Metabolic Adjustments following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture and CB-839 Assessment on Cell Proliferation
2.3. Metabolomics
2.4. U-13C- Glutamine Tracing Experiments
2.5. 15N-Glutamine Tracing Assays
2.6. Statistical Analysis
3. Results
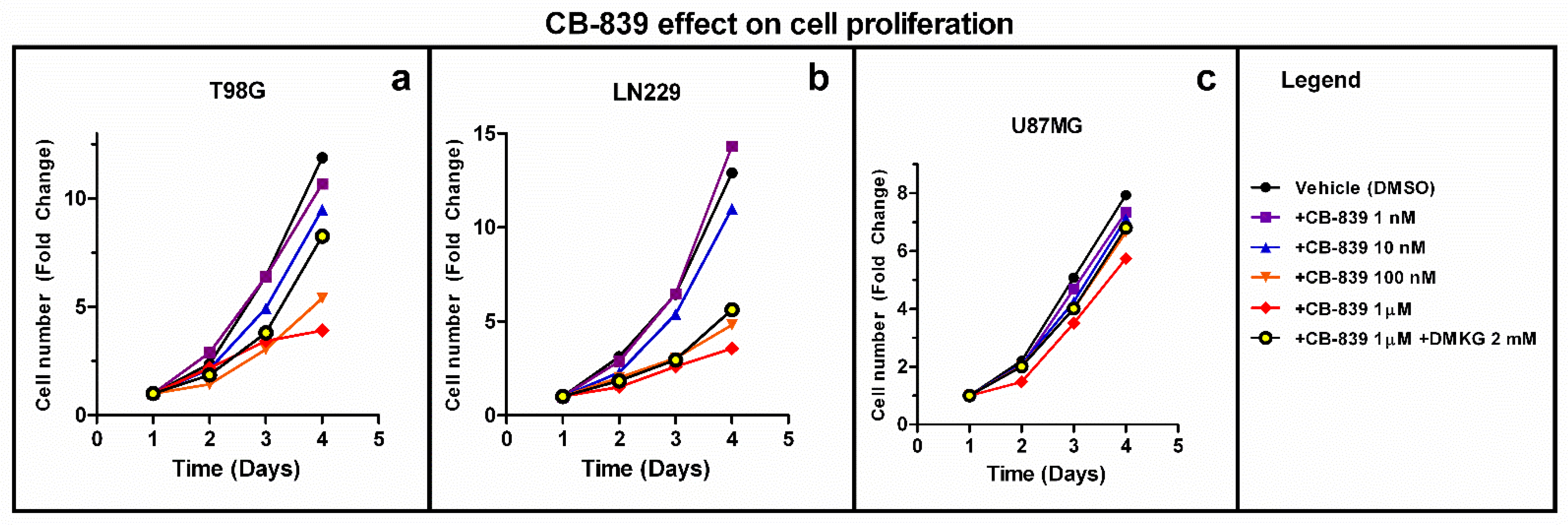
3.1. Effect of CB-839 on Cell Proliferation in Three Human GBM Cell Lines
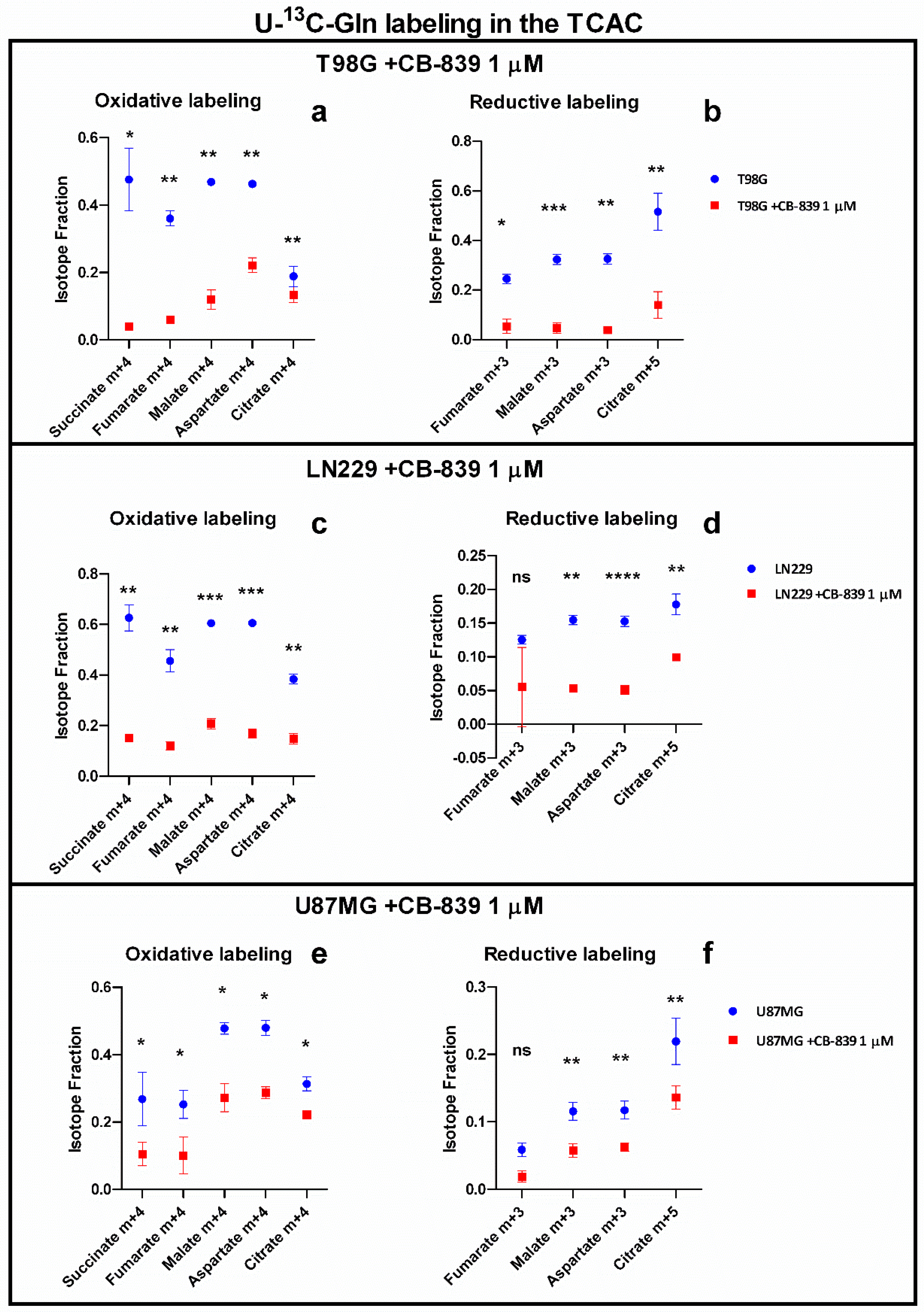
3.2. CB-839 Suppresses Both Oxidative Decarboxylation and Reductive Carboxylation of Gln-Derived αKG in the TCAC
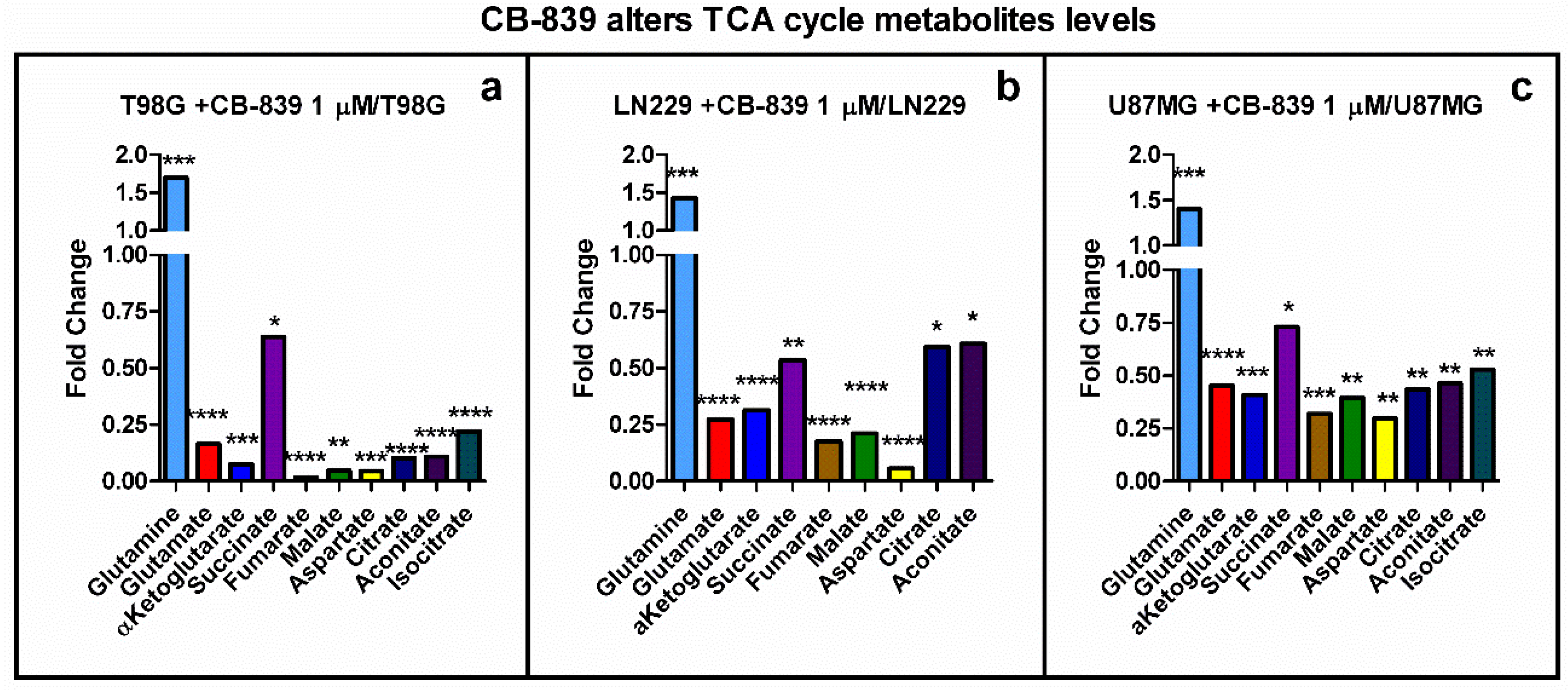
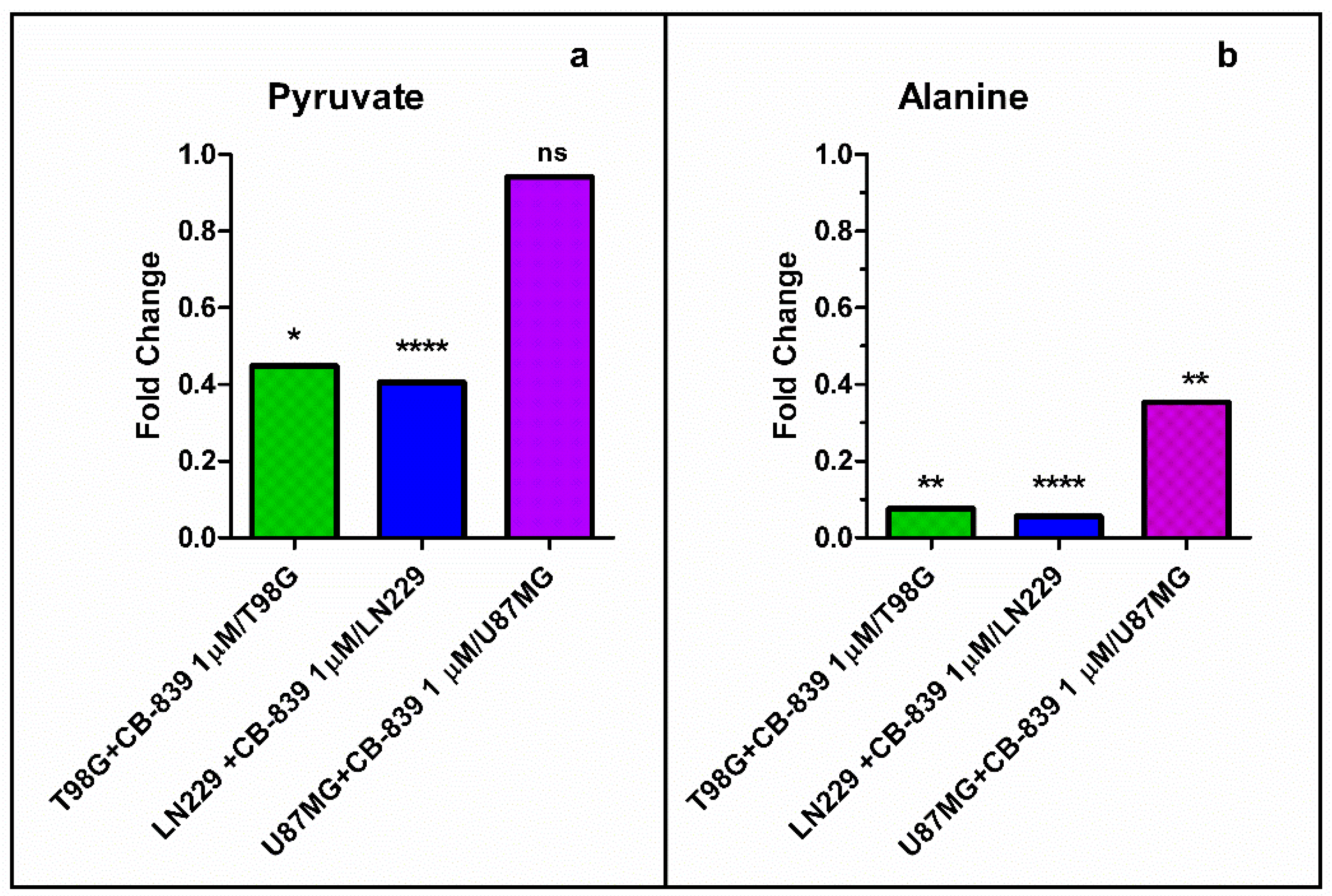
3.3. GLS Inhibition by CB-839 Modifies Key Metabolites
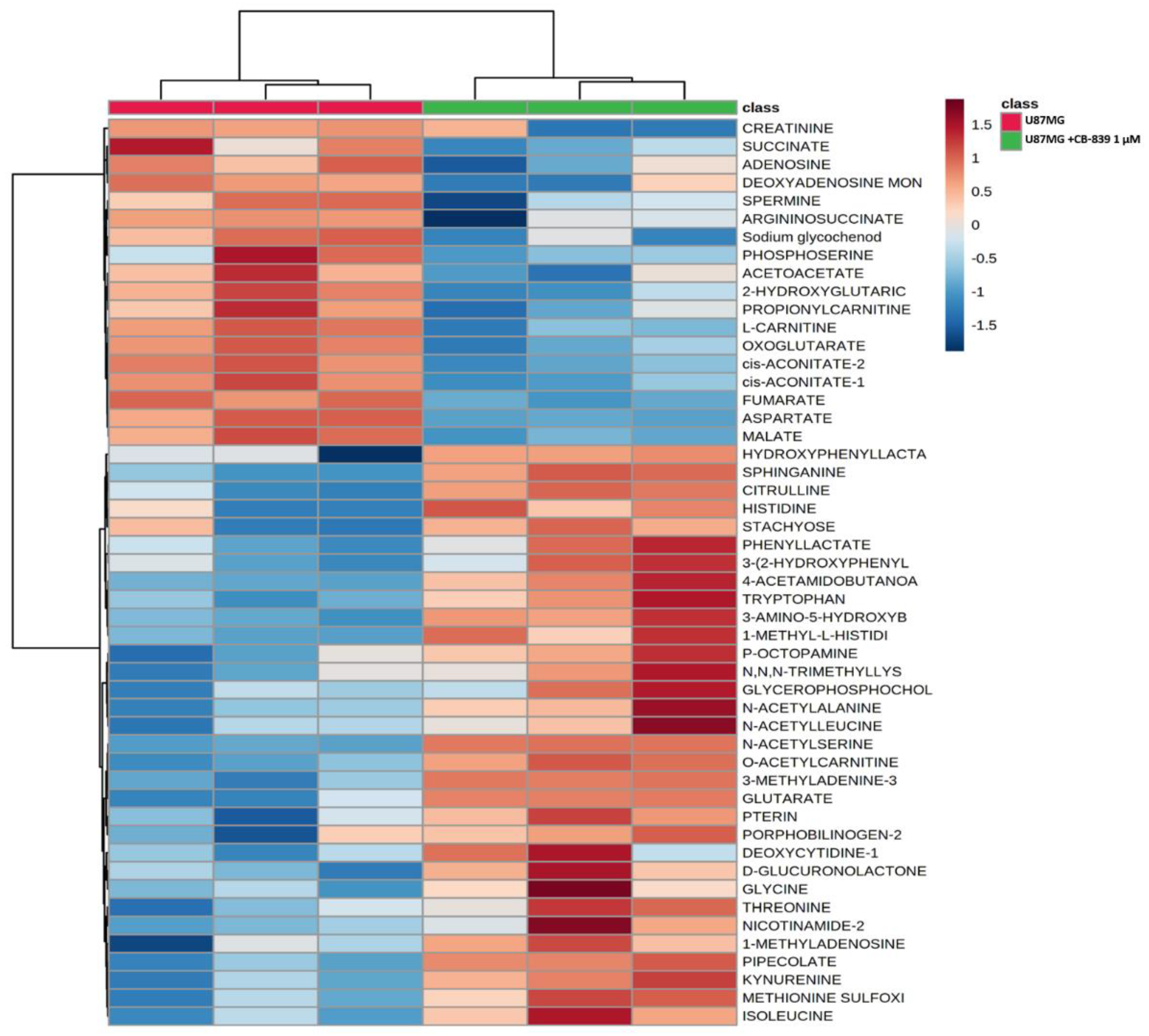
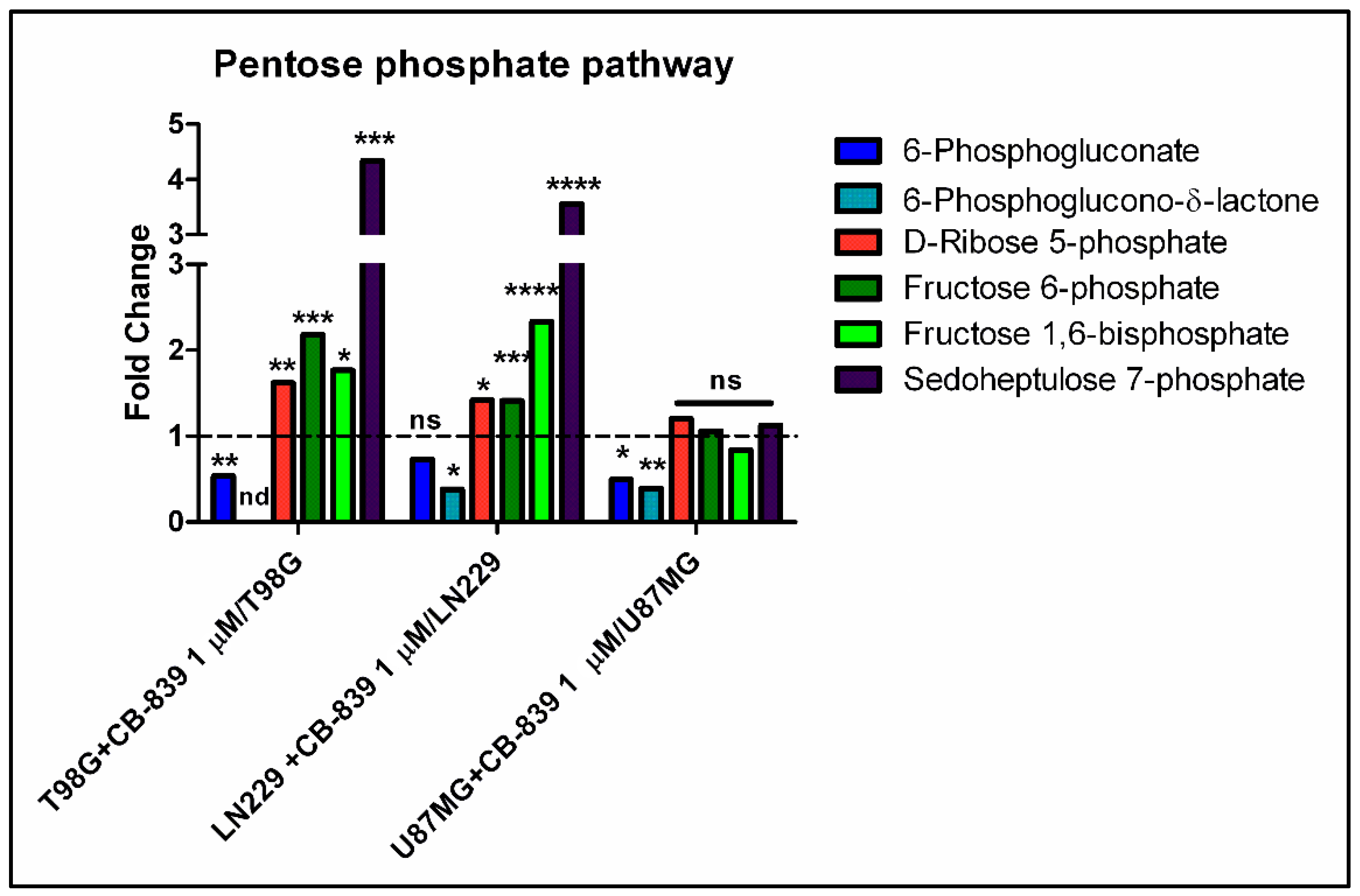
3.4. CB-839 Reshapes Core Metabolic Pathways in Human GBM Cell Lines
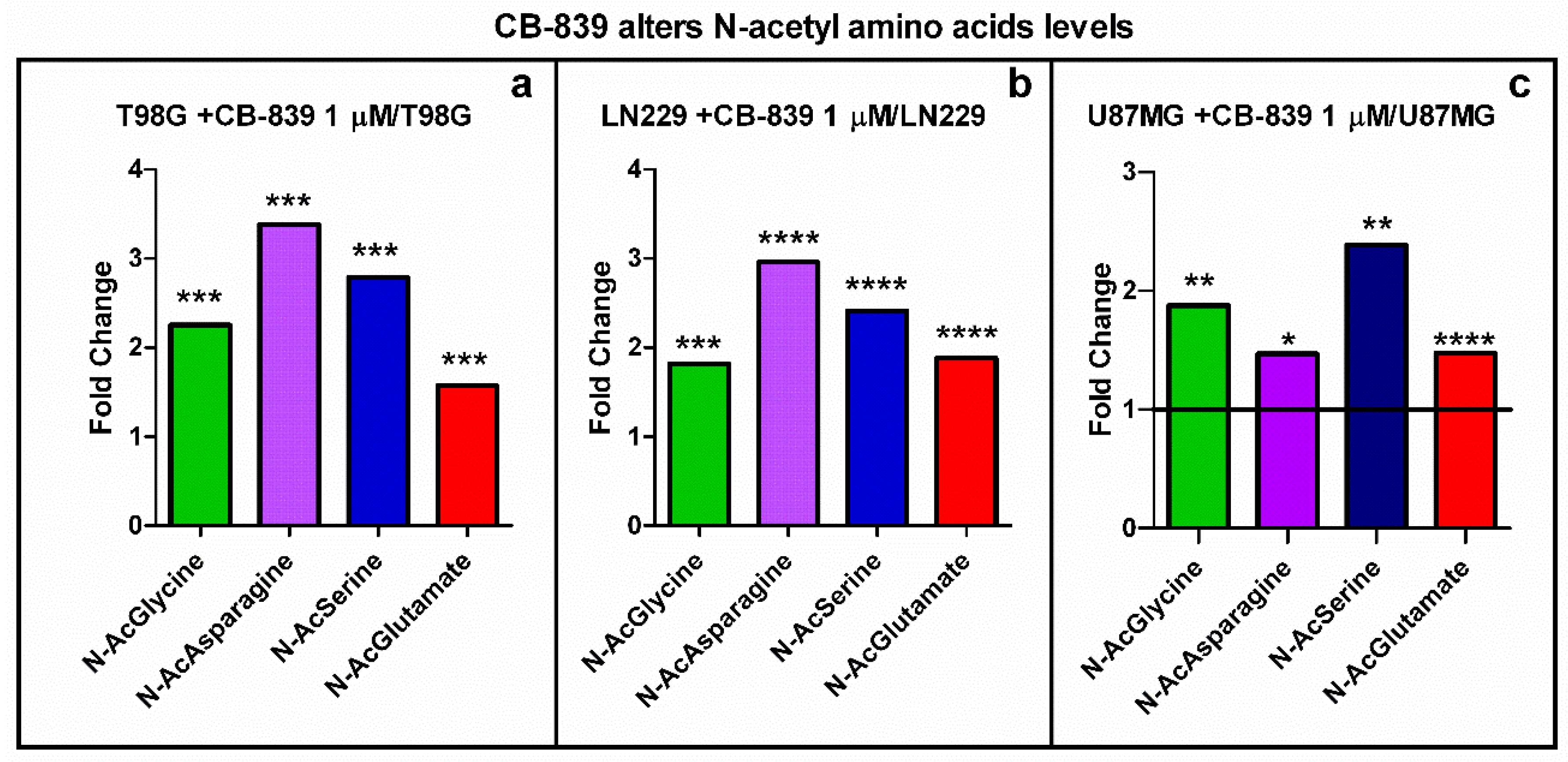
3.5. CB-839 Alters Acetylated Metabolites and Urea Cycle Reactions
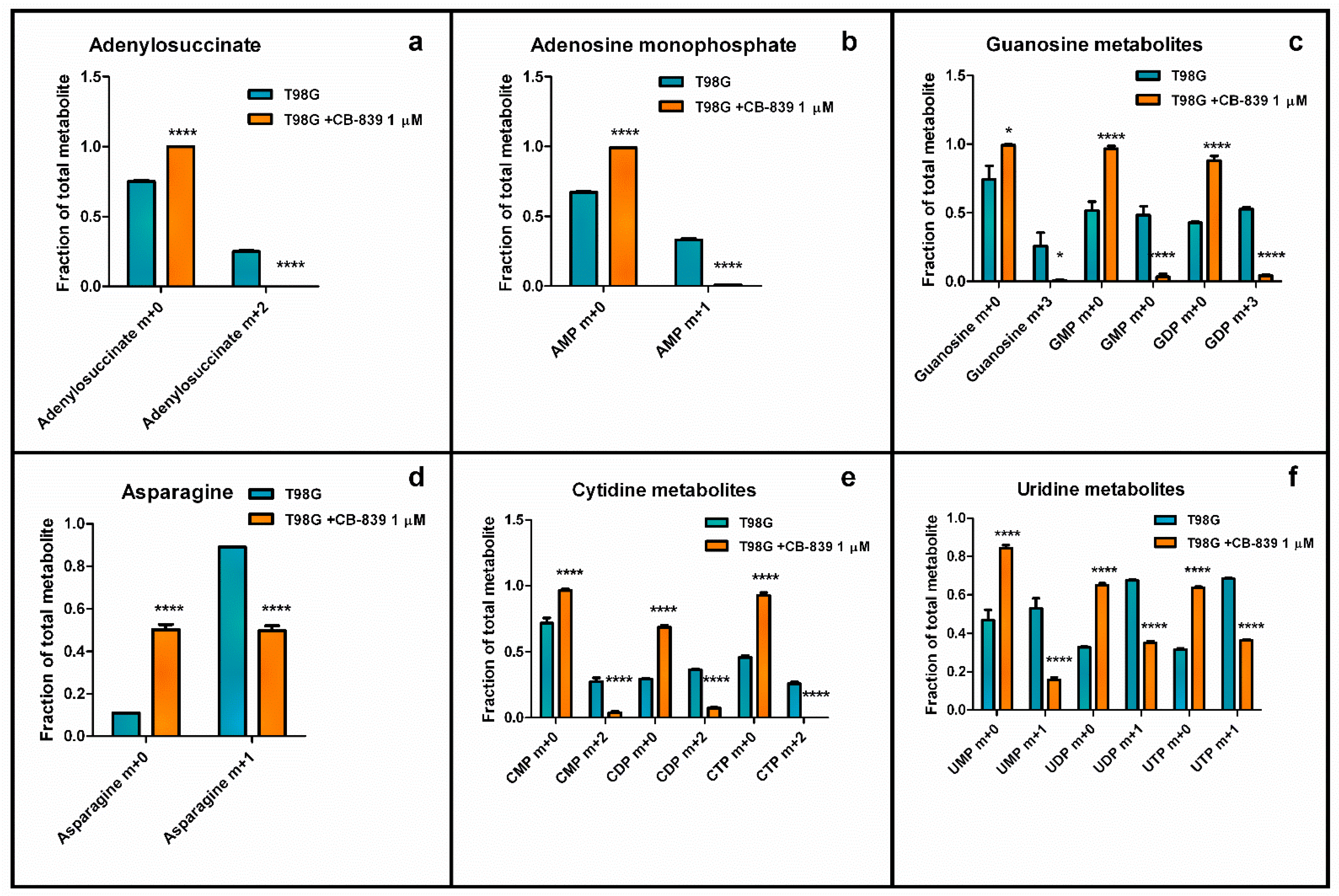
3.6. CB-839 Modifies De Novo Biosynthesis of Nucleotides
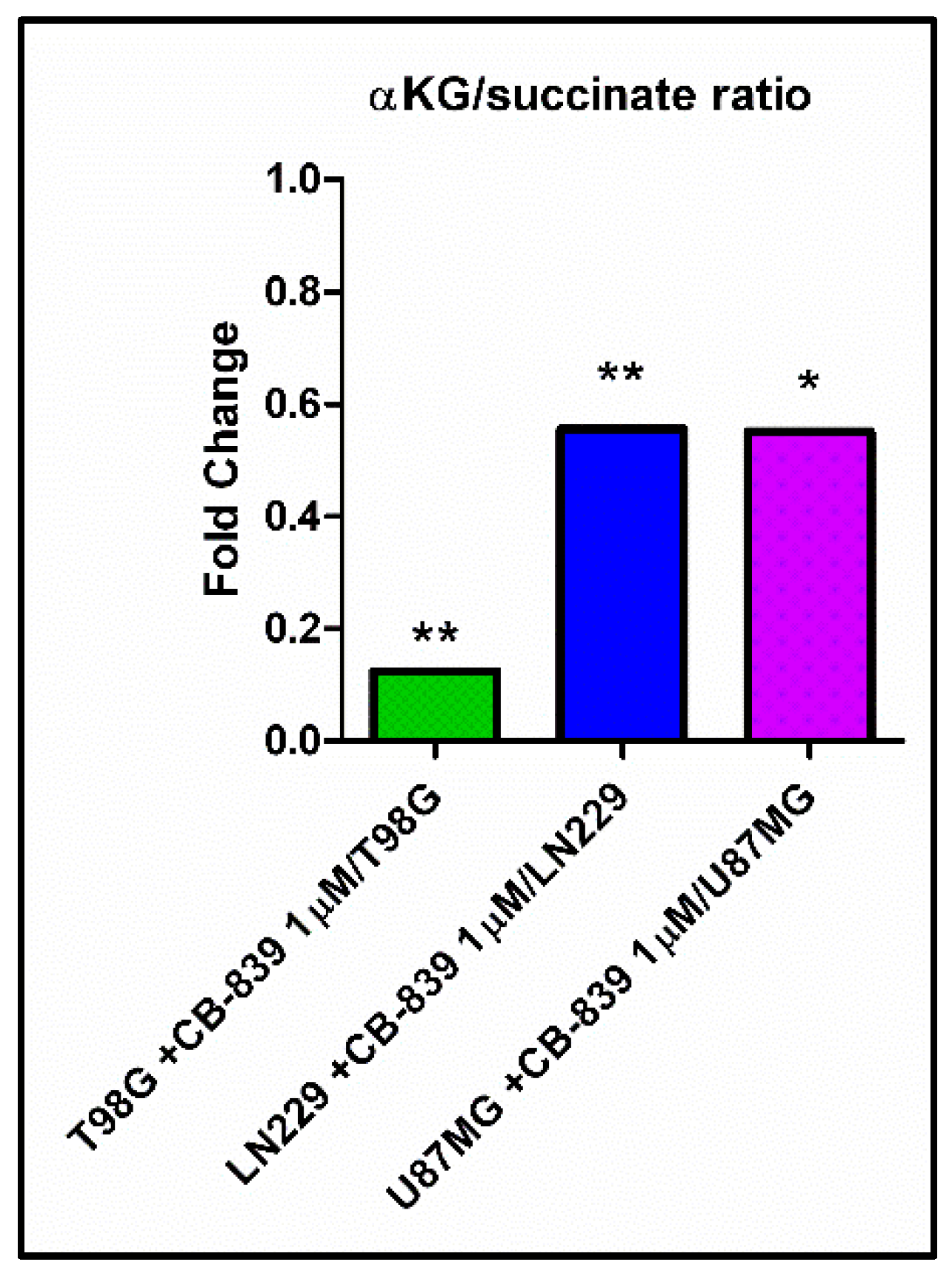
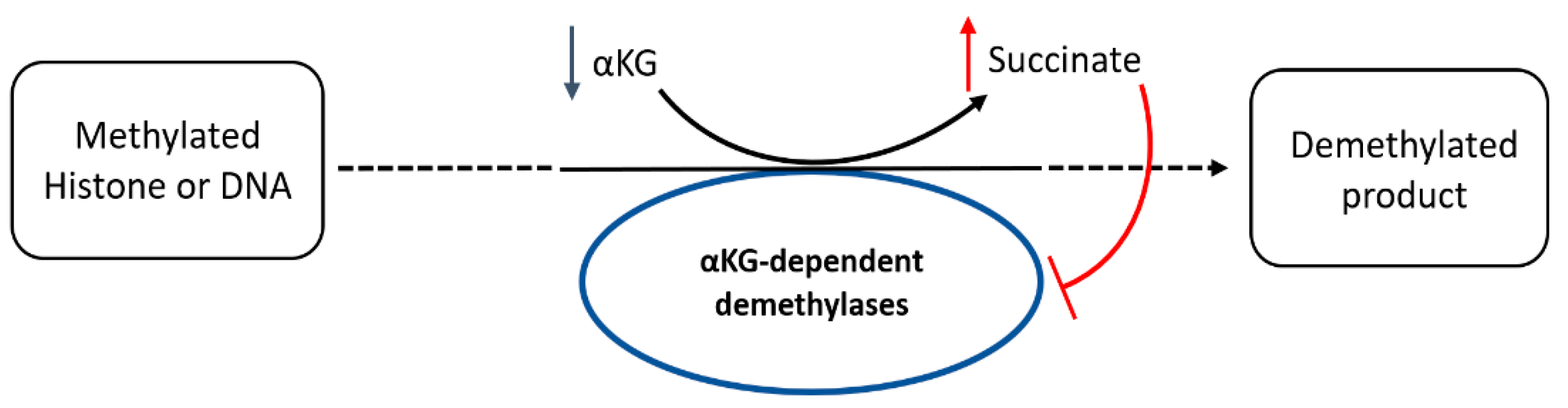
3.7. CB-839 Increases Methylation Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Di Paola, F.J.; Campos-Sandoval, J.A.; Mazurek, S.; Márquez, J. Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin. Cell Dev. Biol. 2020, 98, 34–43. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas (TCGA) Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Campos-Sandoval, J.A.; Gómez-García, M.C.; Santos-Jiménez, J.L.; Matés, J.M.; Alonso, F.J.; Márquez, J. Antioxidant responses related to temozolomide resistance in glioblastoma. Neurochem. Int. 2021, 149, 105136. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Waitkus, M.S.; Diplas, B.H.; Yan, H. Isocitrate dehydrogenase mutations in gliomas. Neuro. Oncol. 2016, 18, 16–26. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Szeliga, M.; Obara-Michlewska, M.; Matyja, E.; Lazarczyk, M.; Lobo, C.; Hilgier, W.; Alonso, F.J.; Márquez, J.; Albrecht, J. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia 2009, 57, 1014. [Google Scholar] [CrossRef]
- de los Santos-Jiménez, J.; Campos-Sandoval, J.A.; Márquez-Torres, C.; Urbano-Polo, N.; Brøndegaard, D.; Martín-Rufián, M.; Lobo, C.; Peñalver, A.; Gómez-García, M.C.; Martín-Campos, J.; et al. Glutaminase isoforms expression switches microRNA levels and oxidative status in glioblastoma cells. J. Biomed. Sci. 2021, 28, 14. [Google Scholar] [CrossRef] [PubMed]
- Márquez, J.; Alonso, F.J.; Matés, J.M.; Segura, J.A.; Martín-Rufián, M.; Campos-Sandoval, J.A. Glutamine addiction in gliomas. Neurochem. Res. 2017, 42, 1735. [Google Scholar] [CrossRef] [PubMed]
- Majewska, E.; Márquez, J.; Albrecht, J.; Szeliga, M. Transfection with GLS2 Glutaminase (GAB) Sensitizes Human Glioblastoma Cell Lines to Oxidative Stress by a Common Mechanism Involving Suppression of the PI3K/AKT Pathway. Cancers 2019, 11, E115. [Google Scholar] [CrossRef] [PubMed]
- López de la Oliva, A.R.; Campos-Sandoval, J.A.; Gómez-García, M.C.; Cardona, C.; Martín-Rufián, M.; Sialana, F.J.; Castilla, L.; Bae, N.; Lobo, C.; Peñalver, A.; et al. Nuclear translocation of glutaminase GLS2 in human cancer cells associates with proliferation arrest and differentiation. Sci. Rep. 2020, 10, 2259. [Google Scholar] [CrossRef] [PubMed]
- Márquez, J.; Matés, J.M.; Campos-Sandoval, J.A. Glutaminases. In The Glutamate/GABA-Glutamine Cycle, Advances in Neurobiology 13, 1st ed.; Schousboe, A., Sonnewald, U., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 133–171. [Google Scholar] [CrossRef]
- Esemen, Y.; Awan, M.; Parwez, R.; Baig, A.; Rahman, S.; Masala, I.; Franchini, S.; Giakoumettis, D. Molecular Pathogenesis of Glioblastoma in Adults and Future Perspectives: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 2607. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Sudderth, J.; Yang, C.; Mullen, A.R.; Jin, E.S.; Matés, J.M.; DeBerardinis, R.J. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8674–8679. [Google Scholar] [CrossRef]
- Restall, I.J.; Cseh, O.; Richards, L.M.; Pugh, T.J.; Luchman, H.A.; Weiss, S. Brain Tumor Stem Cell Dependence on Glutaminase Reveals a Metabolic Vulnerability through the Amino Acid Deprivation Response Pathway. Cancer Res. 2020, 80, 5478–5490. [Google Scholar] [CrossRef]
- Huang, Q.; Lian, C.; Dong, Y.; Zeng, H.; Liu, B.; Xu, N.; He, Z.; Guo, H. SNAP25 Inhibits Glioma Progression by Regulating Synapse Plasticity via GLS-Mediated Glutaminolysis. Front. Oncol. 2021, 11, 698835. [Google Scholar] [CrossRef]
- Obara-Michlewska, M.; Szeliga, M. Targeting Glutamine Addiction in Gliomas. Cancers 2020, 12, 310. [Google Scholar] [CrossRef]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.C.; He, X.; Kamp, M.A.; Sabel, M.; Barker, R.A.; Steiger, H.J.; et al. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef]
- Paes de Araújo, R.; Bertoni, N.; Seneda, A.L.; Felix, T.F.; Carvalho, M.; Lewis, K.E.; Hasimoto, É.N.; Beckmann, M.; Drigo, S.A.; Reis, P.P.; et al. Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma. Metabolites 2019, 9, 47. [Google Scholar] [CrossRef]
- Karvelsson, S.T.; Sigurdsson, A.; Seip, K.; Grinde, M.T.; Wang, Q.; Johannsson, F.; Mælandsmo, G.M.; Moestue, S.A.; Rolfsson, O.; Halldorsson, S. EMT-Derived Alterations in Glutamine Metabolism Sensitize Mesenchymal Breast Cells to mTOR Inhibition. Mol. Cancer Res. 2021, 19, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Poonaki, E.; Nickel, A.C.; Shafiee Ardestani, M.; Rademacher, L.; Kaul, M.; Apartsin, E.; Meuth, S.G.; Gorji, A.; Janiak, C.; Kahlert, U.D. CD133-Functionalized Gold Nanoparticles as a Carrier Platform for Telaglenastat (CB-839) against Tumor Stem Cells. Int. J. Mol. Sci. 2022, 23, 5479. [Google Scholar] [CrossRef]
- Khadka, S.; Arthur, K.; Barekatain, Y.; Behr, E.; Washington, M.; Ackroyd, J.; Crowley, K.; Suriyamongkol, P.; Lin, Y.H.; Pham, C.D.; et al. Impaired anaplerosis is a major contributor to glycolysis inhibitor toxicity in glioma. Cancer Metab. 2021, 9, 27. [Google Scholar] [CrossRef]
- Shah, R.; Chen, S. Metabolic Signaling Cascades Prompted by Glutaminolysis in Cancer. Cancers 2020, 12, 2624. [Google Scholar] [CrossRef] [PubMed]
- Frégeau-Proulx, L.; Lacouture, A.; Berthiaume, L.; Weidmann, C.; Harvey, M.; Gonthier, K.; Pelletier, J.F.; Neveu, B.; Jobin, C.; Bastien, D.; et al. Multiple metabolic pathways fuel the truncated tricarboxylic acid cycle of the prostate to sustain constant citrate production and secretion. Mol. Metab. 2022, 62, 101516. [Google Scholar] [CrossRef]
- Fink, M.A.; Paland, H.; Herzog, S.; Grube, M.; Vogelgesang, S.; Weitmann, K.; Bialke, A.; Hoffmann, W.; Rauch, B.H.; Schroeder, H.W.S.; et al. Overexpression of carnitine palmitoyltransferase 1A promotes mitochondrial fusion and differentiation of glioblastoma stem cells. Lab. Investig. 2022, 102, 722–730. [Google Scholar] [CrossRef]
- Xiong, N.; Gao, X.; Zhao, H.; Cai, F.; Zhang, F.C.; Yuan, Y.; Liu, W.; He, F.; Zacharias, L.G.; Lin, H.; et al. Using arterial-venous analysis to characterize cancer metabolic consumption in patients. Nat. Commun. 2020, 11, 3169. [Google Scholar] [CrossRef]
- Madiraju, P.; Pande, S.V.; Prentki, M.; Madiraju, S.R. Mitochondrial acetylcarnitine provides acetyl groups for nuclear histone acetylation. Epigenetics 2009, 4, 399–403. [Google Scholar] [CrossRef]
- Matés, J.M.; Campos-Sandoval, J.A.; de Los Santos-Jiménez, J.; Márquez, J. Glutaminases regulate glutathione and oxidative stress in cancer. Arch. Toxicol. 2020, 94, 2603–2623. [Google Scholar] [CrossRef]
- Libiad, M.; Vitvitsky, V.; Bostelaar, T.; Bak, D.W.; Lee, H.J.; Sakamoto, N.; Fearon, E.; Lyssiotis, C.A.; Weerapana, E.; Banerjee, R. Hydrogen sulfide perturbs mitochondrial bioenergetics and triggers metabolic reprogramming in colon cells. J. Biol. Chem. 2019, 294, 12077–12090. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.G.; Yang, M.; Prag, H.A.; Blanco, G.R.; Nikitopoulou, E.; Segarra-Mondejar, M.; Powell, C.A.; Young, T.; Burger, N.; Miljkovic, J.L.; et al. Disruption of the TCA cycle reveals an ATF4-dependent integration of redox and amino acid metabolism. Elife 2021, 10, e72593. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, N.M.; Baran, N.; Shanmugavelandy, S.S.; Lee, J.; Lujan, J.V.; Dutta, P.; Millward, S.W.; Cai, T.; Wood, C.G.; Piwnica-Worms, D.; et al. Assessing Metabolic Intervention with a Glutaminase Inhibitor in Real-Time by Hyperpolarized Magnetic Resonance in Acute Myeloid Leukemia. Mol. Cancer Ther. 2019, 18, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Servillo, L.; Giovane, A.; Cautela, D.; Castaldo, D.; Balestrieri, M.L. Where does N(ε)-trimethyllysine for the carnitine biosynthesis in mammals come from? PLoS ONE 2014, 9, e84589. [Google Scholar] [CrossRef] [PubMed]
- Maas, M.N.; Hintzen, J.C.J.; Porzberg, M.R.B.; Mecinović, J. Trimethyllysine: From Carnitine Biosynthesis to Epigenetics. Int. J. Mol. Sci. 2020, 21, 9451. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, N.; Spurlin, G.; Korm, S.; Huang, S.; Anderson, N.M.; Huiting, L.N.; Liu, H.; Feng, H. α-Ketoglutarate-Mediated DNA Demethylation Sustains T-Acute Lymphoblastic Leukemia upon TCA cycle Targeting. Cancers 2022, 14, 2983. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, G.W.; Yoo, J.; Lee, S.W.; Jeon, Y.H.; Kim, S.Y.; Kang, H.G.; Kim, D.H.; Chun, K.H.; Choi, J.; et al. Histone demethylase KDM4C controls tumorigenesis of glioblastoma by epigenetically regulating p53 and c-Myc. Cell Death Dis. 2021, 12, 89. [Google Scholar] [CrossRef]
- Martín-Rufián, M.; Nascimento-Gomes, R.; Higuero, A.; Crisma, A.R.; Campos-Sandoval, J.A.; Gómez-García, M.C.; Cardona, C.; Cheng, T.; Lobo, C.; Segura, J.A.; et al. Both GLS silencing and GLS2 overexpression synergize with oxidative stress against proliferation of glioma cells. J. Mol. Med. 2014, 92, 277–290. [Google Scholar] [CrossRef]
- Lieberman, B.P.; Ploessl, K.; Wang, L.; Qu, W.; Zha, Z.; Wise, D.R.; Chodosh, L.A.; Belka, G.; Thompson, C.B.; Kung, H.F. PET imaging of glutaminolysis in tumors by 18F-(2S,4R)4-fluoroglutamine. J. Nucl. Med. 2011, 52, 1947–1955. [Google Scholar] [CrossRef]
- Ohba, S.; Murayama, K.; Abe, M.; Hasegawa, M.; Hirose, Y. Magnetic Resonance Imaging and Proton Magnetic Resonance Spectroscopy for Differentiating Between Enhanced Gliomas and Malignant Lymphomas. World. Neurosurg. 2019, 127, e779–e787. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.N.; DeBerardinis, R.J. Role of glutamine in cancer: Therapeutic and imaging implications. J. Nucl. Med. 2011, 52, 1005–1008. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Campos-Sandoval, J.A.; de los Santos-Jiménez, J.; Segura, J.A.; Alonso, F.J.; Márquez, J. Metabolic Reprogramming of Cancer by Chemicals that Target Glutaminase Isoenzymes. Curr. Med. Chem. 2020, 27, 5317–5339. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De los Santos-Jiménez, J.; Rosales, T.; Ko, B.; Campos-Sandoval, J.A.; Alonso, F.J.; Márquez, J.; DeBerardinis, R.J.; Matés, J.M. Metabolic Adjustments following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines. Cancers 2023, 15, 531. https://doi.org/10.3390/cancers15020531
De los Santos-Jiménez J, Rosales T, Ko B, Campos-Sandoval JA, Alonso FJ, Márquez J, DeBerardinis RJ, Matés JM. Metabolic Adjustments following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines. Cancers. 2023; 15(2):531. https://doi.org/10.3390/cancers15020531
Chicago/Turabian StyleDe los Santos-Jiménez, Juan, Tracy Rosales, Bookyung Ko, José A. Campos-Sandoval, Francisco J. Alonso, Javier Márquez, Ralph J. DeBerardinis, and José M. Matés. 2023. "Metabolic Adjustments following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines" Cancers 15, no. 2: 531. https://doi.org/10.3390/cancers15020531
APA StyleDe los Santos-Jiménez, J., Rosales, T., Ko, B., Campos-Sandoval, J. A., Alonso, F. J., Márquez, J., DeBerardinis, R. J., & Matés, J. M. (2023). Metabolic Adjustments following Glutaminase Inhibition by CB-839 in Glioblastoma Cell Lines. Cancers, 15(2), 531. https://doi.org/10.3390/cancers15020531








