Hallmarks of Cancer Affected by the MIF Cytokine Family
Abstract
Simple Summary
Abstract
1. Introduction
2. General Characteristics of the MIF Family
2.1. MIF Family Members
2.2. Polymorphisms and Isoforms
2.3. Structure and Secretion
2.4. MIF Receptors: CD74
2.5. MIF Receptors: Chemokine Receptors
2.6. Intracellular MIF-Interacting Proteins
3. MIF and DDT Expressions in Cancer
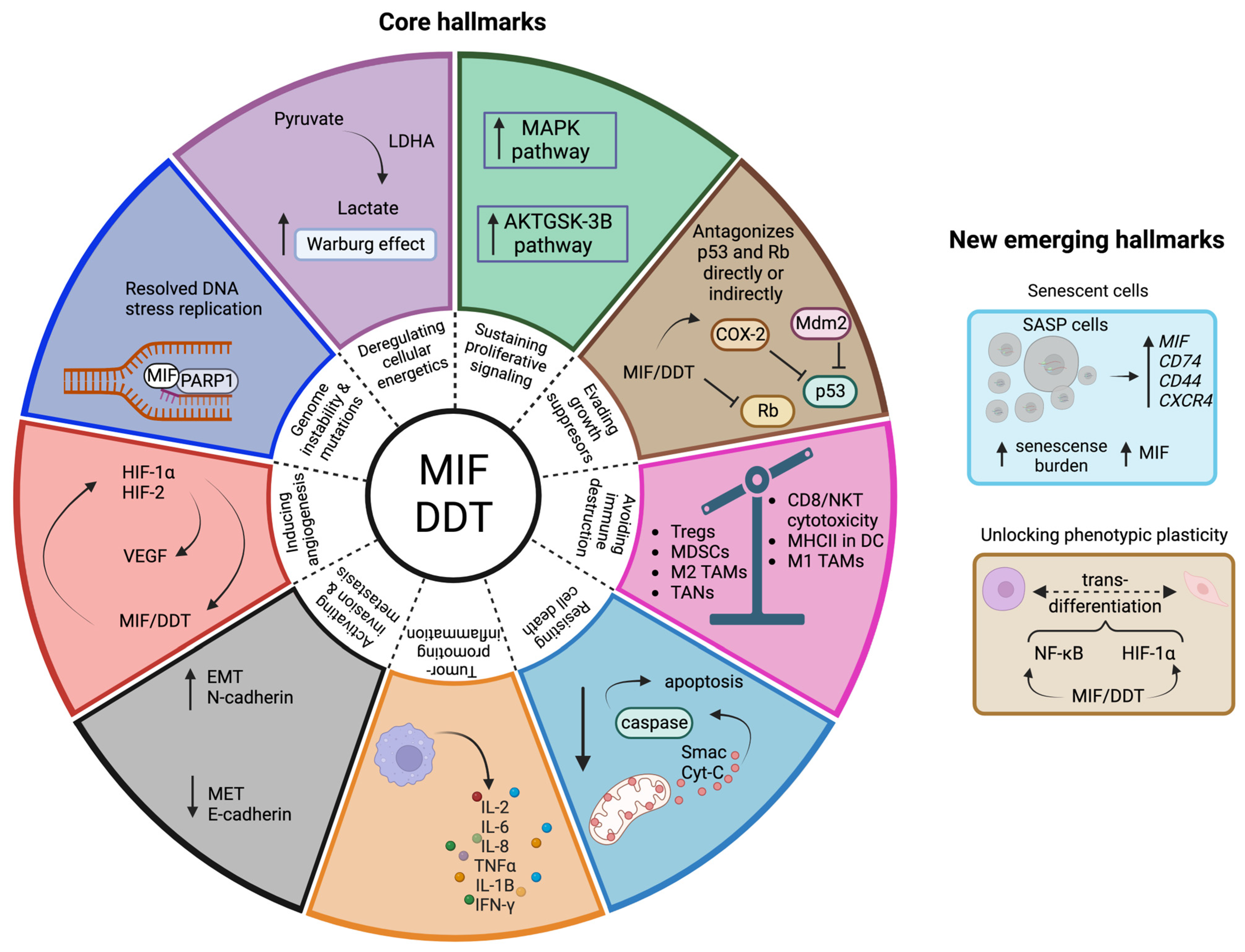
4. Hallmarks of Cancer Linked to the MIF Family
4.1. Evading Growth Suppressors
4.2. Resisting Cell Death
4.3. Genome Instability and Mutation
4.4. Inducing Angiogenesis
4.5. Activating Invasion and Metastasis
4.6. Deregulating Cellular Energetics
4.7. Sustaining Proliferative Signaling
4.8. Tumor-Promoting Inflammation
4.9. Avoiding Immune Destruction
4.10. Unlocking Phenotypic Plasticity
4.11. Senescent Cells
5. MIF and DDT as Novel Targets in Cancer Therapy
6. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front. Pharmacol. 2018, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Fu, X.; Li, Q.; Niu, T. Safety and Efficacy of Anti-PD-1 Monoclonal Antibodies in Patients With Relapsed or Refractory Lymphoma: A Meta-Analysis of Prospective Clinic Trails. Front. Pharmacol. 2019, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health (NIH). Pembrolizumab. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/pembrolizumab (accessed on 7 November 2022).
- National Institutes of Health (NIH). Nivolumab. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/nivolumab (accessed on 7 November 2022).
- National Institutes of Health (NIH). Ipilimumab. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/ipilimumab (accessed on 7 November 2022).
- Moon, W.S.; Rhyu, K.H.; Kang, M.J.; Lee, D.G.; Yu, H.C.; Yeum, J.H.; Koh, G.Y.; Tarnawski, A.S. Overexpression of VEGF and angiopoietin 2: A key to high vascularity of hepatocellular carcinoma? Mod. Pathol. 2003, 16, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.; Riedemann, L.; Amoozgar, Z.; Seano, G.; Susek, K.; Yu, V.; Dalvie, N.; Amelung, R.L.; Datta, M.; Song, J.W.; et al. Ang-2/VEGF bispecific antibody reprograms macrophages and resident microglia to anti-tumor phenotype and prolongs glioblastoma survival. Proc. Natl. Acad. Sci. USA 2016, 113, 4476–4481. [Google Scholar] [CrossRef]
- David, J.R. Delayed hypersensitivity in vitro: Its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc. Natl. Acad. Sci. USA 1966, 56, 72–77. [Google Scholar] [CrossRef]
- Sugimoto, H.; Taniguchi, M.; Nakagawa, A.; Tanaka, I.; Suzuki, M.; Nishihira, J. Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution. Biochemistry 1999, 38, 3268–3279. [Google Scholar] [CrossRef]
- Lee, S.H.; Kwon, H.J.; Park, S.; Kim, C.I.; Ryu, H.; Kim, S.S.; Park, J.B.; Kwon, J.T. Macrophage migration inhibitory factor (MIF) inhibitor 4-IPP downregulates stemness phenotype and mesenchymal trans-differentiation after irradiation in glioblastoma multiforme. PLoS ONE 2021, 16, e0257375. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, Y.; Zhang, J. Combined Knockdown of D-dopachrome Tautomerase and Migration Inhibitory Factor Inhibits the Proliferation, Migration, and Invasion in Human Cervical Cancer. Int. J. Gynecol. Cancer 2017, 27, 634–642. [Google Scholar] [CrossRef]
- Xu, X.; Wang, B.; Ye, C.; Yao, C.; Lin, Y.; Huang, X.; Zhang, Y.; Wang, S. Overexpression of macrophage migration inhibitory factor induces angiogenesis in human breast cancer. Cancer Lett. 2008, 261, 147–157. [Google Scholar] [CrossRef]
- Michelet, C.; Danchin, E.G.J.; Jaouannet, M.; Bernhagen, J.; Panstruga, R.; Kogel, K.H.; Keller, H.; Coustau, C. Cross-Kingdom Analysis of Diversity, Evolutionary History, and Site Selection within the Eukaryotic Macrophage Migration Inhibitory Factor Superfamily. Genes 2019, 10, 740. [Google Scholar] [CrossRef]
- Gunther, S.; Fagone, P.; Jalce, G.; Atanasov, A.G.; Guignabert, C.; Nicoletti, F. Role of MIF and D-DT in immune-inflammatory, autoimmune, and chronic respiratory diseases: From pathogenic factors to therapeutic targets. Drug Discov. Today 2019, 24, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Feng, Z.; Tao, S.; Gao, J.; Lin, Y.; Wei, X.; Zheng, B.; Huang, B.; Zheng, Z.; Zhang, X.; et al. Destabilization of macrophage migration inhibitory factor by 4-IPP reduces NF-kappaB/P-TEFb complex-mediated c-Myb transcription to suppress osteosarcoma tumourigenesis. Clin. Transl. Med. 2022, 12, e652. [Google Scholar] [CrossRef] [PubMed]
- Illescas, O.; Pacheco Fernandez, T.; Laclette, J.P.; Rodriguez, T.; Rodriguez-Sosa, M. Immune modulation by the macrophage migration inhibitory factor (MIF) family: D-dopachrome tautomerase (DDT) is not (always) a backup system. Cytokine 2020, 133, 155121. [Google Scholar] [CrossRef]
- Merk, M.; Mitchell, R.A.; Endres, S.; Bucala, R. D-dopachrome tautomerase (D-DT or MIF-2): Doubling the MIF cytokine family. Cytokine 2012, 59, 10–17. [Google Scholar] [CrossRef]
- Tong, X.; He, J.; Liu, S.; Peng, S.; Yan, Z.; Zhang, Y.; Hong, F. Macrophage migration inhibitory factor -173G/C gene polymorphism increases the risk of renal disease: A meta-analysis. Nephrology 2015, 20, 68–76. [Google Scholar] [CrossRef]
- Zhang, X.; Weng, W.; Xu, W.; Wang, Y.; Yu, W.; Tang, X.; Ma, L.; Pan, Q.; Wang, J.; Sun, F. The association between the migration inhibitory factor -173G/C polymorphism and cancer risk: A meta-analysis. Onco Targets Ther. 2015, 8, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Avalos-Navarro, G.; Del Toro Arreola, A.; Daneri-Navarro, A.; Quintero-Ramos, A.; Bautista-Herrera, L.A.; Franco Topete, R.A.; Anaya Macias, B.U.; Javalera Castro, D.I.; Moran-Mendoza, A.J.; Oceguera-Villanueva, A.; et al. Association of the genetic variants (-794 CATT5-8 and -173 G > C) of macrophage migration inhibitory factor (MIF) with higher soluble levels of MIF and TNFalpha in women with breast cancer. J. Clin. Lab. Anal. 2020, 34, e23209. [Google Scholar] [CrossRef]
- Schinagl, A.; Kerschbaumer, R.J.; Sabarth, N.; Douillard, P.; Scholz, P.; Voelkel, D.; Hollerweger, J.C.; Goettig, P.; Brandstetter, H.; Scheiflinger, F.; et al. Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch. Biochemistry 2018, 57, 1523–1532. [Google Scholar] [CrossRef]
- Thiele, M.; Kerschbaumer, R.J.; Tam, F.W.; Volkel, D.; Douillard, P.; Schinagl, A.; Kuhnel, H.; Smith, J.; McDaid, J.P.; Bhangal, G.; et al. Selective Targeting of a Disease-Related Conformational Isoform of Macrophage Migration Inhibitory Factor Ameliorates Inflammatory Conditions. J. Immunol. 2015, 195, 2343–2352. [Google Scholar] [CrossRef]
- Dickerhof, N.; Schindler, L.; Bernhagen, J.; Kettle, A.J.; Hampton, M.B. Macrophage migration inhibitory factor (MIF) is rendered enzymatically inactive by myeloperoxidase-derived oxidants but retains its immunomodulatory function. Free Radic. Biol. Med. 2015, 89, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Skeens, E.; Gadzuk-Shea, M.; Shah, D.; Bhandari, V.; Schweppe, D.K. Redox-Dependent structure and dynamics of macrophage migration inhibitory factor reveal sites of latent allostery. Structure 2022, 30, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Schinagl, A.; Thiele, M.; Douillard, P.; Völkel, D.; Kenner, L.; Kazemi, Z.; Freissmuth, M.; Scheiflinger, F.; Kerschbaumer, R.J. Oxidized macrophage migration inhibitory factor is a potential new tissue marker and drug target in cancer. Oncotarget 2016, 7, 73486–73496. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, D.; Patel, M.R.; Sachdev, J.C.; Hart, L.L.; Halama, N.; Ramanathan, R.K.; Sarantopoulos, J.; Volkel, D.; Youssef, A.; de Jong, F.A.; et al. Phase I study of imalumab (BAX69), a fully human recombinant antioxidized macrophage migration inhibitory factor antibody in advanced solid tumours. Br. J. Clin. Pharmacol. 2020, 86, 1836–1848. [Google Scholar] [CrossRef]
- Calandra, T.; Roger, T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef]
- Song, S.; Xiao, Z.; Dekker, F.J.; Poelarends, G.J.; Melgert, B.N. Macrophage migration inhibitory factor family proteins are multitasking cytokines in tissue injury. Cell. Mol. Life Sci. 2022, 79, 105. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Wang, C.; Yang, M.; Wang, Y.; Bao, L.; Wang, J.E.; Kim, B.; Chan, K.Y.; Xu, W.; et al. MIF is a 3’ flap nuclease that facilitates DNA replication and promotes tumor growth. Nat. Commun. 2021, 12, 2954. [Google Scholar] [CrossRef]
- Jankauskas, S.S.; Wong, D.W.L.; Bucala, R.; Djudjaj, S.; Boor, P. Evolving complexity of MIF signaling. Cell Signal. 2019, 57, 76–88. [Google Scholar] [CrossRef]
- Roger, T.; David, J.; Glauser, M.P.; Calandra, T. MIF regulates innate immune responses through modulation of TLR-4. Nature 2001, 414, 920–924. [Google Scholar] [CrossRef]
- Merk, M.; Baugh, J.; Zierow, S.; Leng, L.; Pal, U.; Lee, S.J.; Ebert, A.D.; Mizue, Y.; Trent, J.O.; Mitchell, R.; et al. The Golgi-associated protein p115 mediates the secretion of macrophage migration inhibitory factor. J. Immunol. 2009, 182, 6896–6906. [Google Scholar] [CrossRef]
- Flieger, O.; Engling, A.; Bucala, R.; Lue, H.; Nickel, W.; Bernhagen, J. Regulated secretion of macrophage migration inhibitory factor is mediated by a non-classical pathway involving an ABC transporter. FEBS Lett. 2003, 551, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Lee, J.P.W.; Elgass, K.; Pinar, A.A.; Tate, M.D.; Aitken, E.H.; Fan, H.; Creed, S.J.; Deen, N.S.; Traore, D.A.K.; et al. Macrophage migration inhibitory factor is required for NLRP3 inflammasome activation. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.S.; Kang, Y.; Wahl, E.R.; Park, H.J.; Lazova, R.; Leng, L.; Mamula, M.; Krishnaswamy, S.; Bucala, R.; Kang, I. Macrophage migration inhibitory factor regulates U1-snRNP immune complex mediated activation of the NLRP3 inflammasome. Arthritis Rheumatol. 2019, 71, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Dankers, W.; Hasnat, M.A.; Swann, V.; Alharbi, A.; Lee, J.P.; Cristofaro, M.A.; Gantier, M.P.; Jones, S.A.; Morand, E.F.; Flynn, J.K.; et al. Necrotic cell death increases the release of macrophage migration inhibitory factor by monocytes/macrophages. Immunol. Cell Biol. 2020, 98, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, X.; Zhu, W.; Zhang, Y.; Hong, Y.; Liang, X.; Fan, B.; Zhao, H.; He, H.; Zhang, F. Exosomes from mesenchymal stem cells overexpressing MIF enhance myocardial repair. J. Cell. Physiol. 2020, 235, 8010–8022. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zuo, F.; Zhang, K.; Xia, T.; Lei, W.; Zhang, Z.; Bao, L.; You, Y. Exosomal MIF Derived From Nasopharyngeal Carcinoma Promotes Metastasis by Repressing Ferroptosis of Macrophages. Front. Cell Dev. Biol. 2021, 9, 791187. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Merk, M.; Zierow, S.; Leng, L.; Das, R.; Du, X.; Schulte, W.; Fan, J.; Lue, H.; Chen, Y.; Xiong, H.; et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF). Proc. Natl. Acad. Sci. USA 2011, 108, E577–E585. [Google Scholar] [CrossRef]
- Xu, F.; Li, M.X.; Chen, J. D-dopachrome tautomerase from Japanese sea bass (Lateolabrax japonicus) is a chemokine-like cytokine and functional homolog of macrophage migration inhibitory factor. Zool. Res. 2020, 41, 39–50. [Google Scholar]
- Lindner, R. Invariant Chain Complexes and Clusters as Platforms for MIF Signaling. Cells 2017, 6, 6. [Google Scholar] [CrossRef]
- Wu, G.; Sun, Y.; Wang, K.; Chen, Z.; Wang, X.; Chang, F.; Li, T.; Feng, P.; Xia, Z. Relationship between elevated soluble CD74 and severity of experimental and clinical ALI/ARDS. Sci. Rep. 2016, 6, 30067. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Bustos, M.A.; Cho, S.N.; Roszik, J.; Ryu, S.; Lopez, V.M.; Burks, J.K.; Lee, J.E.; Grimm, E.A.; Hoon, D.S.B.; et al. Interplay between soluble CD74 and macrophage-migration inhibitory factor drives tumor growth and influences patient survival in melanoma. Cell Death Dis. 2022, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Metz, C.N.; Fang, Y.; Xu, J.; Donnelly, S.; Baugh, J.; Delohery, T.; Chen, Y.B.; Mitchell, R.A.; Bucala, R. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 2003, 197, 1467–1476. [Google Scholar] [CrossRef]
- Shi, X.R.; Leng, L.; Wang, T.; Wang, W.K.; Du, X.; Li, J.; McDonald, C.; Chen, Z.; Murphy, J.W.; Lolis, E.; et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity 2006, 25, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Jaffer, T.; Ma, D. The emerging role of chemokine receptor CXCR2 in cancer progression. Transl. Cancer Res. 2016, 5, S616–S628. [Google Scholar] [CrossRef]
- Tarnowski, M.; Grymula, K.; Liu, R.; Tarnowska, J.; Drukala, J.; Ratajczak, J.; Mitchell, R.A.; Ratajczak, M.Z.; Kucia, M. Macrophage migration inhibitory factor is secreted by rhabdomyosarcoma cells, modulates tumor metastasis by binding to CXCR4 and CXCR7 receptors and inhibits recruitment of cancer-associated fibroblasts. Mol. Cancer Res. 2010, 8, 1328–1343. [Google Scholar] [CrossRef]
- Kontos, C.; El Bounkari, O.; Krammer, C.; Sinitski, D.; Hille, K.; Zan, C.; Yan, G.; Wang, S.; Gao, Y.; Brandhofer, M.; et al. Designed CXCR4 mimic acts as a soluble chemokine receptor that blocks atherogenic inflammation by agonist-specific targeting. Nat. Commun. 2020, 11, 5981. [Google Scholar] [CrossRef]
- Rajasekaran, D.; Groning, S.; Schmitz, C.; Zierow, S.; Drucker, N.; Bakou, M.; Kohl, K.; Mertens, A.; Lue, H.; Weber, C.; et al. Macrophage Migration Inhibitory Factor-CXCR4 Receptor Interactions: Evidence for partial allosteric agonism in comparison with CXCL12 chemokine. J. Biol. Chem. 2016, 291, 15881–15895. [Google Scholar] [CrossRef]
- Lacy, M.; Kontos, C.; Brandhofer, M.; Hille, K.; Groning, S.; Sinitski, D.; Bourilhon, P.; Rosenberg, E.; Krammer, C.; Thavayogarajah, T.; et al. Identification of an Arg-Leu-Arg tripeptide that contributes to the binding interface between the cytokine MIF and the chemokine receptor CXCR4. Sci. Rep. 2018, 8, 5171. [Google Scholar] [CrossRef]
- Tilstam, P.V.; Schulte, W.; Holowka, T.; Kim, B.S.; Nouws, J.; Sauler, M.; Piecychna, M.; Pantouris, G.; Lolis, E.; Leng, L.; et al. MIF but not MIF-2 recruits inflammatory macrophages in an experimental polymicrobial sepsis model. J. Clin. Investig. 2021, 131, 127171. [Google Scholar] [CrossRef]
- Song, S.; Liu, B.; Habibie, H.; van den Bor, J.; Smit, M.J.; Gosens, R.; Wu, X.; Brandsma, C.A.; Cool, R.H.; Haisma, H.J.; et al. D-dopachrome tautomerase contributes to lung epithelial repair via atypical chemokine receptor 3-dependent Akt signaling. EBioMedicine 2021, 68, 103412. [Google Scholar] [CrossRef] [PubMed]
- Kleemann, R.; Hausser, A.; Geiger, G.; Mischke, R.; Burger-Kentischer, A.; Flieger, O.; Johannes, F.J.; Roger, T.; Calandra, T.; Kapurniotu, A.; et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature 2000, 408, 211–216. [Google Scholar] [CrossRef]
- Burger-Kentischer, A.; Finkelmeier, D.; Thiele, M.; Schmucker, J.; Geiger, G.; Tovar, G.E.; Bernhagen, J. Binding of JAB1/CSN5 to MIF is mediated by the MPN domain but is independent of the JAMM motif. FEBS Lett. 2005, 579, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.M.; Rendon, B.E.; Zhao, M.; Qian, M.W.; Bucala, R.; Xin, D.; Mitchell, R.A. Cooperative regulation of non-small cell lung carcinoma angiogenic potential by macrophage migration inhibitory factor and its homolog, D-dopachrome tautomerase. J. Immunol. 2008, 181, 2330–2337. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hou, J.; Luo, Y.; Wang, D. Functional disruption of macrophage migration inhibitory factor (MIF) suppresses proliferation of human H460 lung cancer cells by caspase-dependent apoptosis. Cancer Cell Int. 2013, 13, 28. [Google Scholar] [CrossRef]
- Zhang, H.; Duan, J.; Wu, O. The expression of macrophage migration inhibitory factor in the non-small cell lung cancer. Saudi J. Biol. Sci. 2020, 27, 1527–1532. [Google Scholar] [CrossRef]
- Xiao, Z.; Osipyan, A.; Song, S.; Chen, D.; Schut, R.A.; van Merkerk, R.; van der Wouden, P.E.; Cool, R.H.; Quax, W.J.; Melgert, B.N.; et al. Thieno[2,3-d]pyrimidine-2,4(1H,3H)-dione Derivative Inhibits d-Dopachrome Tautomerase Activity and Suppresses the Proliferation of Non-Small Cell Lung Cancer Cells. J. Med. Chem. 2022, 65, 2059–2077. [Google Scholar] [CrossRef]
- Charan, M.; Das, S.; Mishra, S.; Chatterjee, N.; Varikuti, S.; Kaul, K.; Misri, S.; Ahirwar, D.K.; Satoskar, A.R.; Ganju, R.K. Macrophage migration inhibitory factor inhibition as a novel therapeutic approach against triple-negative breast cancer. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Balogh, K.N.; Templeton, D.J.; Cross, J.V. Macrophage Migration Inhibitory Factor protects cancer cells from immunogenic cell death and impairs anti-tumor immune responses. PLoS ONE 2018, 13, e0197702. [Google Scholar] [CrossRef]
- Meyer-Siegler, K.L.; Iczkowski, K.A.; Vera, P.L. Further evidence for increased macrophage migration inhibitory factor expression in prostate cancer. BMC Cancer 2005, 5, 73. [Google Scholar] [CrossRef]
- Meyer-Siegler, K.L.; Iczkowski, K.A.; Leng, L.; Bucala, R.; Vera, P.L. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 2006, 177, 8730–8739. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Siegler, K.L.; Bellino, M.A.; Tannenbaum, M. Macrophage migration inhibitory factor evaluation compared with prostate specific antigen as a biomarker in patients with prostate carcinoma. Cancer 2002, 94, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.J.; Deng, W.G.; Niu, C.B.; Li, Y.Y.; Fu, Y. Expression of macrophage migration inhibitory factor and CD74 in cervical squamous cell carcinoma. Int. J. Gynecol. Cancer 2011, 21, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Hira, E.; Ono, T.; Dhar, D.K.; El-Assal, O.N.; Hishikawa, Y.; Yamanoi, A.; Nagasue, N. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer 2005, 103, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Teng, H.; Poon, R.T.; Ng, I.O.; Li, Z.; Chen, Y.; Jiang, G.; Lau, C.; Yu, W.C.; Bacher, M.; et al. Macrophage migration inhibitory factor: Roles in regulating tumor cell migration and expression of angiogenic factors in hepatocellular carcinoma. Int. J. Cancer 2003, 20, 22–29. [Google Scholar] [CrossRef]
- Qin, L.; Qin, J.; Lv, X.; Yin, C.; Zhang, Q.; Zhang, J. MIF promoter polymorphism increases peripheral blood expression levels, contributing to increased susceptibility and poor prognosis in hepatocellular carcinoma. Oncol Lett. 2021, 22, 549. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.H.; Yang, Y.; Chu, K.M.; Gu, Q.; Zhang, Y.Y.; He, H.; Wong, W.M.; Leung, S.Y.; Yuen, S.T.; Yuen, M.F.; et al. Serum macrophage migration-inhibitory factor as a diagnostic and prognostic biomarker for gastric cancer. Cancer 2009, 115, 5441–5449. [Google Scholar] [CrossRef]
- He, L.J.; Xie, D.; Hu, P.J.; Liao, Y.J.; Deng, H.X.; Kung, H.F.; Zhu, S.L. Macrophage migration inhibitory factor as a potential prognostic factor in gastric cancer. World J. Gastroenterol. 2015, 21, 9916–9926. [Google Scholar] [CrossRef]
- He, X.; Yang, J.; Ding, Y.; Liu, W.; Shen, Q.; Xia, H.H. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: Potential role of MIF in gastric carcinogenesis. Gut 2006, 55, 797–802. [Google Scholar] [CrossRef]
- Xia, H.H.-X.; Zhang, S.T.; Lam, S.K.; Lin, M.C.-M.; Kung, H.F.; Wong, B.C.-Y. Expression of macrophage migration inhibitory factor in esophageal squamous cell carcinoma and effects of bile acids and NSAIDs. Carcinogenesis 2005, 26, 11–15. [Google Scholar] [CrossRef]
- Alban, T.J.; Bayik, D.; Otvos, B.; Rabljenovic, A.; Leng, L.; Jia-Shiun, L.; Roversi, G.; Lauko, A.; Momin, A.A.; Mohammadi, A.M.; et al. Glioblastoma Myeloid-Derived Suppressor Cell Subsets Express Differential Macrophage Migration Inhibitory Factor Receptor Profiles That Can Be Targeted to Reduce Immune Suppression. Front. Immunol. 2020, 11, 1191. [Google Scholar] [CrossRef] [PubMed]
- Ghoochani, A.; Schwarz, M.A.; Yakubov, E.; Engelhorn, T.; Doerfler, A.; Buchfelder, M.; Bucala, R.; Savaskan, N.E.; Eyupoglu, I.Y. MIF-CD74 signaling impedes microglial M1 polarization and facilitates brain tumorigenesis. Oncogene 2016, 35, 6246–6261. [Google Scholar] [CrossRef] [PubMed]
- Otvos, B.; Silver, D.J.; Mulkearns-Hubert, E.E.; Alvarado, A.G.; Turaga, S.M.; Sorensen, M.D.; Rayman, P.; Flavahan, W.A.; Hale, J.S.; Stoltz, K.; et al. Cancer Stem Cell-Secreted Macrophage Migration Inhibitory Factor Stimulates Myeloid Derived Suppressor Cell Function and Facilitates Glioblastoma Immune Evasion. Stem Cells 2016, 34, 2026–2039. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, V.; Du, W.; Gupta, Y.; Yeh, I.J.; Montano, M.; Magi-Galuzzi, C.; Welford, S.M. Dysregulated D-dopachrome tautomerase, a hypoxia-inducible factor-dependent gene, cooperates with macrophage migration inhibitory factor in renal tumorigenesis. J. Biol. Chem. 2014, 289, 3713–3723. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Guo, J.; Yao, J.; Jiang, K.; Hu, J.; Wang, B.; Liu, H.; Lin, L.; Sun, W.; Jiang, X. D-dopachrome tautomerase is over-expressed in pancreatic ductal adenocarcinoma and acts cooperatively with macrophage migration inhibitory factor to promote cancer growth. Int. J. Cancer 2016, 139, 2056–2067. [Google Scholar] [CrossRef]
- Cavalli, E.; Mazzon, E.; Mammana, S.; Basile, M.S.; Lombardo, S.D.; Mangano, K.; Bramanti, P.; Nicoletti, F.; Fagone, P.; Petralia, M.C. Overexpression of Macrophage Migration Inhibitory Factor and Its Homologue D-Dopachrome Tautomerase as Negative Prognostic Factor in Neuroblastoma. Brain Sci. 2019, 9, 284. [Google Scholar] [CrossRef]
- Kobold, S.; Merk, M.; Hofer, L.; Peters, P.; Bucala, R.; Endres, S. The macrophage migration inhibitory factor (MIF)-homologue D-dopachrome tautomerase is a therapeutic target in a murine melanoma model.pdf. Oncotarget 2013, 5, 103–107. [Google Scholar] [CrossRef]
- Oliveira, C.S.; E De Bock, C.; Molloy, T.J.; Sadeqzadeh, E.; Geng, X.Y.; Hersey, P.; Zhang, X.D.; Thorne, R.F. Macrophage migration inhibitory factor engages PI3K/Akt signalling and is a prognostic factor in metastatic melanoma. BMC Cancer 2014, 14, 630. [Google Scholar] [CrossRef]
- Krockenberger, M.; Kranke, P.; Häusler, S.; Engel, J.B.; Horn, E.; Nürnberger, K.; Wischhusen, J.; Dietl, J.; Hönig, A. Macrophage Migration-inhibitory Factor Levels in Serum of Patients with Ovarian Cancer Correlates with Poor Prognosis. Anticancer Res. 2012, 32, 5233–5238. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Am. Assoc. Cancer Res. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Feroz, W.; Sheikh, A.M.A. Exploring the multiple roles of guardian of the genome: P53. Egypt. J. Med. Hum. Genet. 2020, 21, 49. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Fingerle-Rowson, G.; Petrenko, O.; Metz, C.N.; Forsthuber, T.G.; Mitchell, R.; Huss, R.; Moll, U.; Muller, W.; Bucala, R. The p53-dependent effects of MIF revealed by gene targeting. Proc. Natl. Acad. Sci. USA 2003, 100, 9354–9359. [Google Scholar] [CrossRef]
- Fukaya, R.; Ohta, S.; Yaguchi, T.; Matsuzaki, Y.; Sugihara, E.; Okano, H.; Saya, H.; Kawakami, Y.; Kawase, T.; Yoshida, K.; et al. MIF Maintains the Tumorigenic Capacity of Brain Tumor-Initiating Cells by Directly Inhibiting p53. Cancer Res. 2016, 76, 2813–2823. [Google Scholar] [CrossRef]
- Costa, A.F.; Gomes, S.Z.; Lorenzon-Ojea, A.R.; Martucci, M.; Faria, M.R.; Pinto Ddos, S., Jr.; Oliveira, S.F.; Ietta, F.; Paulesu, L.; Bevilacqua, E. Macrophage migration inhibitory factor induces phosphorylation of Mdm2 mediated by phosphatidylinositol 3-kinase/Akt kinase: Role of this pathway in decidual cell survival. Placenta 2016, 41, 27–38. [Google Scholar] [CrossRef]
- Jung, H.; Seong, H.A.; Ha, H. Critical role of cysteine residue 81 of macrophage migration inhibitory factor (MIF) in MIF-induced inhibition of p53 activity. J. Biol. Chem. 2008, 283, 20383–20396. [Google Scholar] [CrossRef]
- Corcoran, C.A.; He, Q.; Huang, Y.; Sheikh, M.S. Cyclooxygenase-2 interacts with p53 and interferes with p53-dependent transcription and apoptosis. Oncogene 2005, 24, 1634–1640. [Google Scholar] [CrossRef]
- Xin, D.; Rendon, B.E.; Zhao, M.; Winner, M.; McGhee Coleman, A.; Mitchell, R.A. The MIF homologue D-dopachrome tautomerase promotes COX-2 expression through beta-catenin-dependent and -independent mechanisms. Mol. Cancer Res. 2010, 8, 1601–1609. [Google Scholar] [CrossRef]
- Zhang, S.H.; Postigo, A.A.; Dean, D.C. Active Transcriptional Repression by the Rb–E2F Complex Mediates G1 Arrest Triggered by p16INK4a, TGFb, and Contact Inhibition.pdf. Cell 1999, 97, 53–61. [Google Scholar] [CrossRef]
- Sasaki, Y.; Kasuya, K.; Nishihira, J.; Magami, Y.; Tsuchida, A.; Aoki, T.; Koyanagi, Y. Suppression of tumor growth through introduction of an antisense plasmid of macrophage migration inhibitory factor. Int. J Mol. Med. 2002, 10, 579–583. [Google Scholar] [PubMed]
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a Mitochondrial Protein that Promotes Cytochrome c–Dependent Caspase Activation by Eliminating IAP Inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Bolanos, J.P.; Moro, M.A.; Lizasoain, I.; Almeida, A. Mitochondria and reactive oxygen and nitrogen species in neurological disorders and stroke: Therapeutic implications. Adv. Drug Deliv. Rev. 2009, 61, 1299–1315. [Google Scholar] [CrossRef] [PubMed]
- Elena-Real, C.A.; Diaz-Quintana, A.; Gonzalez-Arzola, K.; Velazquez-Campoy, A.; Orzaez, M.; Lopez-Rivas, A.; Gil-Caballero, S.; De la Rosa, M.A.; Diaz-Moreno, I. Cytochrome c speeds up caspase cascade activation by blocking 14-3-3epsilon-dependent Apaf-1 inhibition. Cell Death Dis. 2018, 9, 365. [Google Scholar] [CrossRef] [PubMed]
- Lue, H.; Thiele, M.; Franz, J.; Dahl, E.; Speckgens, S.; Leng, L.; Fingerle-Rowson, G.; Bucala, R.; Luscher, B.; Bernhagen, J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene 2007, 26, 5046–5059. [Google Scholar] [CrossRef]
- De, R.; Sarkar, S.; Mazumder, S.; Debsharma, S.; Siddiqui, A.A.; Saha, S.J.; Banerjee, C.; Nag, S.; Saha, D.; Pramanik, S.; et al. Macrophage migration inhibitory factor regulates mitochondrial dynamics and cell growth of human cancer cell lines through CD74-NF-kappaB signaling. J. Biol. Chem. 2018, 293, 19740–19760. [Google Scholar] [CrossRef]
- Mitchell, R.A.; Liao, H.; Chesney, J.; Fingerle-Rowson, G.; Baugh, J.; David, J.; Bucala, R. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53- Regulatory role in the innate immune response. Proc. Natl. Acad. Sci. USA 2022, 99, 345–350. [Google Scholar] [CrossRef]
- Baumann, R.; Casaulta, C.; Simon, D.; Conus, S.; Yousefi, S.; Simon, H.U. Macrophage migration inhibitory factor delays apoptosis in neutrophils by inhibiting the mitochondria-dependent death pathway. Faseb J. 2003, 17, 2221–2230. [Google Scholar] [CrossRef]
- Schindler, L.; Zwissler, L.; Krammer, C.; Hendgen-Cotta, U.; Rassaf, T.; Hampton, M.B.; Dickerhof, N.; Bernhagen, J. Macrophage migration inhibitory factor inhibits neutrophil apoptosis by inducing cytokine release from mononuclear cells. J. Leukoc. Biol. 2021, 110, 893–905. [Google Scholar] [CrossRef]
- Thiele, M.; Donnelly, S.C.; Mitchell, R.A. OxMIF: A druggable isoform of macrophage migration inhibitory factor in cancer and inflammatory diseases. J. Immunother. Cancer 2022, 10, e005475. [Google Scholar] [CrossRef]
- Hills, S.A.; Diffley, J.F. DNA replication and oncogene-induced replicative stress. Curr. Biol. 2014, 24, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef] [PubMed]
- Weil, M.K.; Chen, A.P. PARP inhibitor treatment in ovarian and breast cancer. Curr. Probl. Cancer 2011, 35, 7–50. [Google Scholar] [CrossRef] [PubMed]
- Huggins, C.F.; Chafin, D.R.; Aoyagi, S.; Henricksen, L.A.; Bambara, R.A.; Hayes, J.J. Flap Endonuclease 1 Efficiently Cleaves Base Excision Repair and DNA Replication Intermediates Assembled into Nucleosomes. Mol. Cell 2002, 10, 1201–1211. [Google Scholar] [CrossRef]
- Wang, D.; Luo, L.; Chen, W.; Chen, L.Z.; Zeng, W.T.; Li, W.; Huang, X.H. Significance of the vascular endothelial growth factor and the macrophage migration inhibitory factor in the progression of hepatocellular carcinoma. Oncol. Rep. 2014, 31, 1199–1204. [Google Scholar] [CrossRef]
- Manaut, C.; Bonvier, J.; Foidart, J.M.; Deprez, M. Macrophage migration inhibitory factor (MIF) expression in human glioblastomas correlates with vascular endothelial growth factor (VEGF) expression. Neuropathol. Appl. Neurobiol. 2022, 28, 452–460. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, G.; Yang, S.; Qiao, J.; Li, T.; Yang, S.; Hong, Y. Macrophage migration inhibitory factor regulating the expression of VEGF-C through MAPK signal pathways in breast cancer MCF-7 cell. World J. Surg. Oncol. 2016, 14, 51. [Google Scholar] [CrossRef]
- Hagemann, T.; Robinson, S.C.; Thompson, R.G.; Charles, K.; Kulbe, H.; Balkwill, F.R. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression, and angiogenesis. Mol. Cancer Ther. 2007, 6, 1993–2002. [Google Scholar] [CrossRef]
- Chesney, J.; Metz, C.; Bacher, M.; Peng, T.; Meinhardt, A.; Bucala, R. An Essential Role for Macrophage Migration Inhibitory Factor (MIF) in Angiogenesis and the Growth of a Murine Lymphoma. Mol. Med. 1999, 5, 181–191. [Google Scholar] [CrossRef]
- Ma, Y.; Su, K.N.; Pfau, D.; Rao, V.S.; Wu, X.; Hu, X.; Leng, L.; Du, X.; Piecychna, M.; Bedi, K.; et al. Cardiomyocyte d-dopachrome tautomerase protects against heart failure. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Büchler, P.; Reber, H.A.; Büchler, M.; Shrinkante, S.; Büchler, M.W.; Friess, H.; Semenza, G.L.; Hines, O.J. Hypoxia-Inducible Factor 1 Regulates Vascular Endothelial Growth Factor Expression in Human Pancreatic Cancer. Pancreas 2003, 26, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Zis, O.; Zhang, S.; Dorovini-Zis, K.; Wang, L.; Song, W. Hypoxia signaling regulates macrophage migration inhibitory factor (MIF) expression in stroke. Mol. Neurobiol. 2015, 51, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Chiles, K.; Feldser, D.; Laughner, E.; Hanrahan, C.; Georgescu, M.M.; Simons, J.W.; Semenza, G.L. Modulation of Hypoxia-inducible Factor 1a Expression by the Epidermal Growth Factor: Phosphatidylinositol 3-Kinase:PTEN:AKT:FRAP Pathway in Human Prostate Cancer Cells- Implications for Tumor Angiogenesis and Therapeutics. Cancer Res. 2000, 60, 1541–1545. [Google Scholar]
- Oda, S.; Oda, T.; Nishi, K.; Takabuchi, S.; Wakamatsu, T.; Tanaka, T.; Adachi, T.; Fukuda, K.; Semenza, G.L.; Hirota, K. Macrophage migration inhibitory factor activates hypoxia-inducible factor in a p53-dependent manner. PLoS ONE 2008, 3, e2215. [Google Scholar] [CrossRef]
- Yang, S.; He, P.; Wang, J.; Schetter, A.; Tang, W.; Funamizu, N.; Yanaga, K.; Uwagawa, T.; Satoskar, A.R.; Gaedcke, J.; et al. A Novel MIF Signaling Pathway Drives the Malignant Character of Pancreatic Cancer by Targeting NR3C2. Cancer Res. 2016, 76, 3838–3850. [Google Scholar] [CrossRef]
- Lai, X.; Li, Q.; Wu, F.; Lin, J.; Chen, J.; Zheng, H.; Guo, L. Epithelial-Mesenchymal Transition and Metabolic Switching in Cancer: Lessons From Somatic Cell Reprogramming. Front. Cell Dev. Biol. 2020, 8, 760. [Google Scholar] [CrossRef]
- Fontana, F.; Hickman-Brecks, C.L.; Salazar, V.S.; Revollo, L.; Abou-Ezzi, G.; Grimston, S.K.; Jeong, S.Y.; Watkins, M.; Fortunato, M.; Alippe, Y.; et al. N-cadherin Regulation of Bone Growth and Homeostasis Is Osteolineage Stage-Specific. J. Bone Miner. Res. 2017, 32, 1332–1342. [Google Scholar] [CrossRef]
- Huang, G.; Ma, L.; Shen, L.; Lei, Y.; Guo, L.; Deng, Y.; Ding, Y. MIF/SCL3A2 depletion inhibits the proliferation and metastasis of colorectal cancer cells via the AKT/GSK-3beta pathway and cell iron death. J. Cell. Mol. Med. 2022, 26, 3410–3422. [Google Scholar] [CrossRef]
- Zhang, M.; Li, Z.; Wang, H.; Wang, S.; Yu, X.; Wu, J.; Pang, X.; Wu, J.; Yang, X.; Tang, Y.; et al. MIF promotes perineural invasion through EMT in salivary adenoid cystic carcinoma. Mol. Carginog. 2019, 58, 898–912. [Google Scholar] [CrossRef]
- Guo, X.; Xu, S.; Gao, X.; Wang, J.; Xue, H.; Chen, Z.; Zhang, J.; Guo, X.; Qian, M.; Qiu, W.; et al. Macrophage migration inhibitory factor promotes vasculogenic mimicry formation induced by hypoxia via CXCR4:AKT:EMT pathway in human glioblastoma cells. Oncotarget 2017, 8, 80358–80372. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, C.; Hu, X.; Lian, Y.; Ding, C.; Ming, L. Inhibition of LDHA suppresses cell proliferation and increases mitochondrial apoptosis via the JNK signaling pathway in cervical cancer cells. Oncol. Rep. 2022, 47, 1–11. [Google Scholar] [CrossRef]
- Geeraerts, X.; Fernandez-Garcia, J.; Hartmann, F.J.; de Goede, K.E.; Martens, L.; Elkrim, Y.; Debraekeleer, A.; Stijlemans, B.; Vandekeere, A.; Rinaldi, G.; et al. Macrophages are metabolically heterogeneous within the tumor microenvironment. Cell Rep. 2021, 37, 110171. [Google Scholar] [CrossRef]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Xie, F.; Peng, J.; Wu, X. Macrophage migration inhibitory factor promotes Warburg effect via activation of the NFkappaB/HIF1alpha pathway in lung cancer. Int. J. Mol. Med. 2018, 41, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Shu, Q.; Liu, J.; Liu, X.; Zhao, S.; Li, H.; Tan, Y.; Xu, J. GABAB R/GSK-3beta/NF-kappaB signaling pathway regulates the proliferation of colorectal cancer cells. Cancer Med. 2016, 5, 1259–1267. [Google Scholar] [CrossRef]
- Tilstam, P.V.; Pantouris, G.; Corman, M.; Andreoli, M.; Mahboubi, K.; Davis, G.; Du, X.; Leng, L.; Lolis, E.; Bucala, R. A selective small-molecule inhibitor of macrophage migration inhibitory factor-2 (MIF-2), a MIF cytokine superfamily member, inhibits MIF-2 biological activity. J. Biol. Chem. 2019, 294, 18522–18531. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-kappaB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Andreu, P.; Coussens, L.M. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010, 29, 309–316. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Benedek, G.; Meza-Romero, R.; Jordan, K.; Zhang, Y.; Nguyen, H.; Kent, G.; Li, J.; Siu, E.; Frazer, J.; Piecychna, M.; et al. MIF and D-DT are potential disease severity modifiers in male MS subjects. Proc. Natl. Acad. Sci. USA 2017, 114, E8421–E8429. [Google Scholar] [CrossRef] [PubMed]
- Bilsborrow, J.B.; Doherty, E.; Tilstam, P.V.; Bucala, R. Macrophage migration inhibitory factor (MIF) as a therapeutic target for rheumatoid arthritis and systemic lupus erythematosus. Expert Opin. Ther. Targets 2019, 23, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Stoppe, C.; Grieb, G.; Leng, L.; Sauler, M.; Assis, D.; Simons, D.; Boecker, A.H.; Schulte, W.; Piecychna, M.; et al. The clinical significance of the MIF homolog d-dopachrome tautomerase (MIF-2) and its circulating receptor (sCD74) in burn. Burns 2016, 42, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.R.; Azevedo, R.A.; Mousdell, S.; Resende-Lara, P.T.; Ireland, L.; Santos, A.; Girola, N.; Cunha, R.L.O.R.; Schmid, M.C.; Polonelli, L.; et al. Blockade of MIF-CD74 Signalling on Macrophages and Dendritic Cells Restores the Antitumour Immune Response Against Metastatic Melanoma. Front. Immunol. 2018, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, H.R.; Leng, L.; Kang, I.; Jorgensen, W.L.; Cho, C.S.; Bucala, R.; Kim, W.U. Role of Macrophage Migration Inhibitory Factor in the Regulatory T Cell Response of Tumor-Bearing Mice. J. Immunol. 2012, 189, 3905–3913. [Google Scholar] [CrossRef]
- Tessaro, F.H.G.; Ko, E.Y.; De Simone, M.; Piras, R.; Broz, M.T.; Goodridge, H.S.; Balzer, B.; Shiao, S.L.; Guarnerio, J. Single-cell RNA-seq of a soft-tissue sarcoma model reveals the critical role of tumor-expressed MIF in shaping macrophage heterogeneity. Cell Rep. 2022, 39, 110977. [Google Scholar] [CrossRef]
- Dumitru, C.A.; Gholaman, H.; Trellakis, S.; Bruderek, K.; Dominas, N.; Gu, X.; Bankfalvi, A.; Whiteside, T.L.; Lang, S.; Brandau, S. Tumor-derived macrophage migration inhibitory factor modulates the biology of head and neck cancer cells via neutrophil activation. Int. J. Cancer 2011, 129, 859–869. [Google Scholar] [CrossRef]
- Dong, H.H.; Xiang, S.; Chen, X.P.; Liang, H.F.; Zhang, W.; Jing, K.; Zhang, W.; Zhang, W.G.; Chen, L. The epithelial-mesenchymal transition promotes transdifferentiation of subcutaneously implanted hepatic oval cells into mesenchymal tumor tissue. Stem Cells Dev. 2009, 18, 1293–1298. [Google Scholar] [CrossRef]
- Yamini, B. NF-kappaB, Mesenchymal Differentiation and Glioblastoma. Cells 2018, 7, 125. [Google Scholar] [CrossRef]
- Joseph, J.V.; Conroy, S.; Pavlov, K.; Sontakke, P.; Tomar, T.; Eggens-Meijer, E.; Balasubramaniyan, V.; Wagemakers, M.; den Dunnen, W.F.; Kruyt, F.A. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1alpha-ZEB1 axis. Cancer Lett. 2015, 359, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N. The roles and mechanisms of senescence-associated secretory phenotype (SASP): Can it be controlled by senolysis? Inflamm. Regen. 2022, 42, 11. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021, 9, 645593. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L.; Atkinson, E.J.; Doolittle, M.L.; Zhang, X.; LeBrasseur, N.K.; Pignolo, R.J.; Robbins, P.D.; Niedernhofer, L.J.; Ikeno, Y.; et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat. Commun. 2022, 13, 4827. [Google Scholar] [CrossRef]
- Rajasekaran, D.; Zierow, S.; Syed, M.; Bucala, R.; Bhandari, V.; Lolis, E.J. Targeting distinct tautomerase sites of D-DT and MIF with a single molecule for inhibition of neutrophil lung recruitment. Faseb J. 2014, 28, 4961–4971. [Google Scholar] [CrossRef] [PubMed]
- Sinitski, D.; Kontos, C.; Krammer, C.; Asare, Y.; Kapurniotu, A.; Bernhagen, J. Macrophage Migration Inhibitory Factor (MIF)-Based Therapeutic Concepts in Atherosclerosis and Inflammation. Thromb. Haemost. 2019, 119, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Takaoka, A. Comparing antibody and small-molecule therapies for cancer. Nat. Rev. Cancer 2006, 6, 714–727. [Google Scholar] [CrossRef]
- Cheng, B.; Wang, Q.; Song, Y.; Liu, Y.; Liu, Y.; Yang, S.; Li, D.; Zhang, Y.; Zhu, C. MIF inhibitor, ISO-1, attenuates human pancreatic cancer cell proliferation, migration and invasion in vitro, and suppresses xenograft tumour growth in vivo. Sci. Rep. 2020, 10, 6741. [Google Scholar] [CrossRef]
- Fingerle-Rowson, G.; Kaleswarapu, D.R.; Schlander, C.; Kabgani, N.; Brocks, T.; Reinart, N.; Busch, R.; Schutz, A.; Lue, H.; Du, X.; et al. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell Biol. 2009, 29, 1922–1932. [Google Scholar] [CrossRef]
- Pombo Antunes, A.R.; Scheyltjens, I.; Lodi, F.; Messiaen, J.; Antoranz, A.; Duerinck, J.; Kancheva, D.; Martens, L.; De Vlaminck, K.; Van Hove, H.; et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 2021, 24, 595–610. [Google Scholar] [CrossRef]
- Ha, W.; Sevim-Nalkiran, H.; Zaman, A.M.; Matsuda, K.; Khasraw, M.; Nowak, A.K.; Chung, L.; Baxter, R.C.; McDonald, K.L. Ibudilast sensitizes glioblastoma to temozolomide by targeting Macrophage Migration Inhibitory Factor (MIF). Sci. Rep. 2019, 9, 2905. [Google Scholar] [CrossRef] [PubMed]
- Varinelli, L.; Caccia, D.; Volpi, C.C.; Caccia, C.; De Bortoli, M.; Taverna, E.; Gualeni, A.V.; Leoni, V.; Gloghini, A.; Manenti, G.; et al. 4-IPP, a selective MIF inhibitor, causes mitotic catastrophe in thyroid carcinomas. Endocr. Relat. Cancer 2015, 22, 759–775. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, A.; De Baetselier, P.; Brys, L.; Cabrito, I.; Sterckx, Y.G.J.; Schoonooghe, S.; Muyldermans, S.; Raes, G.; Bucala, R.; Vanlandschoot, P.; et al. Novel half-life extended anti-MIF nanobodies protect against endotoxic shock. Faseb J. 2018, 32, 3411–3422. [Google Scholar] [CrossRef] [PubMed]
- Scully, M.; Cataland, S.R.; Peyvandi, F.; Coppo, P.; Knobl, P.; Kremer Hovinga, J.A.; Metjian, A.; de la Rubia, J.; Pavenski, K.; Callewaert, F.; et al. Caplacizumab Treatment for Acquired Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 2019, 380, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.Y.; Shah, K. Nanobodies: Next Generation of Cancer Diagnostics and Therapeutics. Front. Oncol. 2020, 10, 1182. [Google Scholar] [CrossRef]
- Arezumand, R.; Alibakhshi, A.; Ranjbari, J.; Ramazani, A.; Muyldermans, S. Nanobodies As Novel Agents for Targeting Angiogenesis in Solid Cancers. Front. Immunol. 2017, 8, 1746. [Google Scholar] [CrossRef]
- Sleep, D.; Cameron, J.; Evans, L.R. Albumin as a versatile platform for drug half-life extension. Biochim. Biophys. Acta 2013, 1830, 5526–5534. [Google Scholar] [CrossRef]
- Zheng, Y.X.; Yang, M.; Rong, T.T.; Yuan, X.L.; Ma, Y.H.; Wang, Z.H.; Shen, L.S.; Cui, L. CD74 and macrophage migration inhibitory factor as therapeutic targets in gastric cancer. World J. Gastroenterol. 2012, 18, 2253–2261. [Google Scholar] [CrossRef]
- Wirtz, T.H.; Saal, A.; Bergmann, I.; Fischer, P.; Heinrichs, D.; Brandt, E.F.; Koenen, M.T.; Djudjaj, S.; Schneider, K.M.; Boor, P.; et al. Macrophage migration inhibitory factor exerts pro-proliferative and anti-apoptotic effects via CD74 in murine hepatocellular carcinoma. Br. J. Pharmacol. 2021, 178, 4452–4467. [Google Scholar] [CrossRef]
- Kaufman, J.L.; Niesvizky, R.; Stadtmauer, E.A.; Chanan-Khan, A.; Siegel, D.; Horne, H.; Wegener, W.A.; Goldenberg, D.M. Phase I, multicentre, dose-escalation trial of monotherapy with milatuzumab (humanized anti-CD74 monoclonal antibody) in relapsed or refractory multiple myeloma. Br. J. Haematol. 2013, 163, 478–486. [Google Scholar] [CrossRef]
- Haran, M.; Mirkin, V.; Braester, A.; Harpaz, N.; Shevetz, O.; Shtreiter, M.; Greenberg, S.; Mordich, O.; Amram, O.; Binsky-Ehrenreich, I.; et al. A phase I-II clinical trial of the anti-CD74 monoclonal antibody milatuzumab in frail patients with refractory chronic lymphocytic leukaemia: A patient based approach. Br. J. Haematol. 2018, 182, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Meza-Romero, R.; Benedek, G.; Leng, L.; Bucala, R.; Vandenbark, A.A. Predicted structure of MIF/CD74 and RTL1000/CD74 complexes. Metab. Brain Dis. 2016, 31, 249–255. [Google Scholar] [CrossRef]
- Offner, H.; Sinha, S.; Burrows, G.G.; Ferro, A.J.; Vandenbark, A.A. RTL therapy for multiple sclerosis: A Phase I clinical study. J. Neuroimmunol 2011, 231, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.; Lue, H.; Zernecke, A.; Kapurniotu, A.; Andreetto, E.; Frank, R.; Lennartz, B.; Weber, C.; Bernhagen, J. MIF-chemokine receptor interactions in atherogenesis are dependent on an N-loop-based 2-site binding mechanism. FASEB J. 2010, 25, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]

| Cytokine | Type of cancer | Relationship | Reference |
|---|---|---|---|
| MIF and DDT | Lung cancer—NSCLC |
| [57,58,59,60] |
| MIF | Breast cancer |
| [61,62] |
| MIF | Prostate cancer |
| [63,64,65] |
| MIF and DDT | Cervical cancer |
| [12,66] |
| MIF | Hepatocellular carcinoma |
| [67,68,69] |
| MIF | Gastric cancer |
| [70,71,72] |
| MIF | Esophageal cancer |
| [73] |
| MIF and DDT | Glioblastoma |
| [11,74,75,76] |
| MIF and DDT | Renal carcinoma |
| [77] |
| MIF and DDT | Pancreatic ductal adenocarcinoma |
| [78] |
| MIF and DDT | Neuroblastoma |
| [79] |
| MIF and DDT | Melanoma |
| [80,81] |
| MIF and DDT | Ovarian cancer |
| [41,82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora Barthelmess, R.; Stijlemans, B.; Van Ginderachter, J.A. Hallmarks of Cancer Affected by the MIF Cytokine Family. Cancers 2023, 15, 395. https://doi.org/10.3390/cancers15020395
Mora Barthelmess R, Stijlemans B, Van Ginderachter JA. Hallmarks of Cancer Affected by the MIF Cytokine Family. Cancers. 2023; 15(2):395. https://doi.org/10.3390/cancers15020395
Chicago/Turabian StyleMora Barthelmess, Romina, Benoit Stijlemans, and Jo A. Van Ginderachter. 2023. "Hallmarks of Cancer Affected by the MIF Cytokine Family" Cancers 15, no. 2: 395. https://doi.org/10.3390/cancers15020395
APA StyleMora Barthelmess, R., Stijlemans, B., & Van Ginderachter, J. A. (2023). Hallmarks of Cancer Affected by the MIF Cytokine Family. Cancers, 15(2), 395. https://doi.org/10.3390/cancers15020395






