Role of Non-Coding RNAs in Hepatocellular Carcinoma Progression: From Classic to Novel Clinicopathogenetic Implications
Abstract
Simple Summary
Abstract
1. Introduction
2. Small Non-Coding RNAs and Hepatocellular Carcinoma
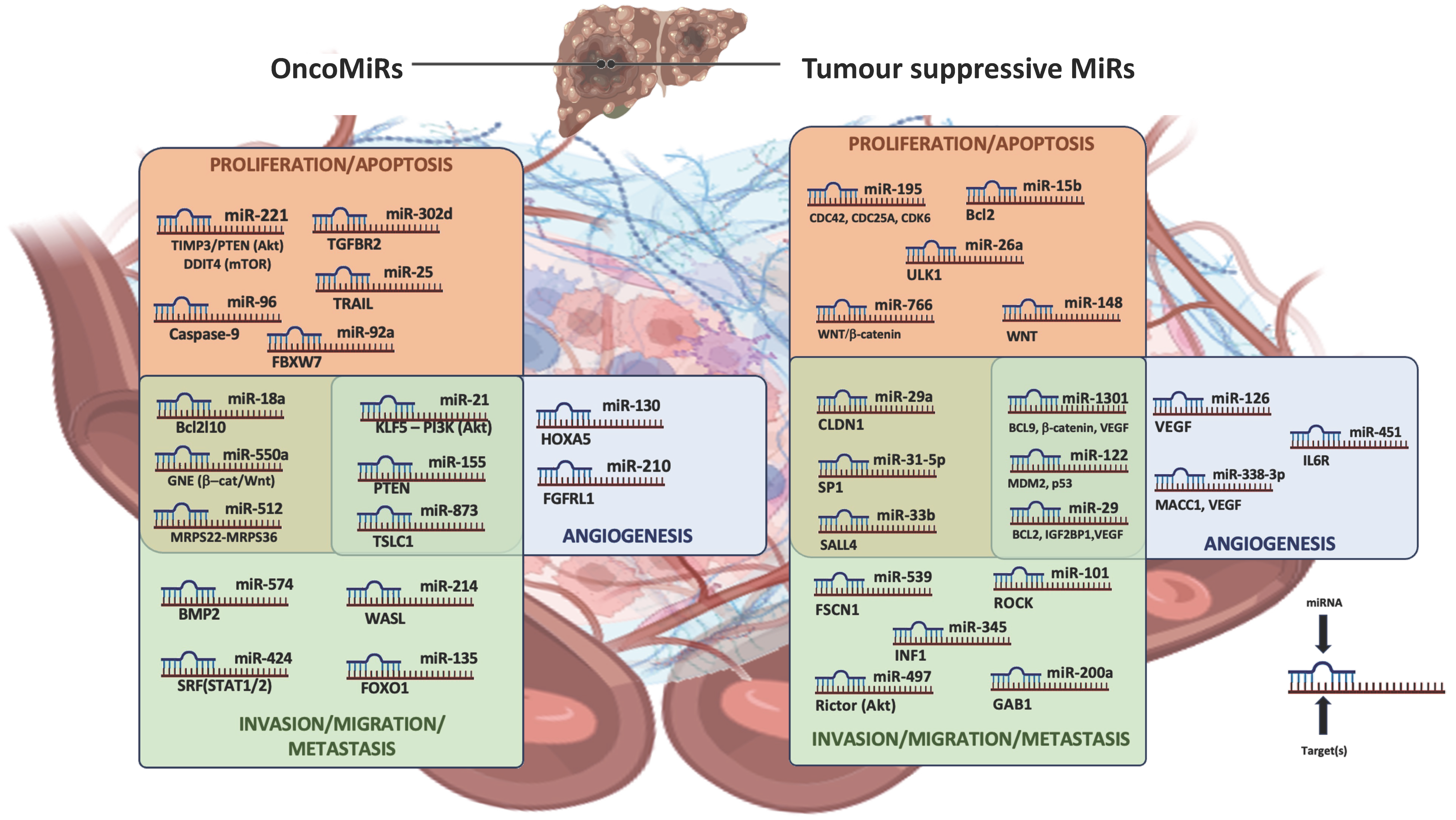
2.1. Role of microRNA Dysregulation in Hepatocellular Carcinoma Progression
2.1.1. Principal TS-miRNAs in Hepatocellular Carcinoma Progression
2.1.2. Principal oncomiRNAs in Hepatocellular Carcinoma Progression
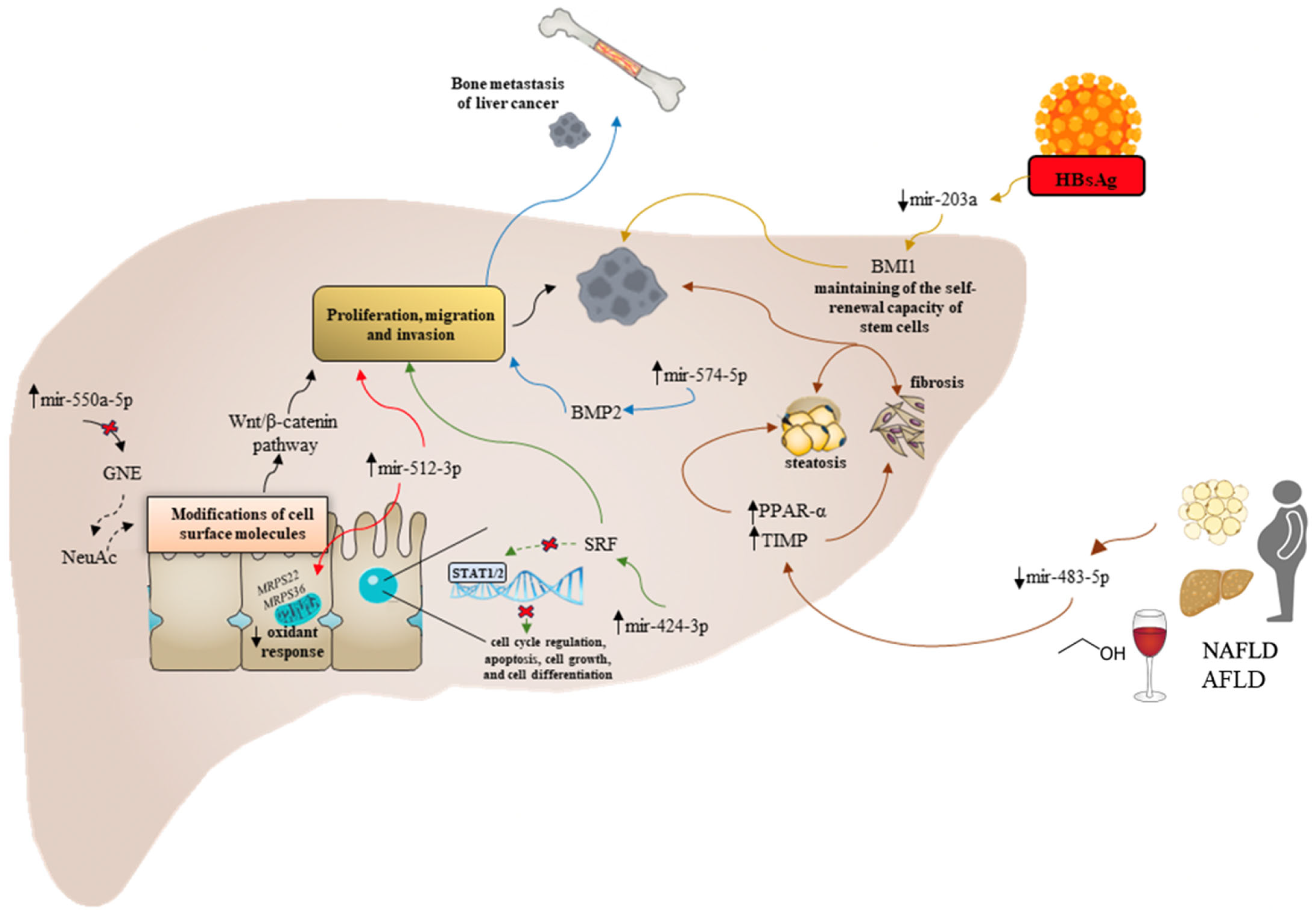
2.2. Etiology-Specific Dysregulated miRNAs in Hepatocellular Carcinoma Progression
3. Long Non-Coding RNAs and Hepatocellular Carcinoma
3.1. Role of Linear Long Non-Coding RNA Dysregulation in Hepatocellular Carcinoma Progression
3.2. Etiology-Specific Dysregulated Linear lncRNAs in Hepatocellular Carcinoma Progression
3.3. Circular RNAs in Liver Cancer Progression: Novel Diagnostic/Prognostic Markers?
3.4. Etiology-Specific Dysregulated Circular RNAs in Hepatocellular Carcinoma Progression
4. Non-Coding RNAs and Novel HCC Pathogenetic Frontiers
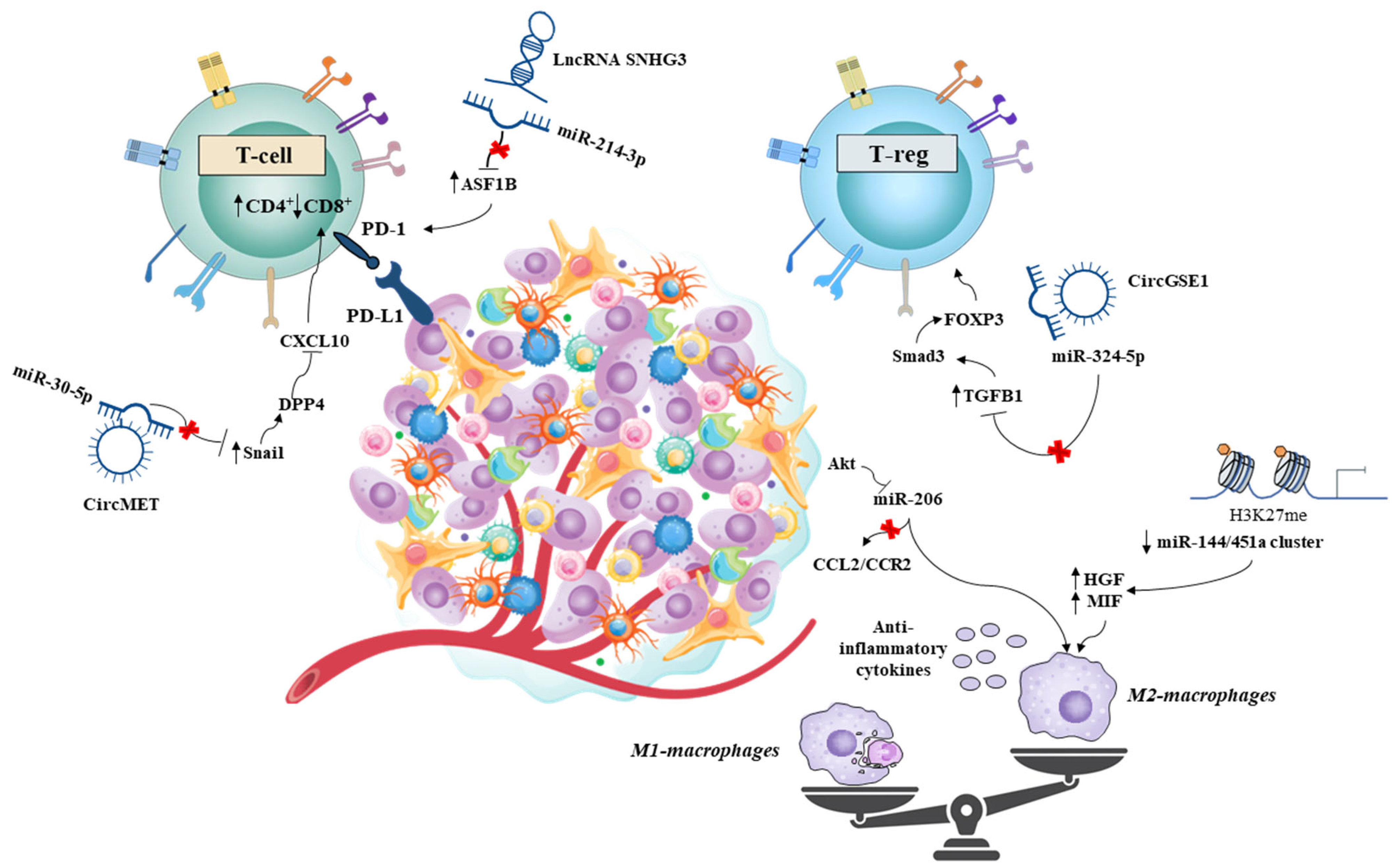
4.1. Role of Non-Coding RNAs in the HCC Microenvironment: A Matter of Immunity?
4.2. Non-Coding RNAs and Oxidative Stress: “Ferroptosis” Influences Immune-Mediated Progression
4.3. Gut Microbiota and Immunity: A Pathogenetic Bridge on HCC Landscape
4.4. NC-RNAs, Gut Microbiota, and Immunity in HCC
5. Potential ncRNA Applications in HCC Treatment: A Close-to-Exploding Time Bomb or an Under-Built Sand Castle?
5.1. SncRNAs’ Therapeutic Applications for HCC
5.2. ncRNAs: Small Molecules Greatly Contribute to the HCC Chemotherapy Resistance
6. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e471. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Otgonsuren, M.; Henry, L.; Venkatesan, C.; Mishra, A.; Erario, M.; Hunt, S. Association of nonalcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004 to 2009. Hepatology 2015, 62, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016, 379, 191–197. [Google Scholar] [CrossRef]
- Wong, C.M.; Ng, I.O. Molecular pathogenesis of hepatocellular carcinoma. Liver Int. 2008, 28, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Losic, B.; Craig, A.J.; Villacorta-Martin, C.; Martins-Filho, S.N.; Akers, N.; Chen, X.; Ahsen, M.E.; von Felden, J.; Labgaa, I.; DʹAvola, D.; et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020, 11, 291. [Google Scholar] [CrossRef]
- Saw, P.E.; Xu, X.; Chen, J.; Song, E.W. Non-coding RNAs: The new central dogma of cancer biology. Sci. China. Life Sci. 2021, 64, 22–50. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Giraud, J.; Chalopin, D.; Blanc, J.F.; Saleh, M. Hepatocellular Carcinoma Immune Landscape and the Potential of Immunotherapies. Front. Immunol. 2021, 12, 655697. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Greten, T.F. Gut microbiome in HCC—Mechanisms, diagnosis and therapy. J. Hepatol. 2020, 72, 230–238. [Google Scholar] [CrossRef]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. MicroRNA 2019, 8, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 2021, 905, 174178. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Kai, A.K.; Tsang, F.H.; Ng, I.O. Regulation of hepatocarcinogenesis by microRNAs. Front. Biosci. 2013, 5, 49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruiz-Manriquez, L.M.; Carrasco-Morales, O.; Sanchez, Z.E.; Osorio-Perez, S.M.; Estrada-Meza, C.; Pathak, S.; Banerjee, A.; Bandyopadhyay, A.; Duttaroy, A.K.; Paul, S. MicroRNA-mediated regulation of key signaling pathways in hepatocellular carcinoma: A mechanistic insight. Front. Genet. 2022, 13, 910733. [Google Scholar] [CrossRef]
- Thakral, S.; Ghoshal, K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef]
- Yang, J.; Gong, X.; Yang, J.; Ouyang, L.; Xiao, R.; You, X.; Ouyang, Y. Suppressive role of microRNA-29 in hepatocellular carcinoma via targeting IGF2BP1. Int. J. Clin. Exp. Pathol. 2018, 11, 1175–1185. [Google Scholar]
- Yang, Y.L.; Tsai, M.C.; Chang, Y.H.; Wang, C.C.; Chu, P.Y.; Lin, H.Y.; Huang, Y.H. MIR29A Impedes Metastatic Behaviors in Hepatocellular Carcinoma via Targeting LOX, LOXL2, and VEGFA. Int. J. Mol. Sci. 2021, 22, 6001. [Google Scholar] [CrossRef]
- Xiong, Y.; Fang, J.H.; Yun, J.P.; Yang, J.; Zhang, Y.; Jia, W.H.; Zhuang, S.M. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010, 51, 836–845. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, N.; Li, S.; Fang, J.H.; Chen, M.X.; Yang, J.; Jia, W.H.; Yuan, Y.; Zhuang, S.M. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology 2013, 58, 642–653. [Google Scholar] [CrossRef]
- Lee, M.H.; Kundu, J.K.; Chae, J.I.; Shim, J.H. Targeting ROCK/LIMK/cofilin signaling pathway in cancer. Arch. Pharm. Res. 2019, 42, 481–491. [Google Scholar] [CrossRef]
- Weidle, U.H.; Schmid, D.; Birzele, F.; Brinkmann, U. MicroRNAs Involved in Metastasis of Hepatocellular Carcinoma: Target Candidates, Functionality and Efficacy in Animal Models and Prognostic Relevance. Cancer Genom. Proteom. 2020, 17, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, J.; Zhang, R.; Yang, J.; Zhang, Q.; Liu, B. miR-497 inhibits the carcinogenesis of hepatocellular carcinoma by targeting the Rictor/Akt signal pathway. Int. J. Clin. Exp. Pathol. 2019, 12, 1992–2000. [Google Scholar] [PubMed]
- Mizuguchi, Y.; Takizawa, T.; Yoshida, H.; Uchida, E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol. Res. 2016, 46, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef]
- Yuan, B.; Dong, R.; Shi, D.; Zhou, Y.; Zhao, Y.; Miao, M.; Jiao, B. Down-regulation of miR-23b may contribute to activation of the TGF-β1/Smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett. 2011, 585, 927–934. [Google Scholar] [CrossRef]
- Wei, J.; Feng, L.; Li, Z.; Xu, G.; Fan, X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed. Pharmacother. Biomed. Pharmacother. 2013, 67, 387–392. [Google Scholar] [CrossRef]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. CR 2018, 37, 324. [Google Scholar] [CrossRef]
- Marquez, R.T.; Bandyopadhyay, S.; Wendlandt, E.B.; Keck, K.; Hoffer, B.A.; Icardi, M.S.; Christensen, R.N.; Schmidt, W.N.; McCaffrey, A.P. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Investig. A J. Technol. Methods Pathol. 2010, 90, 1727–1736. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Lv, X.; Ma, Y.; Chen, L.; Chen, Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017, 8, 44050–44058. [Google Scholar] [CrossRef]
- Yoon, J.S.; Kim, G.; Lee, Y.R.; Park, S.Y.; Tak, W.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Ha, H.T.; et al. Clinical significance of microRNA-21 expression in disease progression of patients with hepatocellular carcinoma. Biomark. Med. 2018, 12, 1105–1114. [Google Scholar] [CrossRef]
- Yerukala Sathipati, S.; Ho, S.Y. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci. Rep. 2020, 10, 14452. [Google Scholar] [CrossRef]
- Zhang, P.; Ma, Y.; Wang, F.; Yang, J.; Liu, Z.; Peng, J.; Qin, H. Comprehensive gene and microRNA expression profiling reveals the crucial role of hsa-let-7i and its target genes in colorectal cancer metastasis. Mol. Biol. Rep. 2012, 39, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, X.; Yu, L. Study of serum miR-518 and its correlation with inflammatory factors in patients with gestational diabetes mellitus complicated with hypertensive disorder complicating pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol. 2022, 272, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Huang, C.H.; Lai, S.J.; Yang, C.S.; Hsiao, T.H.; Lin, C.H.; Fu, P.K.; Ko, T.P.; Chen, Y. Mechanism and inhibition of human UDP-GlcNAc 2-epimerase, the key enzyme in sialic acid biosynthesis. Sci. Rep. 2016, 6, 23274. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Nishida, N.; Aoki, T.; Chishina, H.; Takita, M.; Ida, H.; Hagiwara, S.; Minami, Y.; Ueshima, K.; Kudo, M. Role of β-Catenin Activation in the Tumor Immune Microenvironment and Immunotherapy of Hepatocellular Carcinoma. Cancers 2023, 15, 2311. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Li, C.M.; Wu, W.; Zhu, Z.; Lu, P.L.; Gong, J.P.; Ma, R. miR-550a-5p promotes the proliferation and migration of hepatocellular carcinoma by targeting GNE via the Wnt/β-catenin signaling pathway. Neoplasma 2022, 69, 1359–1372. [Google Scholar] [CrossRef]
- Feng, L.; Chen, X.; Li, P.; Li, Y.; Zhai, Y.; Liu, X.; Jin, Q.; Zhang, H.; Yu, C.; Xing, B.; et al. miR-424-3p promotes metastasis of hepatocellular carcinoma via targeting the SRF-STAT1/2 axis. Carcinogenesis 2023, 44, 610–625. [Google Scholar] [CrossRef]
- Li, T.; Li, M.; Peng, Y.; Li, X.; Chen, Y.; Zhang, J.; Zhang, Z.; Zhang, H.; Du, J. Liver cancer cell-secreted exosomes promote bone metastasis of liver cancer by facilitating osteoclast differentiation through the miR-574-5p/BMP2 axis. Pathol. Res. Pract. 2023, 245, 154485. [Google Scholar] [CrossRef]
- Tessitore, A.; Cicciarelli, G.; Del Vecchio, F.; Gaggiano, A.; Verzella, D.; Fischietti, M.; Mastroiaco, V.; Vetuschi, A.; Sferra, R.; Barnabei, R.; et al. MicroRNA expression analysis in high fat diet-induced NAFLD-NASH-HCC progression: Study on C57BL/6J mice. BMC Cancer 2016, 16, 3. [Google Scholar] [CrossRef]
- De la Cruz-Ojeda, P.; Schmid, T.; Boix, L.; Moreno, M.; Sapena, V.; Praena-Fernández, J.M.; Castell, F.J.; Falcón-Pérez, J.M.; Reig, M.; Brüne, B.; et al. miR-200c-3p, miR-222-5p, and miR-512-3p Constitute a Biomarker Signature of Sorafenib Effectiveness in Advanced Hepatocellular Carcinoma. Cells 2022, 11, 2673. [Google Scholar] [CrossRef] [PubMed]
- Koga, C.; Kobayashi, S.; Nagano, H.; Tomimaru, Y.; Hama, N.; Wada, H.; Kawamoto, K.; Eguchi, H.; Konno, M.; Ishii, H.; et al. Reprogramming using microRNA-302 improves drug sensitivity in hepatocellular carcinoma cells. Ann. Surg. Oncol. 2014, 21 (Suppl. S4), S591–S600. [Google Scholar] [CrossRef] [PubMed]
- Iwai, N.; Yasui, K.; Tomie, A.; Gen, Y.; Terasaki, K.; Kitaichi, T.; Soda, T.; Yamada, N.; Dohi, O.; Seko, Y.; et al. Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int. J. Oncol. 2018, 53, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Jiang, J.; Shi, S.; Xie, H.; Zhou, L.; Zheng, S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int. J. Oncol. 2016, 49, 2600–2610. [Google Scholar] [CrossRef]
- Shigoka, M.; Tsuchida, A.; Matsudo, T.; Nagakawa, Y.; Saito, H.; Suzuki, Y.; Aoki, T.; Murakami, Y.; Toyoda, H.; Kumada, T.; et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol. Int. 2010, 60, 351–357. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, C.; Yang, Z.; Liu, W.; Yuan, Y.; Li, K.; Zhang, Y.; Wang, Y.; Shi, Y.; Qiu, Y.; et al. Dysregulated Sp1/miR-130b-3p/HOXA5 axis contributes to tumor angiogenesis and progression of hepatocellular carcinoma. Theranostics 2020, 10, 5209–5224. [Google Scholar] [CrossRef]
- Saad, A.; Zhu, X.Y.; Herrmann, S.; Hickson, L.; Tang, H.; Dietz, A.B.; van Wijnen, A.J.; Lerman, L.; Textor, S. Adipose-derived mesenchymal stem cells from patients with atherosclerotic renovascular disease have increased DNA damage and reduced angiogenesis that can be modified by hypoxia. Stem Cell Res. Ther. 2016, 7, 128. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Ren, Z. MicroRNA-214-5p Inhibits the Invasion and Migration of Hepatocellular Carcinoma Cells by Targeting Wiskott-Aldrich Syndrome Like. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 757–764. [Google Scholar] [CrossRef]
- Grants, J.M.; Wegrzyn, J.; Hui, T.; O’Neill, K.; Shadbolt, M.; Knapp, D.; Parker, J.; Deng, Y.; Gopal, A.; Docking, T.R.; et al. Altered microRNA expression links IL6 and TNF-induced inflammaging with myeloid malignancy in humans and mice. Blood 2020, 135, 2235–2251. [Google Scholar] [CrossRef]
- Zimta, A.A.; Cenariu, D.; Irimie, A.; Magdo, L.; Nabavi, S.M.; Atanasov, A.G.; Berindan-Neagoe, I. The Role of Nrf2 Activity in Cancer Development and Progression. Cancers 2019, 11, 1755. [Google Scholar] [CrossRef]
- Rui, T.; Zhang, X.; Feng, S.; Huang, H.; Zhan, S.; Xie, H.; Zhou, L.; Ling, Q.; Zheng, S. The Similar Effects of miR-512-3p and miR-519a-2-5p on the Promotion of Hepatocellular Carcinoma: Different Tunes Sung With Equal Skill. Front. Oncol. 2020, 10, 1244. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wen, H.; Jing, L.; Yang, Y.; Wang, W.; Liang, X.; Nan, K.; Yao, Y.; Tian, T. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017, 108, 620–631. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Zhao, Q.; Wei, W.; Dong, Z.; Shao, L.; Li, J.; Wu, W.; Zhang, H.; Huang, H.; et al. The miR-873/NDFIP1 axis promotes hepatocellular carcinoma growth and metastasis through the AKT/mTOR-mediated Warburg effect. Am. J. Cancer Res. 2019, 9, 927–944. [Google Scholar]
- Yang, Y.; Hou, N.; Wang, X.; Wang, L.; Chang, S.; He, K.; Zhao, Z.; Zhao, X.; Song, T.; Huang, C. miR-15b-5p induces endoplasmic reticulum stress and apoptosis in human hepatocellular carcinoma, both in vitro and in vivo, by suppressing Rab1A. Oncotarget 2015, 6, 16227–16238. [Google Scholar] [CrossRef][Green Version]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Que, K.; Zhou, Y.; Zhang, Z.; Zhao, X.; Gong, J.; Liu, Z. MicroRNA-766-3p Inhibits Tumour Progression by Targeting Wnt3a in Hepatocellular Carcinoma. Mol. Cells 2018, 41, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bo, L.; Lu, W.; Zhou, G.; Chen, Q. MicroRNA-148b targets Rho-associated protein kinase 1 to inhibit cell proliferation, migration and invasion in hepatocellular carcinoma. Mol. Med. Rep. 2016, 13, 477–482. [Google Scholar] [CrossRef][Green Version]
- Hu, M.H.; Ma, C.Y.; Wang, X.M.; Ye, C.D.; Zhang, G.X.; Chen, L.; Wang, J.G. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget 2016, 7, 66922–66934. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, A.; Xiang, J.; Lv, Y.; Zhang, X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol. Rep. 2016, 36, 1385–1392. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, W.; Zeng, X.C.; Jiang, N.; Fu, B.S.; Guo, Y.; Yi, H.M.; Li, H.; Zhang, Q.; Chen, W.J.; et al. Down-regulation of microRNA-338-3p promoted angiogenesis in hepatocellular carcinoma. Biomed. Pharmacother. Biomed. Pharmacother. 2016, 84, 583–591. [Google Scholar] [CrossRef]
- Liu, Y.; Hong, W.; Zhou, C.; Jiang, Z.; Wang, G.; Wei, G.; Li, X. miR-539 inhibits FSCN1 expression and suppresses hepatocellular carcinoma migration and invasion. Oncol. Rep. 2017, 37, 2593–2602. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Xue, H.; Wang, Y.; Shen, Q.; Jiang, Q.; Zhang, X.; Li, K.; Jia, M.; Jia, J.; Xu, J.; et al. miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int. J. Oncol. 2017, 50, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Weng, C.; Bai, R.; Sheng, J.; Gao, X.; Li, L.; Xu, Z. The regulatory network of miR-141 in the inhibition of angiogenesis. Angiogenesis 2019, 22, 251–262. [Google Scholar] [CrossRef]
- Zhao, G.; Han, C.; Zhang, Z.; Wang, L.; Xu, J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem. Biophys. Res. Commun. 2017, 490, 371–377. [Google Scholar] [CrossRef]
- Li, Y.; Li, R.; Fu, X.; Zhou, W.; Peng, S.; Fu, L. MicroRNA-33b inhibits cell proliferation in hepatocellular carcinoma via targeting SALL4. Zhong Nan Da Xue Xue Bao Yi Xue Ban J. Cent. South. Univ. Med. Sci. 2016, 41, 905–910. [Google Scholar] [CrossRef]
- Mahati, S.; Xiao, L.; Yang, Y.; Mao, R.; Bao, Y. miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem. Biophys. Res. Commun. 2017, 486, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Giovannini, C.; Veronese, A.; Ferracin, M.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Tavolari, S.; et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009, 69, 5761–5767. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Xu, Y.; Cheng, F.; Hu, Y.; Yang, S.; Rao, J.; Wang, X. miR-1301 inhibits hepatocellular carcinoma cell migration, invasion, and angiogenesis by decreasing Wnt/β-catenin signaling through targeting BCL9. Cell Death Dis. 2017, 8, e2999. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Harricharran, T.; Huaman, J.; Galuza, A.; Odumuwagun, O.; Tan, Y.; Ma, G.X.; Nguyen, M.T. Mechanisms of hepatocellular carcinoma progression. World J. Gastroenterol. 2019, 25, 2279–2293. [Google Scholar] [CrossRef]
- Ito, T.; Nguyen, M.H. Perspectives on the Underlying Etiology of HCC and Its Effects on Treatment Outcomes. J. Hepatocell. Carcinoma 2023, 10, 413–428. [Google Scholar] [CrossRef]
- Palma, R.; Pronio, A.; Romeo, M.; Scognamiglio, F.; Ventriglia, L.; Ormando, V.M.; Lamazza, A.; Pontone, S.; Federico, A.; Dallio, M. The Role of Insulin Resistance in Fueling NAFLD Pathogenesis: From Molecular Mechanisms to Clinical Implications. J. Clin. Med. 2022, 11, 3649. [Google Scholar] [CrossRef] [PubMed]
- Oura, K.; Morishita, A.; Masaki, T. Molecular and Functional Roles of MicroRNAs in the Progression of Hepatocellular Carcinoma-A Review. Int. J. Mol. Sci. 2020, 21, 8362. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.L.; Novo-Veleiro, I.; Manzanedo, L.; Alvela-Suárez, L.; Macías, R.; Laso, F.J.; Marcos, M. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J. Gastroenterol. 2018, 24, 4104–4118. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Majumder, S.; Nuovo, G.; Kutay, H.; Volinia, S.; Patel, T.; Schmittgen, T.D.; Croce, C.; Ghoshal, K.; Jacob, S.T. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 2009, 50, 1152–1161. [Google Scholar] [CrossRef]
- Niture, S.; Gadi, S.; Qi, Q.; Gyamfi, M.A.; Varghese, R.S.; Rios-Colon, L.; Chimeh, U.; Vandana; Ressom, H.W.; Kumar, D. MicroRNA-483-5p Inhibits Hepatocellular Carcinoma Cell Proliferation, Cell Steatosis, and Fibrosis by Targeting PPARα and TIMP2. Cancers 2023, 15, 1715. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, F.Y.; Tan, G.Q.; Chen, W.J.; Huang, B.; Yan, L.; Zhang, H.L.; Chen, S.; Jiao, Y.; Wang, B.L. Down-regulation of EGFL8 regulates migration, invasion and apoptosis of hepatocellular carcinoma through activating Notch signaling pathway. BMC Cancer 2021, 21, 704. [Google Scholar] [CrossRef]
- Mirza, A.Z.; Althagafi, I.I.; Shamshad, H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur. J. Med. Chem. 2019, 166, 502–513. [Google Scholar] [CrossRef]
- Peeney, D.; Liu, Y.; Lazaroff, C.; Gurung, S.; Stetler-Stevenson, W.G. Unravelling the distinct biological functions and potential therapeutic applications of TIMP2 in cancer. Carcinogenesis 2022, 43, 405–418. [Google Scholar] [CrossRef]
- D’Souza, S.; Lau, K.C.; Coffin, C.S.; Patel, T.R. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J. Gastroenterol. 2020, 26, 5759–5783. [Google Scholar] [CrossRef]
- Stroffolini, T.; Stroffolini, G. A Historical Overview on the Role of Hepatitis B and C Viruses as Aetiological Factors for Hepatocellular Carcinoma. Cancers 2023, 15, 2388. [Google Scholar] [CrossRef]
- Qin, Y.F.; Zhou, Z.Y.; Fu, H.W.; Lin, H.M.; Xu, L.B.; Wu, W.R.; Liu, C.; Xu, X.L.; Zhang, R. Hepatitis B Virus Surface Antigen Promotes Stemness of Hepatocellular Carcinoma through Regulating MicroRNA-203a. J. Clin. Transl. Hepatol. 2023, 11, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fan, H.; Sun, M.; Lv, Z.; Yi, W. Roles of BMI1 in the Initiation, Progression, and Treatment of Hepatocellular Carcinoma. Technol. Cancer Res. Treat. 2022, 21, 15330338211070689. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef]
- Wong, C.M.; Tsang, F.H.; Ng, I.O. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef]
- Liang, W.; Shi, C.; Hong, W.; Li, P.; Zhou, X.; Fu, W.; Lin, L.; Zhang, J. Super-enhancer-driven lncRNA-DAW promotes liver cancer cell proliferation through activation of Wnt/β-catenin pathway. Mol. Therapy. Nucleic Acids 2021, 26, 1351–1363. [Google Scholar] [CrossRef]
- Wang, Y.L.; Liu, J.Y.; Yang, J.E.; Yu, X.M.; Chen, Z.L.; Chen, Y.J.; Kuang, M.; Zhu, Y.; Zhuang, S.M. Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation. Hepatology 2019, 70, 259–275. [Google Scholar] [CrossRef]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef]
- Song, H.; Liu, Y.; Li, X.; Chen, S.; Xie, R.; Chen, D.; Gao, H.; Wang, G.; Cai, B.; Yang, X. Long noncoding RNA CASC11 promotes hepatocarcinogenesis and HCC progression through EIF4A3-mediated E2F1 activation. Clin. Transl. Med. 2020, 10, e220. [Google Scholar] [CrossRef]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xiao, Z.; Liu, F.; Cui, M.; Li, W.; Yang, Z.; Li, J.; Ye, L.; Zhang, X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget 2016, 7, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.R.; Feng, J.L.; Liu, X.J.; Wang, J.M. LncRNA HULC promots HCC growth by downregulating miR-29. Zhonghua Zhong Liu Za Zhi Chin. J. Oncol. 2019, 41, 659–666. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Fu, W.M.; Zhu, X.; Wang, W.M.; Lu, Y.F.; Hu, B.G.; Wang, H.; Liang, W.C.; Wang, S.S.; Ko, C.H.; Waye, M.M.; et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J. Hepatol. 2015, 63, 886–895. [Google Scholar] [CrossRef]
- Yuan, Y.; Ye, J.; Zhang, X.; Liu, Z. LncRNA CASC2 Regulate Cell Proliferation and Invasion by Targeting miR-155/SOCS1 Axis in Hepatocellular Carcinoma. J. Oncol. 2023, 2023, 8457112. [Google Scholar] [CrossRef]
- Wang, X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell. Biochem. 2018, 119, 1567–1574. [Google Scholar] [CrossRef]
- Jin, S.S.; Lin, X.F.; Zheng, J.Z.; Wang, Q.; Guan, H.Q. lncRNA NEAT1 regulates fibrosis and inflammatory response induced by nonalcoholic fatty liver by regulating miR-506/GLI3. Eur. Cytokine Netw. 2019, 30, 98–106. [Google Scholar] [CrossRef]
- Xiang, J.; Deng, Y.Y.; Liu, H.X.; Pu, Y. LncRNA MALAT1 Promotes PPARα/CD36-Mediated Hepatic Lipogenesis in Nonalcoholic Fatty Liver Disease by Modulating miR-206/ARNT Axis. Front. Bioeng. Biotechnol. 2022, 10, 858558. [Google Scholar] [CrossRef]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017, 77, 1155–1167. [Google Scholar] [CrossRef]
- Liang, H.W.; Wang, N.; Wang, Y.; Wang, F.; Fu, Z.; Yan, X.; Zhu, H.; Diao, W.; Ding, Y.; Chen, X.; et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 2016, 64, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.C.; Sun, T.; Ching, A.K.; He, M.; Li, J.W.; Wong, A.M.; Co, N.N.; Chan, A.W.; Li, P.S.; Lung, R.W.; et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 2014, 25, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Kong, G.; You, X.; Zhang, S.; Zhang, T.; Gao, Y.; Ye, L.; Zhang, X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J. Biol. Chem. 2012, 287, 26302–26311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, C.; Zhao, Y.; Li, M.; Wu, L.; Yang, X.; Wan, X.; Wang, A.; Zhang, M.Q.; Sang, X.; et al. Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget 2015, 6, 43770–43778. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Li, G.; Liu, H.; Li, T.; Liu, J.; Zhao, Q.; Wang, C. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine 2016, 95, e3811. [Google Scholar] [CrossRef]
- Li, M.F.; Li, Y.H.; He, Y.H.; Wang, Q.; Zhang, Y.; Li, X.F.; Meng, X.M.; Huang, C.; Li, J. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 99, 848–858. [Google Scholar] [CrossRef]
- Pan, L.; Ren, F.; Rong, M.; Dang, Y.; Luo, Y.; Luo, D.; Chen, G. Correlation between down-expression of miR-431 and clinicopathological significance in HCC tissues. Clin. Transl. Oncol. 2015, 17, 557–563. [Google Scholar] [CrossRef]
- Sun, R.; Li, H.; Li, J.; Shen, S.; Cui, G.; Dong, G. CircRNA circ-0038718 promotes hepatocellular carcinoma progression through sponging miR-139-3p. Biochem. Biophys. Res. Commun. 2020, 533, 845–852. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, M.; Zhao, Y.; Pan, Y.; Kan, M.; Li, J.; He, K.; Zhang, X. HNF-4α inhibits hepatocellular carcinoma cell proliferation through mir-122-adam17 pathway. PLoS ONE 2020, 15, e0230450. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, C.; Lin, J.; Song, X.; Wang, H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 1275–1283. [Google Scholar] [CrossRef]
- Jiang, Y.L.; Shang, M.M.; Dong, S.Z.; Chang, Y.C. Abnormally expressed circular RNAs as novel non-invasive biomarkers for hepatocellular carcinoma: A meta-analysis. World J. Gastrointest. Oncol. 2019, 11, 909–924. [Google Scholar] [CrossRef]
- Cao, J.; Wu, L.; Lei, X.; Shi, K.; Shi, L.; Shi, Y. Long non-coding RNA-based signature for predicting prognosis of hepatocellular carcinoma. Bioengineered 2021, 12, 673–681. [Google Scholar] [CrossRef]
- Wang, M.; Gu, B.; Yao, G.; Li, P.; Wang, K. Circular RNA Expression Profiles and the Pro-tumorigenic Function of CircRNA_10156 in Hepatitis B Virus-Related Liver Cancer. Int. J. Med. Sci. 2020, 17, 1351–1365. [Google Scholar] [CrossRef]
- Chien, Y.; Tsai, P.H.; Lai, Y.H.; Lu, K.H.; Liu, C.Y.; Lin, H.F.; Huang, C.S.; Wu, W.W.; Wang, C.Y. CircularRNA as novel biomarkers in liver diseases. J. Chin. Med. Assoc. JCMA 2020, 83, 15–17. [Google Scholar] [CrossRef]
- Yu, L.; Gong, X.; Sun, L.; Zhou, Q.; Lu, B.; Zhu, L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS ONE 2016, 11, e0158347. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, M.; Zheng, X.; Yi, P.; Lan, C.; Xu, M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 17–27. [Google Scholar] [CrossRef]
- Zhu, Q.; Lu, G.; Luo, Z.; Gui, F.; Wu, J.; Zhang, D.; Ni, Y. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem. Biophys. Res. Commun. 2018, 497, 626–632. [Google Scholar] [CrossRef]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef]
- Sun, Z.P.; Tan, Z.G.; Peng, C. Long noncoding RNA LINC01419 promotes hepatocellular carcinoma malignancy by mediating miR-485-5p/LSM4 axis. Kaohsiung J. Med. Sci. 2022, 38, 826–838. [Google Scholar] [CrossRef]
- Dang, H.; Chen, L.; Tang, P.; Cai, X.; Zhang, W.; Zhang, R.; Huang, A.; Tang, H. LINC01419 promotes cell proliferation and metastasis in hepatocellular carcinoma by enhancing NDRG1 promoter activity. Cell. Oncol. 2020, 43, 931–947. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, X.; Fu, Q.; Liu, C.; Luo, Q.; Yu, P.; Chen, S.; Zhang, H.; Qin, T. LINC01419 Promotes the Proliferation of Hepatoma Cells by Recruiting XRCC5 and Regulating Its Phosphorylation to Repair DNA Damage. Dis. Markers 2022, 2022, 9313680. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, X.; Ma, L.; Ding, R.; Zhao, L.; Ma, F.; Deng, X. LINC01419 facilitates hepatocellular carcinoma growth and metastasis through targeting EZH2-regulated RECK. Aging 2020, 12, 11071–11084. [Google Scholar] [CrossRef]
- Li, Y.C.; Wang, D.; Zhu, G.Y. Increased expression of long noncoding RNA AK021443 predicts worse clinical outcome in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4855–4860. [Google Scholar] [CrossRef]
- Yang, J.; Li, J.; Liu, B.; Zhang, R.; Gu, F.; Zhao, J.; Cheng, S. Long Noncoding RNA AK021443 Promotes Cell Proliferation and Migration by Regulating Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells. DNA Cell Biol. 2018, 37, 481–490. [Google Scholar] [CrossRef]
- Chen, T.; Pei, J.; Wang, J.; Luo, R.; Liu, L.; Wang, L.; Jia, H. HBx-related long non-coding RNA 01152 promotes cell proliferation and survival by IL-23 in hepatocellular carcinoma. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 115, 108877. [Google Scholar] [CrossRef]
- Chen, Y.; Li, S.; Wei, Y.; Xu, Z.; Wu, X. Circ-RNF13, as an oncogene, regulates malignant progression of HBV-associated hepatocellular carcinoma cells and HBV infection through ceRNA pathway of circ-RNF13/miR-424-5p/TGIF2. Bosn. J. Basic Med. Sci. 2021, 21, 555–568. [Google Scholar] [CrossRef]
- Zhu, K.; Zhan, H.; Peng, Y.; Yang, L.; Gao, Q.; Jia, H.; Dai, Z.; Tang, Z.; Fan, J.; Zhou, J. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis 2020, 41, 296–302. [Google Scholar] [CrossRef]
- He, W.; Zhu, X.; Tang, X.; Xiang, X.; Yu, J.; Sun, H. Circ_0027089 regulates NACC1 by targeting miR-136-5p to aggravate the development of hepatitis B virus-related hepatocellular carcinoma. Anti-Cancer Drugs 2022, 33, e336–e348. [Google Scholar] [CrossRef]
- Liu, Y.; Xun, Z.; Ma, K.; Liang, S.; Li, X.; Zhou, S.; Sun, L.; Liu, Y.; Du, Y.; Guo, X.; et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol. 2023, 78, 770–782. [Google Scholar] [CrossRef]
- Feng, D.; Xiong, Q.; Wei, Q.; Yang, L. Cellular landscape of tumour microenvironment in prostate cancer. Immunology 2023, 168, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Xiao, W.; Hu, H.; Lu, G.; Chen, L.; Sun, Z.; Lü, M.; Ma, W.; Jiang, T.; Gao, Y.; et al. Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer. Clin. Transl. Med. 2021, 11, e422. [Google Scholar] [CrossRef]
- Sherman, M.H.; Beatty, G.L. Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance. Annu. Rev. Pathol. 2023, 18, 123–148. [Google Scholar] [CrossRef]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef]
- Huang, H.; Tsui, Y.M.; Ng, I.O. Fueling HCC Dynamics: Interplay Between Tumor Microenvironment and Tumor Initiating Cells. Cell. Mol. Gastroenterol. Hepatol. 2023, 15, 1105–1116. [Google Scholar] [CrossRef]
- Zhan, T.; Gao, X.; Wang, G.; Li, F.; Shen, J.; Lu, C.; Xu, L.; Li, Y.; Zhang, J. Construction of Novel lncRNA-miRNA-mRNA Network Associated With Recurrence and Identification of Immune-Related Potential Regulatory Axis in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 626663. [Google Scholar] [CrossRef]
- Ouyang, X.; Lv, L.; Zhao, Y.; Zhang, F.; Hu, Q.; Li, Z.; Zhu, D.; Li, L. ASF1B Serves as a Potential Therapeutic Target by Influencing Cell Cycle and Proliferation in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 801506. [Google Scholar] [CrossRef]
- Shi, J.; Li, J.; Yang, S.; Hu, X.; Chen, J.; Feng, J.; Shi, T.; He, Y.; Mei, Z.; He, W.; et al. LncRNA SNHG3 is activated by E2F1 and promotes proliferation and migration of non-small-cell lung cancer cells through activating TGF-β pathway and IL-6/JAK2/STAT3 pathway. J. Cell. Physiol. 2020, 235, 2891–2900. [Google Scholar] [CrossRef]
- Huang, X.Y.; Zhang, P.F.; Wei, C.Y.; Peng, R.; Lu, J.C.; Gao, C.; Cai, J.B.; Yang, X.; Fan, J.; Ke, A.W.; et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 2020, 19, 92. [Google Scholar] [CrossRef]
- Hollande, C.; Boussier, J.; Ziai, J.; Nozawa, T.; Bondet, V.; Phung, W.; Lu, B.; Duffy, D.; Paradis, V.; Mallet, V.; et al. Inhibition of the dipeptidyl peptidase DPP4 (CD26) reveals IL-33-dependent eosinophil-mediated control of tumor growth. Nat. Immunol. 2019, 20, 257–264. [Google Scholar] [CrossRef]
- Huang, M.; Huang, X.; Huang, N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022, 113, 1968–1983. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018, 19, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Tone, Y.; Furuuchi, K.; Kojima, Y.; Tykocinski, M.L.; Greene, M.I.; Tone, M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008, 9, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, S.; Wang, Y.; Chen, H.; Nie, H.; Liu, L.; Zou, X.; Gong, Q.; Zheng, B. MicroRNA: Role in macrophage polarization and the pathogenesis of the liver fibrosis. Front. Immunol. 2023, 14, 1147710. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Zhao, S.; Wang, E.; Zhu, J.; Feng, D.; Zhu, Y.; Dou, W.; Fan, Q.; Hu, J.; et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol. Cancer 2021, 20, 46. [Google Scholar] [CrossRef]
- Lu, C.; Rong, D.; Zhang, B.; Zheng, W.; Wang, X.; Chen, Z.; Tang, W. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: Challenges and opportunities. Mol. Cancer 2019, 18, 130. [Google Scholar] [CrossRef]
- Han, Q.; Zhao, H.; Jiang, Y.; Yin, C.; Zhang, J. HCC-Derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells 2019, 8, 558. [Google Scholar] [CrossRef]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. [Google Scholar] [CrossRef]
- Kogure, T.; Yan, I.K.; Lin, W.L.; Patel, T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer 2013, 4, 261–272. [Google Scholar] [CrossRef]
- Liu, N.; Wang, X.; Steer, C.J.; Song, G. MicroRNA-206 promotes the recruitment of CD8(+) T cells by driving M1 polarisation of Kupffer cells. Gut 2022, 71, 1642–1655. [Google Scholar] [CrossRef]
- Tang, R.; Hua, J.; Xu, J.; Liang, C.; Meng, Q.; Liu, J.; Zhang, B.; Yu, X.; Shi, S. The role of ferroptosis regulators in the prognosis, immune activity and gemcitabine resistance of pancreatic cancer. Ann. Transl. Med. 2020, 8, 1347. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Zhang, J.; Tan, J.; Li, J.; Song, Z. Development and Validation of a Combined Ferroptosis and Immune Prognostic Classifier for Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2020, 8, 596679. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, S.; Xi, X.; Zhang, M.; Liu, X.; Tang, W.; Cai, P.; Xing, S.; Bao, P.; Jin, Y.; et al. Integrative analysis of long extracellular RNAs reveals a detection panel of noncoding RNAs for liver cancer. Theranostics 2021, 11, 181–193. [Google Scholar] [CrossRef]
- Xu, Z.; Peng, B.; Liang, Q.; Chen, X.; Cai, Y.; Zeng, S.; Gao, K.; Wang, X.; Yi, Q.; Gong, Z.; et al. Construction of a Ferroptosis-Related Nine-lncRNA Signature for Predicting Prognosis and Immune Response in Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 719175. [Google Scholar] [CrossRef]
- Shi, L.; Liu, Y.; Li, M.; Luo, Z. Emerging roles of ferroptosis in the tumor immune landscape: From danger signals to anti-tumor immunity. FEBS J. 2022, 289, 3655–3665. [Google Scholar] [CrossRef]
- Lu, J.; Xu, F.; Lu, H. LncRNA PVT1 regulates ferroptosis through miR-214-mediated TFR1 and p53. Life Sci. 2020, 260, 118305. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, X.; Liu, N.; Shi, Y.; Liu, Y.; Ouyang, L.; Tam, S.; Xiao, D.; Liu, S.; Wen, F.; et al. A Nuclear Long Non-Coding RNA LINC00618 Accelerates Ferroptosis in a Manner Dependent upon Apoptosis. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 263–274. [Google Scholar] [CrossRef]
- Liu, C.; Gao, Y.; Ni, J.; Chen, S.; Hu, Q.; Wang, C.; Hu, M.; Chen, M. The ferroptosis-related long non-coding RNAs signature predicts biochemical recurrence and immune cell infiltration in prostate cancer. BMC Cancer 2022, 22, 788. [Google Scholar] [CrossRef]
- Yang, X.; Mei, M.; Yang, J.; Guo, J.; Du, F.; Liu, S. Ferroptosis-related long non-coding RNA signature predicts the prognosis of hepatocellular carcinoma. Aging 2022, 14, 4069–4084. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef]
- Huycke, M.M.; Gaskins, H.R. Commensal bacteria, redox stress, and colorectal cancer: Mechanisms and models. Exp. Biol. Med. 2004, 229, 586–597. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Wan, M.L.Y.; El-Nezami, H. Targeting gut microbiota in hepatocellular carcinoma: Probiotics as a novel therapy. Hepatobiliary Surg. Nutr. 2018, 7, 11–20. [Google Scholar] [CrossRef]
- Grąt, M.; Wronka, K.M.; Krasnodębski, M.; Masior, Ł.; Lewandowski, Z.; Kosińska, I.; Grąt, K.; Stypułkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of Gut Microbiota Associated with the Presence of Hepatocellular Cancer in Patients with Liver Cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef]
- Lu, H.; Ren, Z.; Li, A.; Zhang, H.; Jiang, J.; Xu, S.; Luo, Q.; Zhou, K.; Sun, X.; Zheng, S.; et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci. Rep. 2016, 6, 33142. [Google Scholar] [CrossRef] [PubMed]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Shepard, C.R. TLR9 in MAFLD and NASH: At the Intersection of Inflammation and Metabolism. Front. Endocrinol. 2020, 11, 613639. [Google Scholar] [CrossRef] [PubMed]
- Shanab, A.A.; Scully, P.; Crosbie, O.; Buckley, M.; O’Mahony, L.; Shanahan, F.; Gazareen, S.; Murphy, E.; Quigley, E.M. Small intestinal bacterial overgrowth in nonalcoholic steatohepatitis: Association with toll-like receptor 4 expression and plasma levels of interleukin 8. Dig. Dis. Sci. 2011, 56, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Chu, H.; Williams, B.; Schnabl, B. Gut microbiota, fatty liver disease, and hepatocellular carcinoma. Liver Res. 2018, 2, 43–51. [Google Scholar] [CrossRef]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef]
- Zhu, Z.; Huang, J.; Li, X.; Xing, J.; Chen, Q.; Liu, R.; Hua, F.; Qiu, Z.; Song, Y.; Bai, C.; et al. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes 2020, 12, 1788891. [Google Scholar] [CrossRef] [PubMed]
- Arunima, A.; van Schaik, E.J.; Samuel, J.E. The emerging roles of long non-coding RNA in host immune response and intracellular bacterial infections. Front. Cell. Infect. Microbiol. 2023, 13, 1160198. [Google Scholar] [CrossRef]
- Wen, Y.; Chen, H.; Luo, F.; Zhou, H.; Li, Z. Roles of long noncoding RNAs in bacterial infection. Life Sci. 2020, 263, 118579. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ni, Y.Q.; Xu, H.; Xiang, Q.Y.; Zhao, Y.; Zhan, J.K.; He, J.Y.; Li, S.; Liu, Y.S. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 2021, 6, 383. [Google Scholar] [CrossRef] [PubMed]
- Dávalos, A.; Goedeke, L.; Smibert, P.; Ramírez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L.; et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11, eaav1892. [Google Scholar] [CrossRef]
- Farra, R.; Grassi, M.; Grassi, G.; Dapas, B. Therapeutic potential of small interfering RNAs/micro interfering RNA in hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 8994–9001. [Google Scholar] [CrossRef] [PubMed]
- Koustas, E.; Trifylli, E.M.; Sarantis, P.; Papadopoulos, N.; Papanikolopoulos, K.; Aloizos, G.; Damaskos, C.; Garmpis, N.; Garmpi, A.; Matthaios, D.; et al. An Insight into the Arising Role of MicroRNAs in Hepatocellular Carcinoma: Future Diagnostic and Therapeutic Approaches. Int. J. Mol. Sci. 2023, 24, 7168. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bao, H.; Huang, Z.; Liang, Z.; Wang, M.; Lin, N.; Ni, C.; Xu, Y. Little things with significant impact: miRNAs in hepatocellular carcinoma. Front. Oncol. 2023, 13, 1191070. [Google Scholar] [CrossRef]
- Zheng, G.; Bouamar, H.; Cserhati, M.; Zeballos, C.R.; Mehta, I.; Zare, H.; Broome, L.; Hu, R.; Lai, Z.; Chen, Y.; et al. Integrin alpha 6 is upregulated and drives hepatocellular carcinoma progression through integrin α6β4 complex. Int. J. Cancer 2022, 151, 930–943. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Hao, M.; Tong, H.; Yang, H.; Huang, B.; Zhang, Z.; Luo, K.Q. The interactions between integrin α(5)β(1) of liver cancer cells and fibronectin of fibroblasts promote tumor growth and angiogenesis. Int. J. Biol. Sci. 2022, 18, 5019–5037. [Google Scholar] [CrossRef] [PubMed]
- Bogorad, R.L.; Yin, H.; Zeigerer, A.; Nonaka, H.; Ruda, V.M.; Zerial, M.; Anderson, D.G.; Koteliansky, V. Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nat. Commun. 2014, 5, 3869. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xia, H.; Zhang, X.; Karthik, S.; Pratap, S.V.; Ooi, L.L.; Hong, W.; Hui, K.M. ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J. Hepatol. 2015, 62, 1287–1295. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Zhang, H.; Zhu, M.; Chen, F.; Hu, Y.; Liu, H.; Zhu, H. Interferon-stimulated gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget 2014, 5, 8429–8441. [Google Scholar] [CrossRef]
- Huang, C.H.; Lujambio, A.; Zuber, J.; Tschaharganeh, D.F.; Doran, M.G.; Evans, M.J.; Kitzing, T.; Zhu, N.; de Stanchina, E.; Sawyers, C.L.; et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014, 28, 1800–1814. [Google Scholar] [CrossRef]
- Zuckerman, J.E.; Davis, M.E. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov. 2015, 14, 843–856. [Google Scholar] [CrossRef]
- Yamada, S.; Ohira, M.; Horie, H.; Ando, K.; Takayasu, H.; Suzuki, Y.; Sugano, S.; Hirata, T.; Goto, T.; Matsunaga, T.; et al. Expression profiling and differential screening between hepatoblastomas and the corresponding normal livers: Identification of high expression of the PLK1 oncogene as a poor-prognostic indicator of hepatoblastomas. Oncogene 2004, 23, 5901–5911. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Elzallat, M.; Aboushousha, T.; Elhusseny, Y.; El-Ahwany, E. MicroRNA-122 mimic/microRNA-221 inhibitor combination as a novel therapeutic tool against hepatocellular carcinoma. Non-Coding RNA Res. 2023, 8, 126–134. [Google Scholar] [CrossRef]
- Hsu, S.H.; Yu, B.; Wang, X.; Lu, Y.; Schmidt, C.R.; Lee, R.J.; Lee, L.J.; Jacob, S.T.; Ghoshal, K. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 1169–1180. [Google Scholar] [CrossRef]
- Bozzato, A.M.; Martingano, P.; Pozzi Mucelli, R.A.; Cavallaro, M.F.M.; Cesarotto, M.; Marcello, C.; Tiribelli, C.; Pascut, D.; Pizzolato, R.; Pozzi Mucelli, F.; et al. MicroRNAs Related to TACE Treatment Response: A Review of the Literature from a Radiological Point of View. Diagnostics 2022, 12, 374. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. CR 2019, 38, 53. [Google Scholar] [CrossRef]
- Sousa, D.P.; Conde, J. Gold Nanoconjugates for miRNA Modulation in Cancer Therapy: From miRNA Silencing to miRNA Mimics. ACS Mater. Au 2022, 2, 626–640. [Google Scholar] [CrossRef]
- Mo, Y.; He, L.; Lai, Z.; Wan, Z.; Chen, Q.; Pan, S.; Li, L.; Li, D.; Huang, J.; Xue, F.; et al. Gold nano-particles (AuNPs) carrying miR-326 targets PDK1/AKT/c-myc axis in hepatocellular carcinoma. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2830–2837. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef]
- Ladd, A.D.; Duarte, S.; Sahin, I.; Zarrinpar, A. Mechanisms of drug resistance in HCC. Hepatology 2023. [Google Scholar] [CrossRef]
- Abdelgalil, A.A.; Alkahtani, H.M.; Al-Jenoobi, F.I. Sorafenib. Profiles Drug Subst. Excip. Relat. Methodol. 2019, 44, 239–266. [Google Scholar] [CrossRef] [PubMed]
- Azumi, J.; Tsubota, T.; Sakabe, T.; Shiota, G. miR-181a induces sorafenib resistance of hepatocellular carcinoma cells through downregulation of RASSF1 expression. Cancer Sci. 2016, 107, 1256–1262. [Google Scholar] [CrossRef]
- Liu, K.; Liu, S.; Zhang, W.; Jia, B.; Tan, L.; Jin, Z.; Liu, Y. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol. Rep. 2015, 34, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Ooi, L.L.; Hui, K.M. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology 2013, 58, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Tang, G.; Wu, X.; Wu, C. LncRNA NEAT1 modulates sorafenib resistance in hepatocellular carcinoma through regulating the miR-149-5p/AKT1 axis. Saudi J. Gastroenterol. 2020, 26, 194–203. [Google Scholar] [CrossRef]
- Suyama, K.; Iwase, H. Lenvatinib: A Promising Molecular Targeted Agent for Multiple Cancers. Cancer Control. J. Moffitt Cancer Cent. 2018, 25, 1073274818789361. [Google Scholar] [CrossRef]
- Yamashita, T.; Kudo, M.; Ikeda, K.; Izumi, N.; Tateishi, R.; Ikeda, M.; Aikata, H.; Kawaguchi, Y.; Wada, Y.; Numata, K.; et al. REFLECT-a phase 3 trial comparing efficacy and safety of lenvatinib to sorafenib for the treatment of unresectable hepatocellular carcinoma: An analysis of Japanese subset. J. Gastroenterol. 2020, 55, 113–122. [Google Scholar] [CrossRef]
- Yu, T.; Yu, J.; Lu, L.; Zhang, Y.; Zhou, Y.; Zhou, Y.; Huang, F.; Sun, L.; Guo, Z.; Hou, G.; et al. MT1JP-mediated miR-24-3p/BCL2L2 axis promotes Lenvatinib resistance in hepatocellular carcinoma cells by inhibiting apoptosis. Cell. Oncol. 2021, 44, 821–834. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, W.; Han, P.; Zhang, J.; Tong, L.; Sun, X. MicroRNA-128-3p Mediates Lenvatinib Resistance of Hepatocellular Carcinoma Cells by Downregulating c-Met. J. Hepatocell. Carcinoma 2022, 9, 113–126. [Google Scholar] [CrossRef]
- Han, T.; Zheng, H.; Zhang, J.; Yang, P.; Li, H.; Cheng, Z.; Xiang, D.; Wang, R. Downregulation of MUC15 by miR-183-5p.1 promotes liver tumor-initiating cells properties and tumorigenesis via regulating c-MET/PI3K/AKT/SOX2 axis. Cell Death Dis. 2022, 13, 200. [Google Scholar] [CrossRef]
- Gu, D.; Tong, M.; Wang, J.; Zhang, B.; Liu, J.; Song, G.; Zhu, B. Overexpression of the lncRNA HOTAIRM1 promotes lenvatinib resistance by downregulating miR-34a and activating autophagy in hepatocellular carcinoma. Discover. Oncol. 2023, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Rudalska, R.; Dauch, D.; Longerich, T.; McJunkin, K.; Wuestefeld, T.; Kang, T.W.; Hohmeyer, A.; Pesic, M.; Leibold, J.; von Thun, A.; et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat. Med. 2014, 20, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, J.; Chen, D.; Huang, J.; Feng, B.; Han, S.; Chen, Y.; Song, H.; De, W.; Zhu, Z.; et al. Aurora-A promotes chemoresistance in hepatocelluar carcinoma by targeting NF-kappaB/microRNA-21/PTEN signaling pathway. Oncotarget 2014, 5, 12916–12935. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.B.; Wu, H.M.; He, Y.C.; Huang, Z.T.; Weng, Y.H.; Li, H.; Liang, C.; Yu, W.M.; Chen, W. MiRNA-124-3p.1 sensitizes hepatocellular carcinoma cells to sorafenib by regulating FOXO3a by targeting AKT2 and SIRT1. Cell Death Dis. 2022, 13, 35. [Google Scholar] [CrossRef]
- Chen, P.; Li, Y.; Wei, P.; Liang, L.; Li, B.; Cao, Y.; Han, X.; Wang, Y.; Duan, X.; Jia, H.; et al. siRNA targeting PD-L1 delivered with attenuated Salmonella enhanced the anti-tumor effect of lenvatinib on mice bearing Hepatocellular carcinoma. Int. Immunopharmacol. 2022, 111, 109127. [Google Scholar] [CrossRef]
- Guo, X.; Wang, W.; Zhou, F.; Lu, Z.; Fang, R.; Jia, F.; Bu, X.; Li, R.; Zhang, B.; Wu, M.; et al. siRNA-mediated inhibition of hTERT enhances chemosensitivity of hepatocellular carcinoma. Cancer Biol. Ther. 2008, 7, 1555–1560. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.; Su, H.; Su, L. Non-coding RNAs as biomarkers for hepatocellular carcinoma-A systematic review. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101736. [Google Scholar] [CrossRef]
- Wei, L.; Wang, X.; Lv, L.; Liu, J.; Xing, H.; Song, Y.; Xie, M.; Lei, T.; Zhang, N.; Yang, M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol. Cancer 2019, 18, 147. [Google Scholar] [CrossRef]
- Ding, B.; Lou, W.; Xu, L.; Fan, W. Non-coding RNA in drug resistance of hepatocellular carcinoma. Biosci. Rep. 2018, 38, BSR20180915. [Google Scholar] [CrossRef]
| Molecules’ Name | Biogenesis/ Expression Status | Influence on Molecular Pathways Promoting HCC Progression Mechanisms | microRNA(s) Targeted | ||
|---|---|---|---|---|---|
| Non-alcoholic Fatty Liver Disease (NAFLD)—HCC-related context | Linear Long Non-coding RNAs | NEAT1 | Overexpression [97,98] | Promotion of NAFLD-related fibrosis worsening and HCC cell proliferation by sponging miR-506 [normally down-regulating acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS) expression] [97]. | miR-506 [98] |
| MALAT1 | Overexpression [99] | Upregulation of the splicing factor “SRSF1” and activation of the β-catenin/Wnt pathway [99]. | Undefined/NA in this context | ||
| CASC2 | Downregulation [96] | Regulation of cell proliferation and dysregulation by sponging the oncoMiR miR-155 [96] | miR-155 [96] | ||
| Circular Long-Non-coding RNAs | circRNA CDR1-AS | Overexpression [117] | Promotion of invasion/proliferation mechanisms of liver cancer cells by sponging miR-7 (normally inhibiting spindle checkpoint protein “SPC24”) [117]. | miR-7 [116] | |
| circRNA_0067934 | Overexpression [118] | Promotion of invasion and metastasis-related mechanisms by sponging miR-1234 (normally regulating the β-catenin/Wnt signaling pathway) [118]. | miR-1234 [118] | ||
| circRNA MTO1 | Downregulation [119] | Promotion of HCC progression by sponging the oncomiRNA “miR-9” [119]. | miR-9 [119] | ||
| Viral-hepatitis—HCC-related context | HBx-LINE1 lncRNA | Integration of HBV DNA in ch.8p11.21 [102] | In HBV context: activation of the Wnt signaling pathway; acting as a decoy to sequester miR-122 [101,102]. | miR-122 [101] | |
| HULC | Overexpression induced by HBx protein [91,103] | In HBV context: activation of sphingosine-kinase-1-mediated angiogenesis, functioning as a molecular decoy of miR-107 (normally repressing the expression of the transcription factor E2F1) [103]. | miR-107 [92] | ||
| Linear Long Non-coding RNAs | Linc01419 | Overexpression [104] | In HCV context:
| miR-485-5p [120] | |
| LncAK021443 | Overexpression [124] | In HCV context: promotion of cell proliferation, invasion, and metastasis by repressing epithelial-mesenchymal transition (EMT) [104,125] | Unidentified/NA in this context | ||
| Linc01152 | Overexpression [126] | In HBV context: increases cell proliferation by activating the STAT3 pathway [126]. | Unidentified | ||
| Viral-hepatitis—HCC-related context | Circular Long-Non-coding RNAs | circRNA_10156 | Overexpression [114] | In HBV context: promotes cell proliferation by sponging miR-149-3p (usually down-regulating the AKT1/mTOR pathway) [114]. | miR-149-3p [114] |
| circ-RNF13 [circ_0067717] | Downregulation [127] | In HBV context: promotes HCC progression by sponging miR-424-5p (usually regulating TGFβ-induced factor homeobox 2 (TGIF2) [127]. | miR-424-5p [127] | ||
| circ_0027089 | Overexpression [128] | In HBV contexts: acts as an oncogene and promotes the development of HBV-related HCC by regulating nucleus accumbens associated protein 1 (NACC1) via competitively targeting miR-136-5p [129]. | miR-136-5p [129] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, M.; Dallio, M.; Scognamiglio, F.; Ventriglia, L.; Cipullo, M.; Coppola, A.; Tammaro, C.; Scafuro, G.; Iodice, P.; Federico, A. Role of Non-Coding RNAs in Hepatocellular Carcinoma Progression: From Classic to Novel Clinicopathogenetic Implications. Cancers 2023, 15, 5178. https://doi.org/10.3390/cancers15215178
Romeo M, Dallio M, Scognamiglio F, Ventriglia L, Cipullo M, Coppola A, Tammaro C, Scafuro G, Iodice P, Federico A. Role of Non-Coding RNAs in Hepatocellular Carcinoma Progression: From Classic to Novel Clinicopathogenetic Implications. Cancers. 2023; 15(21):5178. https://doi.org/10.3390/cancers15215178
Chicago/Turabian StyleRomeo, Mario, Marcello Dallio, Flavia Scognamiglio, Lorenzo Ventriglia, Marina Cipullo, Annachiara Coppola, Chiara Tammaro, Giuseppe Scafuro, Patrizia Iodice, and Alessandro Federico. 2023. "Role of Non-Coding RNAs in Hepatocellular Carcinoma Progression: From Classic to Novel Clinicopathogenetic Implications" Cancers 15, no. 21: 5178. https://doi.org/10.3390/cancers15215178
APA StyleRomeo, M., Dallio, M., Scognamiglio, F., Ventriglia, L., Cipullo, M., Coppola, A., Tammaro, C., Scafuro, G., Iodice, P., & Federico, A. (2023). Role of Non-Coding RNAs in Hepatocellular Carcinoma Progression: From Classic to Novel Clinicopathogenetic Implications. Cancers, 15(21), 5178. https://doi.org/10.3390/cancers15215178








