Clinical Significance of Non-Coding RNA Regulation of Programmed Cell Death in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Diagnosis and Survival Prognosis
| ncRNAs | Targets | Effect | Mechanism | Refs. |
|---|---|---|---|---|
| miR-101 | - | Pro-apoptosis Anti-autophagy Better OS | The function of miR-101 is inhibiting autophagy, inducing apoptosis, and suppressing tumorigenicity. Downregulation of miR-101 associated with poorer OS of patients. | [29,30,31] |
| miR-122 | ↓Bcl-w | Pro-apoptosis Better OS and DFS | miR-122 targets the downregulation of the anti-apoptotic gene Bcl-w, thereby triggering apoptosis and predicting better OS and DFS in HCC patients. | [32,33] |
| miR-221 | ↓P53 ↑Bcl-2 | Anti-apoptosis Poorer OS, RFS, and PFS | Silencing of miR-221 induces apoptosis and G2/M phase arrest and inhibits cellular proliferation by upregulating P53 and downregulating Bcl-2 in HCC cells, leading to increased OS, RFS, and PFS in patients. | [34] |
| miR-23a-3p | ↓ACSL4 | Anti-ferroptosis Poorer OS and RFS | An elevated level of miR-23a-3p in HCC patients is associated with OS and RFS, potentially due to the epigenetic downregulation of ferroptosis by inhibiting ACSL4. | [35] |
| lncRNA PVT1 | ↑EZH2 | Anti-apoptosis Poorer OS | PVT1 recruits EZH2 to modulate HCC cell proliferation and apoptosis, leading to lower survival rates in patients with high PVT1 levels. | [36,37] |
| lncRNA PLAC2 | ↑P53 | Pro-apoptosis Better OS | The ability of PLAC2 to upregulate P53 and induce apoptosis in HCC cells leads to significantly higher OS rates in HCC patients with low levels of PLAC2. | [38] |
| lncRNA HCG18 | ↑GPX4 | Anti-ferroptosis Poorer OS | Silencing HCG18 inhibits GPX4 by binding to miR-450b-5p, promotes GPX4-inhibited ferroptosis, and leads to a better prognosis in HCC patients | [39] |
| circRNA circ_0021093 | ↑MTA3 | Anti-apoptosis Poorer OS | Downregulation of circ_0021093 markedly suppresses HCC cell proliferation through enhanced cell apoptosis, impaired migration, and invasion abilities, while overexpression of circ_0021093 predicts lower survival rates by targeting the miR-766-3p/MTA3 pathway in HCC. | [40] |
| circRNA circ-FOXP1 | - | Anti-apoptosis Poorer OS | The circRNA circ-FOXP1 plays a cell-protective role by sequestering miR-875-3p and miR-421, thereby mitigating apoptosis. HCC patients with elevated circ-FOXP1 expression exhibited reduced OS rates. | [41] |
| circRNA circMDK | - | Anti-apoptosis Poorer OS | Knockdown of circRNA circMDK enhances the apoptosis ratio of HCC cells, suggesting its potential tumor suppressor role, while increased circMDK expression is associated with poorer 5-year survival probability in HCC patients. | [27] |
| m7G-tRNA | ↑EGFR | Anti-apoptosis Poorer OS | METTL1/WDR4-mediated m7G tRNA modification in promoting translation of EGFR pathway genes to reduce apoptosis and trigger drug resistance in HCC cells | [43] |
| m1G37-tRNA | ↑HIF-1 | Anti-apoptosis Poorer OS | Knockdown of TRMT5 inactivated the HIF-1 signaling pathway by preventing HIF-1α stability through the enhancement of cellular oxygen content. | [44] |
3. Drug Resistance
3.1. Targeted Therapy and Drug Resistance
3.1.1. Sorafenib Resistance (SR)
Apoptosis
Autophagy
Ferroptosis
3.1.2. Lenvatinib Resistance (LR)
3.2. Immune Therapy Drug Resistance
3.3. Chemotherapy Resistance
4. Drug Side Effects
4.1. Anthracycline
4.2. Platinum Drugs
4.3. Fluorouracil
5. Challenges in RNA Therapeutic Delivery
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular Carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Song, T. Changes in and Challenges Regarding the Surgical Treatment of Hepatocellular Carcinoma in China. Biosci. Trends 2021, 15, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Chen, L.-L. The Biogenesis and Emerging Roles of Circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A CeRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Karreth, F.A.; Pandolfi, P.P. CeRNA Cross-Talk in Cancer: When Ce-Bling Rivalries Go Awry. Cancer Discov. 2013, 3, 1113–1121. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-Coding RNA in Cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef]
- Wong, C.-M.; Tsang, F.H.-C.; Ng, I.O.-L. Non-Coding RNAs in Hepatocellular Carcinoma: Molecular Functions and Pathological Implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Hänggi, K.; Ruffell, B. Cell Death, Therapeutics, and the Immune Response in Cancer. Trends Cancer 2023, 9, 381–396. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Liao, M.; Qin, R.; Zhu, S.; Peng, C.; Fu, L.; Chen, Y.; Han, B. Regulated Cell Death (RCD) in Cancer: Key Pathways and Targeted Therapies. Signal Transduct. Target. Ther. 2022, 7, 1–66. [Google Scholar] [CrossRef] [PubMed]
- Jasirwan, C.O.M.; Fahira, A.; Siregar, L.; Loho, I. The Alpha-Fetoprotein Serum Is Still Reliable as a Biomarker for the Surveillance of Hepatocellular Carcinoma in Indonesia. BMC Gastroenterol. 2020, 20, 215. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M.; American Association for the Study of Liver Diseases. Management of Hepatocellular Carcinoma: An Update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. European Organisation For Research And Treatment Of Cancer. EASL-EORTC Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef]
- Ghosh, S.; Bhowmik, S.; Majumdar, S.; Goswami, A.; Chakraborty, J.; Gupta, S.; Aggarwal, S.; Ray, S.; Chatterjee, R.; Bhattacharyya, S.; et al. The Exosome Encapsulated microRNAs as Circulating Diagnostic Marker for Hepatocellular Carcinoma with Low Alpha-fetoprotein. Int. J. Cancer 2020, 147, 2934–2947. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum Exosomal MicroRNAs Combined with Alphafetoprotein as Diagnostic Markers of Hepatocellular Carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef]
- Lim, L.J.; Wong, S.Y.S.; Huang, F.; Lim, S.; Chong, S.S.; Ooi, L.L.; Kon, O.L.; Lee, C.G. Roles and Regulation of Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Res. 2019, 79, 5131–5139. [Google Scholar] [CrossRef]
- Kamel, M.M.; Matboli, M.; Sallam, M.; Montasser, I.F.; Saad, A.S.; El-Tawdi, A.H.F. Investigation of Long Noncoding RNAs Expression Profile as Potential Serum Biomarkers in Patients with Hepatocellular Carcinoma. Transl. Res. 2016, 168, 134–145. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, X.; Liang, C.; Ling, Y.; Yang, X.; Ye, X.; Zhang, H.; Yang, P.; Cui, X.; Ren, Y.; et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology 2020, 71, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Nafar, S.; Nouri, N.; Alipour, M.; Fallahi, J.; Zare, F.; Tabei, S.M.B. Exosome as a Target for Cancer Treatment. J. Investig. Med. 2022, 70, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Harricharran, T.; Huaman, J.; Galuza, A.; Odumuwagun, O.; Tan, Y.; Ma, G.X.; Nguyen, M.T. Mechanisms of Hepatocellular Carcinoma Progression. World J. Gastroenterol. 2019, 25, 2279–2293. [Google Scholar] [CrossRef]
- Chidambaranathan-Reghupaty, S.; Fisher, P.B.; Sarkar, D. Hepatocellular Carcinoma (HCC): Epidemiology, Etiology and Molecular Classification. Adv. Cancer Res. 2021, 149, 1–61. [Google Scholar] [CrossRef]
- Guo, R.; Wu, Z.; Wang, J.; Li, Q.; Shen, S.; Wang, W.; Zhou, L.; Wang, W.; Cao, Z.; Guo, Y. Development of a Non-Coding-RNA-Based EMT/CSC Inhibitory Nanomedicine for In Vivo Treatment and Monitoring of HCC. Adv. Sci. 2019, 6, 1801885. [Google Scholar] [CrossRef]
- Du, A.; Li, S.; Zhou, Y.; Disoma, C.; Liao, Y.; Zhang, Y.; Chen, Z.; Yang, Q.; Liu, P.; Liu, S.; et al. M6A-Mediated Upregulation of CircMDK Promotes Tumorigenesis and Acts as a Nanotherapeutic Target in Hepatocellular Carcinoma. Mol. Cancer 2022, 21, 109. [Google Scholar] [CrossRef]
- Nakagawa, H.; Fujita, M. Whole Genome Sequencing Analysis for Cancer Genomics and Precision Medicine. Cancer Sci. 2018, 109, 513–522. [Google Scholar] [CrossRef]
- Su, H.; Yang, J.-R.; Xu, T.; Huang, J.; Xu, L.; Yuan, Y.; Zhuang, S.-M. MicroRNA-101, Down-Regulated in Hepatocellular Carcinoma, Promotes Apoptosis and Suppresses Tumorigenicity. Cancer Res. 2009, 69, 1135–1142. [Google Scholar] [CrossRef]
- Xu, L.; Beckebaum, S.; Iacob, S.; Wu, G.; Kaiser, G.M.; Radtke, A.; Liu, C.; Kabar, I.; Schmidt, H.H.; Zhang, X.; et al. MicroRNA-101 Inhibits Human Hepatocellular Carcinoma Progression through EZH2 Downregulation and Increased Cytostatic Drug Sensitivityq. J. Hepatol. 2014, 60, 590–598. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Xiong, L.; Kong, X.; Xu, Y.; Liu, C.; Zou, L.; Li, Z.; Zhao, J.; Lin, N. MicroRNA-101 Suppresses SOX9-Dependent Tumorigenicity and Promotes Favorable Prognosis of Human Hepatocellular Carcinoma. FEBS Lett. 2012, 586, 4362–4370. [Google Scholar] [CrossRef]
- Lin, C.J.-F.; Gong, H.-Y.; Tseng, H.-C.; Wang, W.-L.; Wu, J.-L. MiR-122 Targets an Anti-Apoptotic Gene, Bcl-w, in Human Hepatocellular Carcinoma Cell Lines. Biochem. Biophys. Res. Commun. 2008, 375, 315–320. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, J.; Han, J.; Luo, D.; Sun, Z. MiR-122 Inhibits Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma by Targeting Snail1 and Snail2 and Suppressing WNT/β-Cadherin Signaling Pathway. Exp. Cell Res. 2017, 360, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.; Jayamohan, S.; Moorthy, R.K.; Chabattula, S.C.; Ganeshan, M.; Arockiam, A.J.V. Dysregulation of MiRISC Regulatory Network Promotes Hepatocellular Carcinoma by Targeting PI3K/Akt Signaling Pathway. Int. J. Med. Sci. 2022, 23, 11300. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Zhang, C.; Guo, W.; Xu, Y.; Sharma, R.; Chen, Z.-S.; Zheng, Y.-C.; Wang, N.; et al. Epigenetic Regulation of Ferroptosis via ETS1/MiR-23a-3p/ACSL4 Axis Mediates Sorafenib Resistance in Human Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 3. [Google Scholar] [CrossRef]
- Guo, J.; Hao, C.; Wang, C.; Li, L. Long Noncoding RNA PVT1 Modulates Hepatocellular Carcinoma Cell Proliferation and Apoptosis by Recruiting EZH2. Cancer Cell Int. 2018, 18, 98. [Google Scholar] [CrossRef]
- Lan, T.; Yan, X.; Li, Z.; Xu, X.; Mao, Q.; Ma, W.; Hong, Z.; Chen, X.; Yuan, Y. Long Non-Coding RNA PVT1 Serves as a Competing Endogenous RNA for MiR-186-5p to Promote the Tumorigenesis and Metastasis of Hepatocellular Carcinoma. Tumour Biol. 2017, 39, 101042831770533. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lv, P.; Wang, S.; Cai, Q.; Zhang, B.; Huo, F. LncRNA PLAC2 Upregulates P53 to Induce Hepatocellular Carcinoma Cell Apoptosis. Gene 2019, 712, 143944. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Lian, P.; Lv, Q.; Liu, F. Silencing LncRNA HCG18 Regulates GPX4-Inhibited Ferroptosis by Adsorbing MiR-450b-5p to Avert Sorafenib Resistance in Hepatocellular Carcinoma. Hum. Exp. Toxicol. 2023, 42, 9603271221142818. [Google Scholar] [CrossRef]
- Liu, L.; Qi, X.; Gui, Y.; Huo, H.; Yang, X.; Yang, L. Overexpression of Circ_0021093 Circular RNA Forecasts an Unfavorable Prognosis and Facilitates Cell Progression by Targeting the MiR-766-3p/MTA3 Pathway in Hepatocellular Carcinoma. Gene 2019, 714, 143992. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Li, X.; Liu, B.; Han, S.; Li, X.; Zhang, B.; Li, J.; Sun, S. Circular RNA Circ-FOXP1 Induced by SOX9 Promotes Hepatocellular Carcinoma Progression via Sponging MiR-875-3p and MiR-421. Biomed. Pharmacother. 2020, 121, 109517. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Zhao, D.; Jiang, J.; Tang, F.; Zhang, C.; Duan, C. TRNA Modifications and Modifying Enzymes in Disease, the Potential Therapeutic Targets. Int. J. Biol. Sci. 2023, 19, 1146–1162. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Long, J.; Yao, Z.; Zhao, Y.; Zhao, Y.; Liao, J.; Lei, K.; Xiao, H.; Dai, Z.; Peng, S.; et al. METTL1-Mediated M7G TRNA Modification Promotes Lenvatinib Resistance in Hepatocellular Carcinoma. Cancer Res. 2023, 83, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhang, L.; He, Q.; Chang, H.; Wang, Z.; Cao, H.; Zhou, Y.; Pan, R.; Chen, Y. Targeting TRMT5 Suppresses Hepatocellular Carcinoma Progression via Inhibiting the HIF-1α Pathways. J. Zhejiang Univ. Sci. B 2023, 24, 50–63. [Google Scholar] [CrossRef]
- Bu, F.; Yin, S.; Guan, R.; Xiao, Y.; Zeng, S.; Zhao, Y. Ferroptosis-Related Long Non-Coding RNA Signature Predicts the Prognosis of Hepatocellular Carcinoma: A Review. Medicine 2022, 101, e31747. [Google Scholar] [CrossRef]
- Li, G.; Liu, Y.; Zhang, Y.; Xu, Y.; Zhang, J.; Wei, X.; Zhang, Z.; Zhang, C.; Feng, J.; Li, Q.; et al. A Novel Ferroptosis-Related Long Non-Coding RNA Prognostic Signature Correlates With Genomic Heterogeneity, Immunosuppressive Phenotype, and Drug Sensitivity in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 929089. [Google Scholar] [CrossRef]
- Lian, J.; Zhang, C.; Lu, H. A Ferroptosis-Related LncRNA Signature Associated with Prognosis, Tumor Immune Environment, and Genome Instability in Hepatocellular Carcinoma. Comput. Math. Methods Med. 2022, 2022, 6284540. [Google Scholar] [CrossRef]
- Lin, X.; Yang, S. A Prognostic Signature Based on the Expression Profile of the Ferroptosis-Related Long Non-Coding RNAs in Hepatocellular Carcinoma. Adv. Clin. Exp. Med. 2022, 31, 1099–1109. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, L.; Zhang, Y.; Zhang, Y.; Xu, S. A Ferroptosis-Associated LncRNAs Signature Predicts the Prognosis of Hepatocellular Carcinoma. Medicine 2022, 101, e29546. [Google Scholar] [CrossRef]
- Xu, Z.; Peng, B.; Liang, Q.; Chen, X.; Cai, Y.; Zeng, S.; Gao, K.; Wang, X.; Yi, Q.; Gong, Z.; et al. Construction of a Ferroptosis-Related Nine-LncRNA Signature for Predicting Prognosis and Immune Response in Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 719175. [Google Scholar] [CrossRef]
- Yang, X.; Mei, M.; Yang, J.; Guo, J.; Du, F.; Liu, S. Ferroptosis-Related Long Non-Coding RNA Signature Predicts the Prognosis of Hepatocellular Carcinoma. Aging 2022, 14, 4069–4084. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, P.; Zhao, L.; Han, P.; Liu, J.; Yang, H.; Li, J. A Novel Cuproptosis-Related Prognostic LncRNA Signature for Predicting Immune and Drug Therapy Response in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 954653. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, T.; Wang, G.; Zhang, M.; Zhu, Y.; Shi, X.; Ding, Z. Cuproptosis-Related LncRNA Signature for Predicting Prognosis of Hepatocellular Carcinoma: A Comprehensive Analysis. Dis. Markers 2022, 2022, 3265212. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.-F.; Fan, L.-W.; Zeng, H.-H.; Huang, C.-B.; Wu, X.-H. Establishment and Validation of a Cuproptosis-Related LncRNA Signature That Predicts Prognosis and Potential Targeted Therapy in Hepatocellular Carcinoma. Biotechnol. Genet. Eng. Rev. 2023, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Jin, S.; Chen, P.; Zhang, Y.; Li, Y.; Zhong, C.; Fan, X.; Lin, H. Comprehensive Analysis of Cuproptosis-Related LncRNAs for Prognostic Significance and Immune Microenvironment Characterization in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 991604. [Google Scholar] [CrossRef]
- Liu, C.; Wu, S.; Lai, L.; Liu, J.; Guo, Z.; Ye, Z.; Chen, X. Comprehensive Analysis of Cuproptosis-Related LncRNAs in Immune Infiltration and Prognosis in Hepatocellular Carcinoma. BMC Bioinform. 2023, 24, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, J. A Novel Cuproptosis-Related LncRNA Signature Predicts the Prognosis and Immunotherapy for Hepatocellular Carcinoma. CBM 2023, 37, 13–26. [Google Scholar] [CrossRef]
- Zhang, G.; Sun, J.; Zhang, X. A Novel Cuproptosis-Related LncRNA Signature to Predict Prognosis in Hepatocellular Carcinoma. Sci. Rep. 2022, 12, 11325. [Google Scholar] [CrossRef]
- Cheng, Z.; Han, J.; Jiang, F.; Chen, W.; Ma, X. Prognostic Pyroptosis-Related LncRNA Signature Predicts the Efficacy of Immunotherapy in Hepatocellular Carcinoma. Biochem. Biophys. Rep. 2022, 32, 101389. [Google Scholar] [CrossRef]
- Wang, T.; Yang, Y.; Sun, T.; Qiu, H.; Wang, J.; Ding, C.; Lan, R.; He, Q.; Wang, W. The Pyroptosis-Related Long Noncoding RNA Signature Predicts Prognosis and Indicates Immunotherapeutic Efficiency in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2022, 10, 779269. [Google Scholar] [CrossRef]
- Zhang, Z.; Xia, F.; Xu, Z.; Peng, J.; Kang, F.; Li, J.; Zhang, W.; Hong, Q. Identification and Validation of a Novel Pyroptosis-Related LncRNAs Signature Associated with Prognosis and Immune Regulation of Hepatocellular Carcinoma. Sci. Rep. 2022, 12, 8886. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Qin, S.; Kudo, M.; Su, Y.; Hudgens, S.; Yamashita, T.; Yoon, J.-H.; Fartoux, L.; Simon, K.; López, C.; et al. Lenvatinib versus Sorafenib for First-Line Treatment of Unresectable Hepatocellular Carcinoma: Patient-Reported Outcomes from a Randomised, Open-Label, Non-Inferiority, Phase 3 Trial. Lancet Gastroenterol. Hepatol. 2021, 6, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Matsui, J.; Funahashi, Y.; Uenaka, T.; Watanabe, T.; Tsuruoka, A.; Asada, M. Multi-Kinase Inhibitor E7080 Suppresses Lymph Node and Lung Metastases of Human Mammary Breast Tumor MDA-MB-231 via Inhibition of Vascular Endothelial Growth Factor-Receptor (VEGF-R) 2 and VEGF-R3 Kinase. Clin. Cancer Res. 2008, 14, 5459–5465. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting Apoptosis in Cancer Therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Chen, S.; Cao, Q.; Wen, W.; Wang, H. Targeted Therapy for Hepatocellular Carcinoma: Challenges and Opportunities. Cancer Lett. 2019, 460, 1–9. [Google Scholar] [CrossRef]
- Cheng, Z.; Ni, Q.; Qin, L.; Shi, Y. MicroRNA-92b Augments Sorafenib Resistance in Hepatocellular Carcinoma via Targeting PTEN to Activate PI3K/AKT/MTOR Signaling. Braz. J. Med. Biol. Res. 2021, 54, e10390. [Google Scholar] [CrossRef]
- Li, W.; Dong, X.; He, C.; Tan, G.; Li, Z.; Zhai, B.; Feng, J.; Jiang, X.; Liu, C.; Jiang, H.; et al. LncRNA SNHG1 Contributes to Sorafenib Resistance by Activating the Akt Pathway and Is Positively Regulated by MiR-21 in Hepatocellular Carcinoma Cells. J. Exp. Clin. Cancer Res. 2019, 38, 183. [Google Scholar] [CrossRef]
- He, C.; Dong, X.; Zhai, B.; Jiang, X.; Dong, D.; Li, B.; Jiang, H.; Xu, S.; Sun, X. MiR-21 Mediates Sorafenib Resistance of Hepatocellular Carcinoma Cells by Inhibiting Autophagy via the PTEN/Akt Pathway. Oncotarget 2015, 6, 28867–28881. [Google Scholar] [CrossRef]
- Chen, S.; Xia, X. Long Noncoding RNA NEAT1 Suppresses Sorafenib Sensitivity of Hepatocellular Carcinoma Cells via Regulating MiR-335–C-Met. J. Cell. Physiol. 2019, 234, 14999–15009. [Google Scholar] [CrossRef]
- Fan, L.; Huang, X.; Chen, J.; Zhang, K.; Gu, Y.-H.; Sun, J.; Cui, S.-Y. Long Noncoding RNA MALAT1 Contributes to Sorafenib Resistance by Targeting MiR-140-5p/Aurora-A Signaling in Hepatocellular Carcinoma. Mol. Cancer Ther. 2020, 19, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, P.; Koch, A.; Fritz, V.; Hartmann, A.; Bosserhoff, A.K.; Hellerbrand, C. Wild Type Kirsten Rat Sarcoma Is a Novel MicroRNA-622-Regulated Therapeutic Target for Hepatocellular Carcinoma and Contributes to Sorafenib Resistance. Gut 2018, 67, 1328–1341. [Google Scholar] [CrossRef]
- Shen, Q.; Jiang, S.; Wu, M.; Zhang, L.; Su, X.; Zhao, D. LncRNA HEIH Confers Cell Sorafenib Resistance in Hepatocellular Carcinoma by Regulating MiR-98-5p/PI3K/AKT Pathway. Cancer Manag. Res. 2020, 12, 6585–6595. [Google Scholar] [CrossRef]
- Li, X.; Yin, X.; Bao, H.; Liu, C. Circular RNA ITCH Increases Sorafenib-Sensitivity in Hepatocellular Carcinoma via Sequestering MiR-20b-5p and Modulating the Downstream PTEN-PI3K/Akt Pathway. Mol. Cell Probes 2023, 67, 101877. [Google Scholar] [CrossRef]
- Ji, L.; Lin, Z.; Wan, Z.; Xia, S.; Jiang, S.; Cen, D.; Cai, L.; Xu, J.; Cai, X. MiR-486-3p Mediates Hepatocellular Carcinoma Sorafenib Resistance by Targeting FGFR4 and EGFR. Cell Death Dis. 2020, 11, 250. [Google Scholar] [CrossRef]
- Shao, Y.-Y.; Chen, P.-S.; Lin, L.-I.; Lee, B.-S.; Ling, A.; Cheng, A.-L.; Hsu, C.; Ou, D.-L. Low MiR-10b-3p Associated with Sorafenib Resistance in Hepatocellular Carcinoma. Br. J. Cancer 2022, 126, 1806–1814. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, X.; Xue, X.; Sun, D.; Cai, P.; Song, Q.; Zhang, B.; Qin, L. HANR Enhances Autophagy-Associated Sorafenib Resistance Through MiR-29b/ATG9A Axis in Hepatocellular Carcinoma. OncoTargets Ther. 2020, 13, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zou, B.; Nan, T.; Zheng, X.; Zheng, L.; Lan, J.; Chen, W.; Yu, J. MiR-25 Enhances Autophagy and Promotes Sorafenib Resistance of Hepatocellular Carcinoma via Targeting FBXW7. Int. J. Med. Sci. 2022, 19, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-P.; Liu, J.-P.; Feng, J.-F.; Zhu, C.-P.; Yang, Y.; Zhou, W.-P.; Ding, J.; Huang, C.-K.; Cui, Y.-L.; Ding, C.-H.; et al. MiR-541 Potentiates the Response of Human Hepatocellular Carcinoma to Sorafenib Treatment by Inhibiting Autophagy. Gut 2020, 69, 1309–1321. [Google Scholar] [CrossRef]
- Yuan, J.; Lv, T.; Yang, J.; Wu, Z.; Yan, L.; Yang, J.; Shi, Y. HDLBP-Stabilized LncFAL Inhibits Ferroptosis Vulnerability by Diminishing Trim69-Dependent FSP1 Degradation in Hepatocellular Carcinoma. Redox Biol. 2022, 58, 102546. [Google Scholar] [CrossRef]
- Feng, J.; Lu, P.-Z.; Zhu, G.-Z.; Hooi, S.C.; Wu, Y.; Huang, X.-W.; Dai, H.-Q.; Chen, P.-H.; Li, Z.-J.; Su, W.-J.; et al. ACSL4 Is a Predictive Biomarker of Sorafenib Sensitivity in Hepatocellular Carcinoma. Acta Pharmacol. Sin. 2021, 42, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Yu, J.; Lu, L.; Zhang, Y.; Zhou, Y.; Zhou, Y.; Huang, F.; Sun, L.; Guo, Z.; Hou, G.; et al. MT1JP-Mediated MiR-24-3p/BCL2L2 Axis Promotes Lenvatinib Resistance in Hepatocellular Carcinoma Cells by Inhibiting Apoptosis. Cell Oncol. 2021, 44, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Fu, Y.; Zhu, N.; Wang, Z.; Hong, J.-L.; Li, Y.; Li, W.-J.; Zhang, H.-B.; Song, J.-H. Repression of LncRNA NEAT1 Enhances the Antitumor Activity of CD8+T Cells against Hepatocellular Carcinoma via Regulating MiR-155/Tim-3. Int. J. Biochem. Cell Biol. 2019, 110, 1–8. [Google Scholar] [CrossRef]
- Ye, R.; Lu, X.; Liu, J.; Duan, Q.; Xiao, J.; Duan, X.; Yue, Z.; Liu, F. CircSOD2 Contributes to Tumor Progression, Immune Evasion and Anti-PD-1 Resistance in Hepatocellular Carcinoma by Targeting MiR-497-5p/ANXA11 Axis. Biochem. Genet. 2023, 61, 597–614. [Google Scholar] [CrossRef]
- Liu, Z.; Ning, F.; Cai, Y.; Sheng, H.; Zheng, R.; Yin, X.; Lu, Z.; Su, L.; Chen, X.; Zeng, C.; et al. The EGFR-P38 MAPK Axis up-Regulates PD-L1 through MiR-675-5p and down-Regulates HLA-ABC via Hexokinase-2 in Hepatocellular Carcinoma Cells. Cancer Commun. 2021, 41, 62–78. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, T.; Cheng, X.; Li, D.; Zhao, M.; Zheng, W.V. MicroRNA-378a-3p Regulates the Progression of Hepatocellular Carcinoma by Regulating PD-L1 and STAT3. Bioengineered 2022, 13, 4730–4743. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Pan, B.; Zhang, X.; Wang, Z.; Qiu, J.; Wang, X.; Tang, N. Lipopolysaccharide Facilitates Immune Escape of Hepatocellular Carcinoma Cells via M6A Modification of LncRNA MIR155HG to Upregulate PD-L1 Expression. Cell Biol. Toxicol. 2022, 38, 1159–1173. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, B.; Qiu, J.; Ke, X.; Shen, S.; Wang, X.; Tang, N. LncRNA MIAT Targets MiR-411-5p/STAT3/PD-L1 Axis Mediating Hepatocellular Carcinoma Immune Response. Int. J. Exp. Pathol. 2022, 103, 102–111. [Google Scholar] [CrossRef]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y.; et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal MiR-23a-3p and Up-Regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, 241–258. [Google Scholar] [CrossRef]
- Kong, X.; Zheng, Z.; Song, G.; Zhang, Z.; Liu, H.; Kang, J.; Sun, G.; Sun, G.; Huang, T.; Li, X.; et al. Over-Expression of GUSB Leads to Primary Resistance of Anti-PD1 Therapy in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 876048. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhang, G.; Li, T.; Zhou, C.; Bai, L.; Zhao, J.; Tursun, T. LINC00657 Knockdown Suppresses Hepatocellular Carcinoma Progression by Sponging MiR-424 to Regulate PD-L1 Expression. Genes Genom. 2020, 42, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Xue, C.; Li, J.; Zhao, H.; Du, Y.; Du, N. LINC00244 Suppresses Cell Growth and Metastasis in Hepatocellular Carcinoma by Downregulating Programmed Cell Death Ligand 1. Bioengineered 2022, 13, 7635–7647. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Ye, S.; Chen, Y.; Zhang, Q.; Luo, Y.; Gai, L.; Luo, B. HOXA-AS3 Promotes Proliferation and Migration of Hepatocellular Carcinoma Cells via the MiR-455-5p/PD-L1 Axis. J. Immunol. Res. 2021, 2021, 9289719. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liao, R.; Wu, Z.; Du, C.; You, Y.; Que, K.; Duan, Y.; Yin, K.; Ye, W. Hepatic Stellate Cell Exosome-Derived CircWDR25 Promotes the Progression of Hepatocellular Carcinoma via the MiRNA-4474-3P-ALOX-15 and EMT Axes. Biosci. Trends 2022, 16, 267–281. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, E.; Tang, Y.; Mao, J.; Shen, J.; Zheng, X.; Xie, S.; Zhang, S.; Wu, Y.; Liu, H.; et al. MiR-223 Overexpression Inhibits Doxorubicin-Induced Autophagy by Targeting FOXO3a and Reverses Chemoresistance in Hepatocellular Carcinoma Cells. Cell Death Dis. 2019, 10, 843. [Google Scholar] [CrossRef]
- Lee, H.; Kim, C.; Kang, H.; Tak, H.; Ahn, S.; Yoon, S.K.; Kuh, H.-J.; Kim, W.; Lee, E.K. MicroRNA-200a-3p Increases 5-Fluorouracil Resistance by Regulating Dual Specificity Phosphatase 6 Expression. Exp. Mol. Med. 2017, 49, e327. [Google Scholar] [CrossRef]
- Ma, L.; Xu, A.; Kang, L.; Cong, R.; Fan, Z.; Zhu, X.; Huo, N.; Liu, W.; Xue, C.; Ji, Q.; et al. LSD1-Demethylated LINC01134 Confers Oxaliplatin Resistance Through SP1-Induced P62 Transcription in HCC. Hepatology 2021, 74, 3213–3234. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Zhang, S.; Huang, D.; Li, B.; Liang, G.; Wu, Y.; Jiang, Q.; Li, L.; Lin, C.; et al. CircRNA CircARNT2 Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting the MiR-155-5p/PDK1 Axis. Mol. Ther. Nucleic Acids 2021, 23, 244–254. [Google Scholar] [CrossRef]
- Li, P.; Song, R.; Yin, F.; Liu, M.; Liu, H.; Ma, S.; Jia, X.; Lu, X.; Zhong, Y.; Yu, L.; et al. CircMRPS35 Promotes Malignant Progression and Cisplatin Resistance in Hepatocellular Carcinoma. Mol. Ther. 2022, 30, 431–447. [Google Scholar] [CrossRef]
- Tang, W.; Chen, Z.; Zhang, W.; Cheng, Y.; Zhang, B.; Wu, F.; Wang, Q.; Wang, S.; Rong, D.; Reiter, F.P.; et al. The Mechanisms of Sorafenib Resistance in Hepatocellular Carcinoma: Theoretical Basis and Therapeutic Aspects. Signal Transduct. Target. Ther. 2020, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Shariati, M.; Meric-Bernstam, F. Targeting AKT for Cancer Therapy. Expert Opin. Investig. Drugs 2019, 28, 977–988. [Google Scholar] [CrossRef]
- Li, Y.-J.; Lei, Y.-H.; Yao, N.; Wang, C.-R.; Hu, N.; Ye, W.-C.; Zhang, D.-M.; Chen, Z.-S. Autophagy and Multidrug Resistance in Cancer. Chin. J. Cancer 2017, 36, 52. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, Biology and Role in Disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Niu, X.; Chen, R.; He, W.; Chen, D.; Kang, R.; Tang, D. Metallothionein-1G Facilitates Sorafenib Resistance through Inhibition of Ferroptosis. Hepatology 2016, 64, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Houessinon, A.; François, C.; Sauzay, C.; Louandre, C.; Mongelard, G.; Godin, C.; Bodeau, S.; Takahashi, S.; Saidak, Z.; Gutierrez, L.; et al. Metallothionein-1 as a Biomarker of Altered Redox Metabolism in Hepatocellular Carcinoma Cells Exposed to Sorafenib. Mol. Cancer 2016, 15, 38. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, J.; Lin, Y.; Sun, X.-M.; Zhang, J.-N.; Cheng, Z.-Q. Identification of MiR-211-5p as a Tumor Suppressor by Targeting ACSL4 in Hepatocellular Carcinoma. J. Transl. Med. 2020, 18, 326. [Google Scholar] [CrossRef]
- Tang, S.; Tan, G.; Jiang, X.; Han, P.; Zhai, B.; Dong, X.; Qiao, H.; Jiang, H.; Sun, X. An Artificial LncRNA Targeting Multiple MiRNAs Overcomes Sorafenib Resistance in Hepatocellular Carcinoma Cells. Oncotarget 2016, 7, 73257–73269. [Google Scholar] [CrossRef]
- Al-Salama, Z.T.; Syed, Y.Y.; Scott, L.J. Lenvatinib: A Review in Hepatocellular Carcinoma. Drugs 2019, 79, 665–674. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.-N.; Wang, K.-T.; Chen, L. Lenvatinib for Hepatocellular Carcinoma: From Preclinical Mechanisms to Anti-Cancer Therapy. Biochim. Biophys. Acta BBA-Rev. Cancer 2020, 1874, 188391. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus Sorafenib in First-Line Treatment of Patients with Unresectable Hepatocellular Carcinoma: A Randomised Phase 3 Non-Inferiority Trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Luo, J.; Gao, B.; Lin, Z.; Fan, H.; Ma, W.; Yu, D.; Yang, Q.; Tian, J.; Yang, X.; Li, B. Efficacy and Safety of Lenvatinib versus Sorafenib in First-Line Treatment of Advanced Hepatocellular Carcinoma: A Meta-Analysis. Front. Oncol. 2022, 12, 1010726. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, J.; Xu, Q.; Huang, D. Resistance of Lenvatinib in Hepatocellular Carcinoma. Curr. Cancer Drug Targets 2022, 22, 865–878. [Google Scholar] [CrossRef]
- Xie, C.; Li, S.-Y.; Fang, J.-H.; Zhu, Y.; Yang, J.-E. Functional Long Non-Coding RNAs in Hepatocellular Carcinoma. Cancer Lett. 2021, 500, 281–291. [Google Scholar] [CrossRef]
- Shen, H.; Liu, B.; Xu, J.; Zhang, B.; Wang, Y.; Shi, L.; Cai, X. Circular RNAs: Characteristics, Biogenesis, Mechanisms and Functions in Liver Cancer. J. Hematol. Oncol. 2021, 14, 134. [Google Scholar] [CrossRef]
- Tan, W.; Zhang, K.; Chen, X.; Yang, L.; Zhu, S.; Wei, Y.; Xie, Z.; Chen, Y.; Shang, C. GPX2 Is a Potential Therapeutic Target to Induce Cell Apoptosis in Lenvatinib against Hepatocellular Carcinoma. J. Adv. Res. 2023, 44, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Iseda, N.; Itoh, S.; Toshida, K.; Tomiyama, T.; Morinaga, A.; Shimokawa, M.; Shimagaki, T.; Wang, H.; Kurihara, T.; Toshima, T.; et al. Ferroptosis Is Induced by Lenvatinib through Fibroblast Growth Factor Receptor-4 Inhibition in Hepatocellular Carcinoma. Cancer Sci. 2022, 113, 2272–2287. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, S.; Zeng, S.; Shen, H. From Bench to Bed: The Tumor Immune Microenvironment and Current Immunotherapeutic Strategies for Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 396. [Google Scholar] [CrossRef]
- Jiang, Y.; Han, Q.-J.; Zhang, J. Hepatocellular Carcinoma: Mechanisms of Progression and Immunotherapy. World J. Gastroenterol. 2019, 25, 3151–3167. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, H.; Yuan, X.; Fan, X.; Zhang, C. Advances in Immune Checkpoint Inhibitors for Advanced Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 896752. [Google Scholar] [CrossRef]
- Gordan, J.D.; Kennedy, E.B.; Abou-Alfa, G.K.; Beg, M.S.; Brower, S.T.; Gade, T.P.; Goff, L.; Gupta, S.; Guy, J.; Harris, W.P.; et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline. JCO 2020, 38, 4317–4345. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Cardoso, A.P.; Gonçalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Han, Q.; Wang, M.; Dong, X.; Wei, F.; Luo, Y.; Sun, X. Non-Coding RNAs in Hepatocellular Carcinoma: Insights into Regulatory Mechanisms, Clinical Significance, and Therapeutic Potential. Front. Immunol. 2022, 13, 985815. [Google Scholar] [CrossRef]
- Ji, J.; Yin, Y.; Ju, H.; Xu, X.; Liu, W.; Fu, Q.; Hu, J.; Zhang, X.; Sun, B. Long Non-Coding RNA Lnc-Tim3 Exacerbates CD8 T Cell Exhaustion via Binding to Tim-3 and Inducing Nuclear Translocation of Bat3 in HCC. Cell Death Dis. 2018, 9, 478. [Google Scholar] [CrossRef]
- Kuol, N.; Stojanovska, L.; Nurgali, K.; Apostolopoulos, V. PD-1/PD-L1 in Disease. Immunotherapy 2018, 10, 149–160. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in Cancer Immunotherapy: Clinical Implications and Future Considerations. Hum. Vaccin. Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. 2021, 16, 223–249. [Google Scholar] [CrossRef]
- Liu, M.; Xu, M.; Tang, T. Association between Chemotherapy and Prognostic Factors of Survival in Hepatocellular Carcinoma: A SEER Population-Based Cohort Study. Sci. Rep. 2021, 11, 23754. [Google Scholar] [CrossRef]
- Raoul, J.-L.; Forner, A.; Bolondi, L.; Cheung, T.T.; Kloeckner, R.; de Baere, T. Updated Use of TACE for Hepatocellular Carcinoma Treatment: How and When to Use It Based on Clinical Evidence. Cancer Treat. Rev. 2019, 72, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC Strategy for Prognosis Prediction and Treatment Recommendation: The 2022 Update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.D.; Zhang, B.; Tywonek, K.; Meyers, B.M.; Serrano, P.E. Toxicity Profiles of Systemic Therapies for Advanced Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2022, 5, e2222721. [Google Scholar] [CrossRef] [PubMed]
- Fa, H.-G.; Chang, W.-G.; Zhang, X.-J.; Xiao, D.-D.; Wang, J.-X. Noncoding RNAs in Doxorubicin-Induced Cardiotoxicity and Their Potential as Biomarkers and Therapeutic Targets. Acta Pharmacol. Sin. 2021, 42, 499–507. [Google Scholar] [CrossRef]
- Arola, O.J.; Saraste, A.; Pulkki, K.; Kallajoki, M.; Parvinen, M.; Voipio-Pulkki, L.M. Acute Doxorubicin Cardiotoxicity Involves Cardiomyocyte Apoptosis. Cancer Res. 2000, 60, 1789–1792. [Google Scholar]
- Andrieu-Abadie, N.; Jaffrezou, J.P.; Hatem, S.; Laurent, G.; Levade, T.; Mercadier, J.J. L-Carnitine Prevents Doxorubicin-Induced Apoptosis of Cardiac Myocytes: Role of Inhibition of Ceramide Generation. FASEB J. 1999, 13, 1501–1510. [Google Scholar] [CrossRef]
- Fu, Q.; Pan, H.; Tang, Y.; Rong, J.; Zheng, Z. MiR-200a-3p Aggravates DOX-Induced Cardiotoxicity by Targeting PEG3 Through SIRT1/NF-ΚB Signal Pathway. Cardiovasc. Toxicol. 2021, 21, 302–313. [Google Scholar] [CrossRef]
- Xiong, X.; He, Q.; Liu, J.; Dai, R.; Zhang, H.; Cao, Z.; Liao, Y.; Liu, B.; Zhou, Y.; Chen, J.; et al. MicroRNA MiR-215-5p Regulates Doxorubicin-Induced Cardiomyocyte Injury by Targeting ZEB2. J. Cardiovasc. Pharmacol. 2021, 78, 622–629. [Google Scholar] [CrossRef]
- Hu, X.; Liao, W.; Teng, L.; Ma, R.; Li, H. Circ_0001312 Silencing Suppresses Doxorubicin-Induced Cardiotoxicity via MiR-409-3p/HMGB1 Axis. Int. Heart J. 2023, 64, 71–80. [Google Scholar] [CrossRef]
- Christidi, E.; Brunham, L.R. Regulated Cell Death Pathways in Doxorubicin-Induced Cardiotoxicity. Cell Death Dis. 2021, 12, 339. [Google Scholar] [CrossRef]
- Fan, D.; Chen, H.-B.; Leng, Y.; Yang, S.-J. MiR-24-3p Attenuates Doxorubicin-Induced Cardiotoxicity via the Nrf2 Pathway in Mice. Curr. Med. Sci. 2022, 42, 48–55. [Google Scholar] [CrossRef]
- Li, Z.; Ye, Z.; Ma, J.; Gu, Q.; Teng, J.; Gong, X. MicroRNA-133b Alleviates Doxorubicin-induced Cardiomyocyte Apoptosis and Cardiac Fibrosis by Targeting PTBP1 and TAGLN2. Int. J. Mol. Med. 2021, 48, 125. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Cao, Y.; Wang, Q.; Xu, K.; Dou, L.; Huang, X.; Chen, B.; Tang, W.; Lan, M.; Liu, B.; et al. MiR-488-3p Protects Cardiomyocytes against Doxorubicin-Induced Cardiotoxicity by Inhibiting CyclinG1. Oxid. Med. Cell. Longev. 2022, 2022, 5184135. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ruan, Y.; Shen, T.; Qiu, Q.; Yan, M.; Sun, S.; Dou, L.; Huang, X.; Wang, Q.; Zhang, X.; et al. Dexrazoxane Protects Cardiomyocyte from Doxorubicin-Induced Apoptosis by Modulating MiR-17-5p. Biomed. Res. Int. 2020, 2020, 5107193. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.H. Can Dexrazoxane Reduce Myocardial Injury in Anthracycline-Treated Children with Acute Lymphoblastic Leukemia? Nat. Clin. Pract. Oncol. 2004, 1, 16–17. [Google Scholar] [CrossRef]
- Li, Q.; Qin, M.; Li, T.; Gu, Z.; Tan, Q.; Huang, P.; Ren, L. Rutin Protects against Pirarubicin-Induced Cardiotoxicity by Adjusting MicroRNA-125b-1-3p-Mediated JunD Signaling Pathway. Mol. Cell Biochem. 2020, 466, 139–148. [Google Scholar] [CrossRef]
- Guo, L.; Zheng, X.; Wang, E.; Jia, X.; Wang, G.; Wen, J. Irigenin Treatment Alleviates Doxorubicin (DOX)-Induced Cardiotoxicity by Suppressing Apoptosis, Inflammation and Oxidative Stress via the Increase of MiR-425. Biomed. Pharmacother. 2020, 125, 109784. [Google Scholar] [CrossRef]
- Kelland, L. The Resurgence of Platinum-Based Cancer Chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef]
- Kaseb, A.O.; Shindoh, J.; Patt, Y.Z.; Roses, R.E.; Zimmitti, G.; Lozano, R.D.; Hassan, M.M.; Hassabo, H.M.; Curley, S.A.; Aloia, T.A.; et al. Modified Cisplatin/Interferon α-2b/Doxorubicin/5-Fluorouracil (PIAF) Chemotherapy in Patients with No Hepatitis or Cirrhosis Is Associated with Improved Response Rate, Resectability, and Survival of Initially Unresectable Hepatocellular Carcinoma. Cancer 2013, 119, 3334–3342. [Google Scholar] [CrossRef]
- Pabla, N.; Dong, Z. Cisplatin Nephrotoxicity: Mechanisms and Renoprotective Strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, K.; Luo, H.; Wu, C.; Yu, W.; Cheng, F. Novel LncRNA XLOC_032768 Alleviates Cisplatin-Induced Apoptosis and Inflammatory Response of Renal Tubular Epithelial Cells through TNF-α. Int. Immunopharmacol. 2020, 83, 106472. [Google Scholar] [CrossRef]
- Ma, M.; Luo, Q.; Fan, L.; Li, W.; Li, Q.; Meng, Y.; Yun, C.; Wu, H.; Lu, Y.; Cui, S.; et al. The Urinary Exosomes Derived from Premature Infants Attenuate Cisplatin-Induced Acute Kidney Injury in Mice via MicroRNA-30a-5p/ Mitogen-Activated Protein Kinase 8 (MAPK8). Bioengineered 2022, 13, 1650–1665. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.; Zhang, F.; Yang, J.; He, J. Resveratrol Upregulates MiR-455-5p to Antagonize Cisplatin Ototoxicity via Modulating the PTEN-PI3K-AKT Axis. Biochem. Cell Biol. 2021, 99, 385–395. [Google Scholar] [CrossRef]
- Sara, J.D.; Kaur, J.; Khodadadi, R.; Rehman, M.; Lobo, R.; Chakrabarti, S.; Herrmann, J.; Lerman, A.; Grothey, A. 5-Fluorouracil and Cardiotoxicity: A Review. Ther. Adv. Med. Oncol. 2018, 10, 1758835918780140. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, G.; Ren, W.; Wang, B.; Yang, C.; Li, M. LncRNA NEAT1 Regulates 5-Fu Sensitivity, Apoptosis and Invasion in Colorectal Cancer Through the MiR-150-5p/CPSF4 Axis. OncoTargets Ther. 2020, 13, 6373–6383. [Google Scholar] [CrossRef] [PubMed]
- Amrovani, M.; Mohammadtaghizadeh, M.; Aghaali, M.K.; Zamanifard, S.; Alqasi, A.; Sanei, M. Long Non-Coding RNAs: Potential Players in Cardiotoxicity Induced by Chemotherapy Drugs. Cardiovasc. Toxicol. 2022, 22, 191–206. [Google Scholar] [CrossRef]
- Chen, H.; Xia, W.; Hou, M. LncRNA-NEAT1 from the Competing Endogenous RNA Network Promotes Cardioprotective Efficacy of Mesenchymal Stem Cell-Derived Exosomes Induced by Macrophage Migration Inhibitory Factor via the MiR-142-3p/FOXO1 Signaling Pathway. Stem Cell Res. Ther. 2020, 11, 31. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Dowdy, S.F. Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [Google Scholar] [CrossRef]
- Hao, M.; Zhang, L.; Chen, P. Membrane Internalization Mechanisms and Design Strategies of Arginine-Rich Cell-Penetrating Peptides. Int. J. Mol. Sci. 2022, 23, 9038. [Google Scholar] [CrossRef]
- Dzierlega, K.; Yokota, T. Optimization of Antisense-Mediated Exon Skipping for Duchenne Muscular Dystrophy. Gene Ther. 2020, 27, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Ladokhin, A.S. PH-Triggered Conformational Switching along the Membrane Insertion Pathway of the Diphtheria Toxin T-Domain. Toxins 2013, 5, 1362–1380. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.S.; Dakwar, G.R.; Zagato, E.; Brans, T.; Figueiredo, C.; Raemdonck, K.; Azevedo, N.F.; De Smedt, S.C.; Braeckmans, K. Intracellular Delivery of Oligonucleotides in Helicobacter Pylori by Fusogenic Liposomes in the Presence of Gastric Mucus. Biomaterials 2017, 138, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nogrady, B. The Challenge of Delivering RNA-Interference Therapeutics to Their Target Cells. Nature 2019, 574, S8–S9. [Google Scholar] [CrossRef]
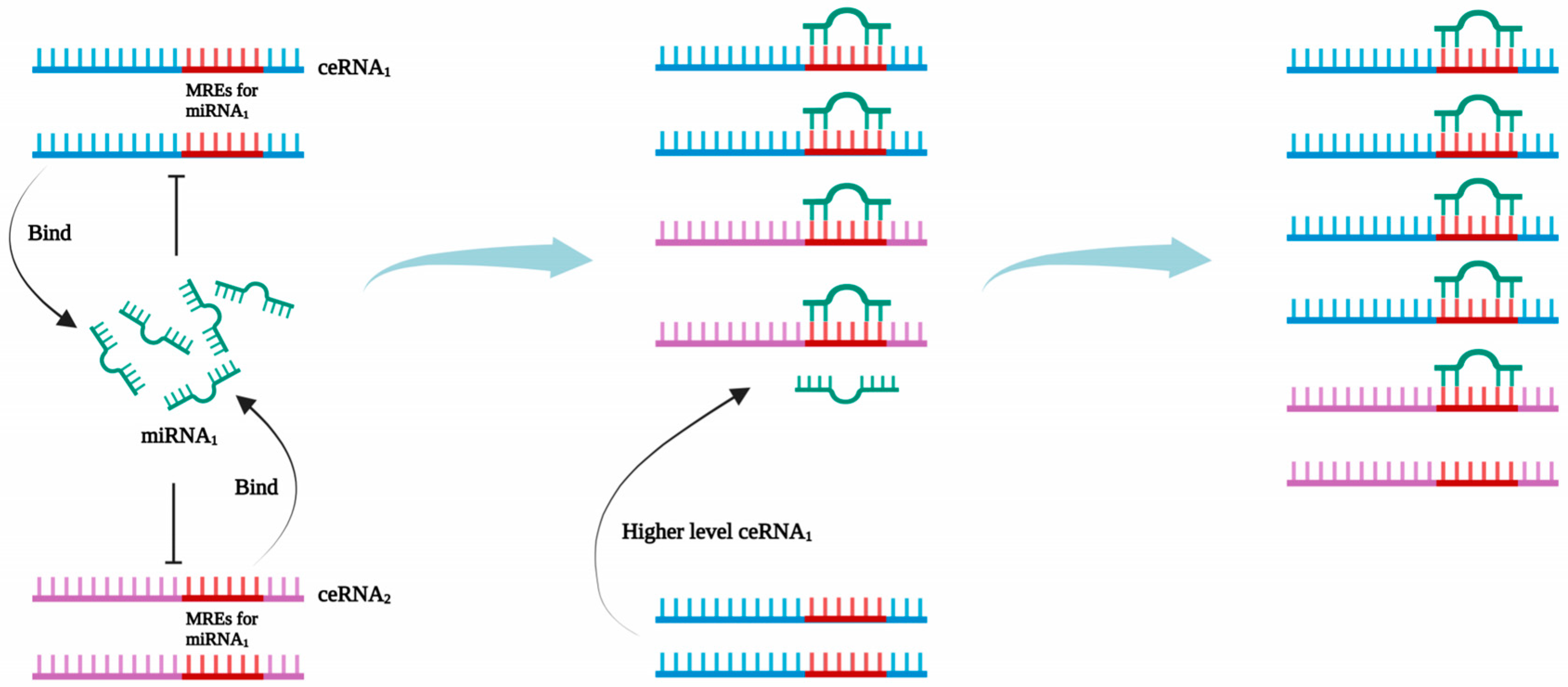


| Therapy | ncRNAs | Targets | Effect | Mechanism | Refs. |
|---|---|---|---|---|---|
| Sorafenib | miR-92b | ↓PTEN | Anti-apoptosis Promote SR | miR-92b reduces sorafenib-induced apoptosis and promotes SR by targeting PTEN and activating the PI3K/AKT/mTOR pathway. | [68] |
| miR-21 | ↓PTEN | Anti-apoptosis Anti-autophagy Promote SR | miR-21 promotes the expression of SNHG1 while simultaneously downregulating the expression of PTEN, which leads to the activation of the AKT pathway, independent of SNHG1. | [69,70] | |
| lncRNA SNHG1 | ↑SLC3A2 | Anti-apoptosis Promote SR | SNHG1 activates the AKT pathway by regulating SLC3A2, and its depletion enhances sorafenib-induced apoptosis. | [69] | |
| lncRNA NEAT1 | ↑c-Met-AKT | Anti-apoptotic Promote SR | NEAT1 can negatively regulate miR-335, which further suppresses the c-Met-AKT pathway. | [71] | |
| lncRNA MALAT1 | ↑Aurora-A | Anti-apoptosis Promote SR | MALAT1 regulated Aurora-A expression by sponging miR-140-5p, thus promoting SR in HCC cells. | [72] | |
| miR-622 | ↓KRAS | Pro-apoptosis Reduce SR | KRAS, which leads to the suppression of RAF/ERK and PI3K/AKT signaling pathways, is directly targeted by miR-622. | [73] | |
| lncRNA HEIH | ↓PI3K/AKT | Pro-apoptotic Reduce SR | HEIH can act as a sponge for miR-98-5p, and its inhibition activates the PI3K/AKT pathway, promoting SR. | [74] | |
| circRNA ITCH | ↑PTEN | Pro-apoptotic Reduce SR | ITCH increases PTEN expression by sponging miR-20b-5p and then inactivates PI3K/Akt signals. | [75] | |
| miR-486-3p | ↓FGFR4 ↓EGFR | Pro-apoptotic Reduce SR | miR-486-3p induces apoptosis by targeting FGFR4 and EGFR, effectively overcoming SR and inhibiting tumor growth when combined with sorafenib treatment. | [76] | |
| miR-10b-3p | ↓CyclinE1 | Pro-apoptotic Reduce SR | Overexpressing miR-10b-3p enhances sorafenib-induced apoptosis in HCC cells, while depletion of miR-10b-3p partially abrogates this effect. | [77] | |
| lncRNA HANR | ↑ATG9A | Pro-autophagy Promote SR | HANR promotes autophagy and contributes to SR. miR-29b targets ATG9A and counteracts HANR-induced SR by suppressing autophagy. | [78] | |
| miR-25 | ↓FBXW7 | Pro-autophagy Promote SR | By inducing autophagy, miR-25 enhances SR in HCC and concurrently downregulates the expression of FBXW7 protein to modulate autophagy. | [79] | |
| miR-541 | ↓ATG2A ↓RAB1B | Anti-autophagy Reduce SR | miR-541 directly acts on ATG2A and RAB1B, thereby inhibiting the malignant phenotype and autophagy of HCC cells. Higher miR-541 expression predicts a better response to sorafenib. | [80] | |
| lncRNA lncFAL | ↑FSP1 | Anti-ferroptosis Promote SR | lncFAL reduces ferroptosis vulnerability by directly binding to FSP1 and competitively inhibiting Trim69-mediated FSP1 polyubiquitination degradation, thereby diminishing the anti-cancer effect of ferroptosis inducers like sorafenib. | [81] | |
| lncRNA HCG18 | ↑GPX4 | Anti-ferroptosis Promote SR | Silencing HCG18 inhibits GPX4 by binding to miR-450b-5p, promotes GPX4-inhibited ferroptosis, and averts SR in HCC. | [39] | |
| miR-211-5p | ↓ACSL4 | Anti-ferroptosis Promote SR | Decreasing ACSL4 significantly reduces sorafenib-induced lipid peroxidation and ferroptosis in HCC cells. Higher ACSL4 level indicates improved response to sorafenib treatment | [82] | |
| Lenvatinib | lncRNA MT1JP | ↑BCL2L2 | Anti-apoptosis Promote LR | The inhibition of apoptosis by the MT1JP-mediated miR-24-3p/BCL2L2 axis promotes resistance to lenvatinib in HCC cells. | [83] |
| m7G-tRNA | ↑EGFR | Anti-apoptosis Promote LR | METTL1/WDR4-mediated m7G tRNA modification in promoting translation of EGFR pathway genes to reduce apoptosis and trigger drug resistance in HCC cells. | [43] | |
| Immune therapy | LncRNA NEAT1 | ↓Tim3 | Promote CD8+ T cell apoptosis | The increased expression of lncRNA NEAT1 in peripheral blood monocytes of HCC patients can disrupt the expression of Tim-3 by interacting with miR-155. The reduction of NEAT1 suppresses apoptosis in CD8+ T cells. | [84] |
| circRNA circSOD2 | ↑ANXA11 | Promote CD8+ T cell apoptosis | High circSOD2 expression induces CD8+ T cell death, leading to CD8+ T cell dysfunction and immune escape in HCC, thereby hindering the effectiveness of anti-PD-1 drugs. | [85] | |
| miR-675-5p | - | ↓PD-L1 | Downregulating miR-675-5p stabilizes PD-L1 mRNA, leading to the accumulation of PD-L1 in HCC cells. | [86] | |
| miR-378a-3p | - | ↓PD-L1 | PD-L1 3′-UTR is a target of miR-378a-3p. miR-378a-3p suppresses PD-L1 expression in HCC cells. | [87] | |
| lncRNA MIR155HG | ↑STAT1 | ↑PD-L1 | MIR155HG functions as a sponge for miR-233, thereby upregulating PD-L1 expression through the miR-223/STAT1 axis. | [88] | |
| lncRNA MIAT | ↑STAT3 | ↑PD-L1 | MIAT negatively regulates miR-411-5p, leading to an upregulation of STAT3 and ultimately increasing PD-L1 expression at the transcriptional level | [89] | |
| mir-23a-3p | ↓PTEN ↑PI3K/AKT | ↑PD-L1 (Macrophage) | Extracellular vesicle-mediated transfer of miR-23a-3p from HCC cells to macrophages upregulates PD-L1 expression by inhibiting PTEN and activating the PI3K-AKT pathway. This results in increased T cell apoptosis and decreased CD8+ T cell proportion. | [90] | |
| miR-513a-5p | - | ↓PD-L1 | GUSB downregulates PD-L1 expression by promoting miR-513a-5p. GUSB inhibitor can improve the sensitivity of anti-PD1 therapy. | [91] | |
| lncRNA LINC00657 | ↓miR-424 | ↑PD-L1 | LINC00657 exerts regulatory control over PD-L1 expression by acting as a miR-424 sponge, consequently influencing the progression of HCC. | [92] | |
| lncRNA LINC00244 | - | ↓PD-L1 | LINC00244 downregulates PD-L1 and suppresses cell growth and metastasis in HCC. | [93] | |
| lncRNA HOXA-AS3 | ↓miR-455-5p | ↑PD-L1 | HOXA-AS3 increased the expression of PD-L1 by sponging miR-455-5p. | [94] | |
| circRNA circWDR25 | ↑ALOX15 | ↑PD-L1 | circWDR25 regulates the expression of ALOX15 by sponging miR-4474-3p, ultimately inducing an epithelial-to-mesenchymal transition, and then promoting the expression of PD-L1 in HCC cells. | [95] | |
| Doxorubicin | miR-223 | ↓FOXO3a | Anti-autophagy Promote resistance | miR-223 can inhibit DOX-induced autophagy by targeting FOXO3a and contributes to DOX-resistance in HCC cells. | [96] |
| 5-fluorouracil | miR-200a-3p | ↓DUSP6 | Anti-apoptosis Promote resistance | microRNA-200a-3p increases 5-fluorouracil resistance by regulating DUSP6 expression. | [97] |
| Oxaliplatin | lncRNA LINC01134 | ↑P62 ↑LSD1 | Anti-apoptosis Promote resistance | LINC01134 reduces cell apoptosis through the SP1/P62 pathway, thereby enhancing oxaliplatin resistance in HCC. | [98] |
| Cisplatin | circRNA circARNT2 | ↑PDK1 | Pro-autophagy Reduce resistance | circARNT2 functions as a competing molecule against miR-155-5p, resulting in the upregulation of PDK1-induced autophagy and increasing sensitivity of HCC cells to cisplatin. | [99] |
| circRNA circMRPS35 | ↑STX3 | Anti-apoptosis Promote resistance | circMRPS35 acts as a sponge for miR-148a-3p, resulting in the upregulation of STX3 expression. STX3, in turn, induces the ubiquitination and degradation of PTEN to reduce apoptosis and promote cisplatin resistance. | [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Ruan, M.; Zou, M.; Liu, F.; Liu, H. Clinical Significance of Non-Coding RNA Regulation of Programmed Cell Death in Hepatocellular Carcinoma. Cancers 2023, 15, 4187. https://doi.org/10.3390/cancers15164187
Chen W, Ruan M, Zou M, Liu F, Liu H. Clinical Significance of Non-Coding RNA Regulation of Programmed Cell Death in Hepatocellular Carcinoma. Cancers. 2023; 15(16):4187. https://doi.org/10.3390/cancers15164187
Chicago/Turabian StyleChen, Wuyu, Minghao Ruan, Minghao Zou, Fuchen Liu, and Hui Liu. 2023. "Clinical Significance of Non-Coding RNA Regulation of Programmed Cell Death in Hepatocellular Carcinoma" Cancers 15, no. 16: 4187. https://doi.org/10.3390/cancers15164187
APA StyleChen, W., Ruan, M., Zou, M., Liu, F., & Liu, H. (2023). Clinical Significance of Non-Coding RNA Regulation of Programmed Cell Death in Hepatocellular Carcinoma. Cancers, 15(16), 4187. https://doi.org/10.3390/cancers15164187




