trevisi, G.; Mangiola, A. Current Knowledge about the Peritumoral Microenvironment in Glioblastoma. Cancers 2023, 15, 5460. https://doi.org/10.3390/cancers15225460
Trevisi G, Mangiola A. Current Knowledge about the Peritumoral Microenvironment in Glioblastoma. Cancers. 2023; 15(22):5460. https://doi.org/10.3390/cancers15225460
Chicago/Turabian StyleTrevisi, Gianluca, and Annunziato Mangiola. 2023. "Current Knowledge about the Peritumoral Microenvironment in Glioblastoma" Cancers 15, no. 22: 5460. https://doi.org/10.3390/cancers15225460
APA StyleTrevisi, G., & Mangiola, A. (2023). Current Knowledge about the Peritumoral Microenvironment in Glioblastoma. Cancers, 15(22), 5460. https://doi.org/10.3390/cancers15225460





