Modulation of the Tumor Microenvironment by Ellagic Acid in Rat Model for Hepatocellular Carcinoma: A Potential Target against Hepatic Cancer Stem Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
- Group 1 (CON): Five rats were kept untreated to act as a negative control.
- Group 2 (DMSO): Five rats were given the DMSO same quantity in which the EA was dissolved.
- Group 3 (OVO): Five rats were administered olive oil, the same amount in which the CCL4 was dissolved.
- Group 4 (EA): Five rats were administered EA at a dose of 50 mg/kg b.wt dissolved in DMSO following manufacturer laboratory procedures.
- Group 5 (HCC-M): Thirty rats were used for induction of HCC by a single dose of CCl4 (0.5 mL/kg i.p in olive oil) twice weekly for eight weeks. The duration was modified from that applied by [23] based on what was declared that the administration of a different amount (up to 0.5 mL/kg) might affect tumor numbers [24].
- Group 6 (EA + HCC): Fifteen rats confirmed with HCC from group HCC-M were administered EA orally at 50 mg/kg bw [18] daily for five weeks.
2.2. Materials
2.2.1. Chemicals
2.2.2. Biochemical Analysis
2.2.3. Morphology
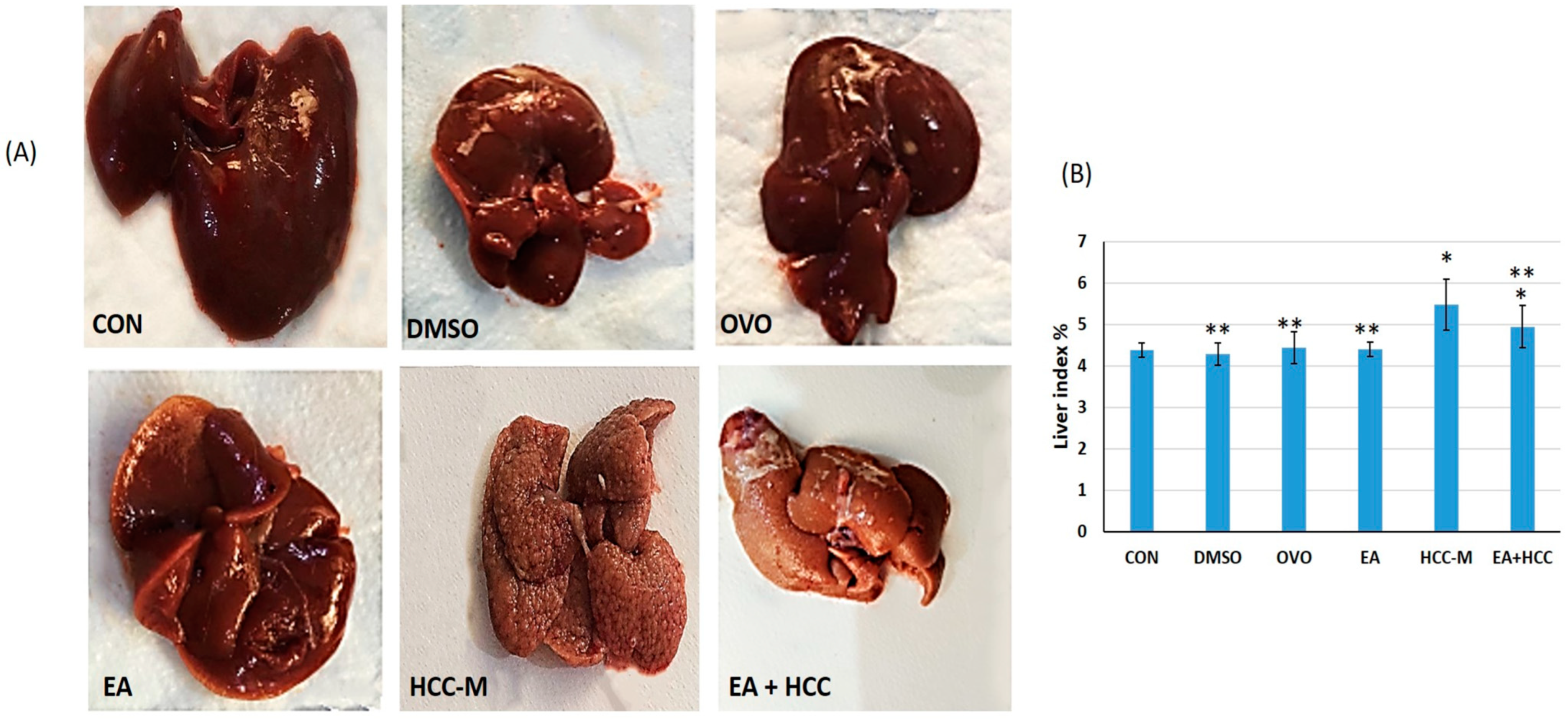
2.2.4. Liver Index %
2.2.5. Histopathology
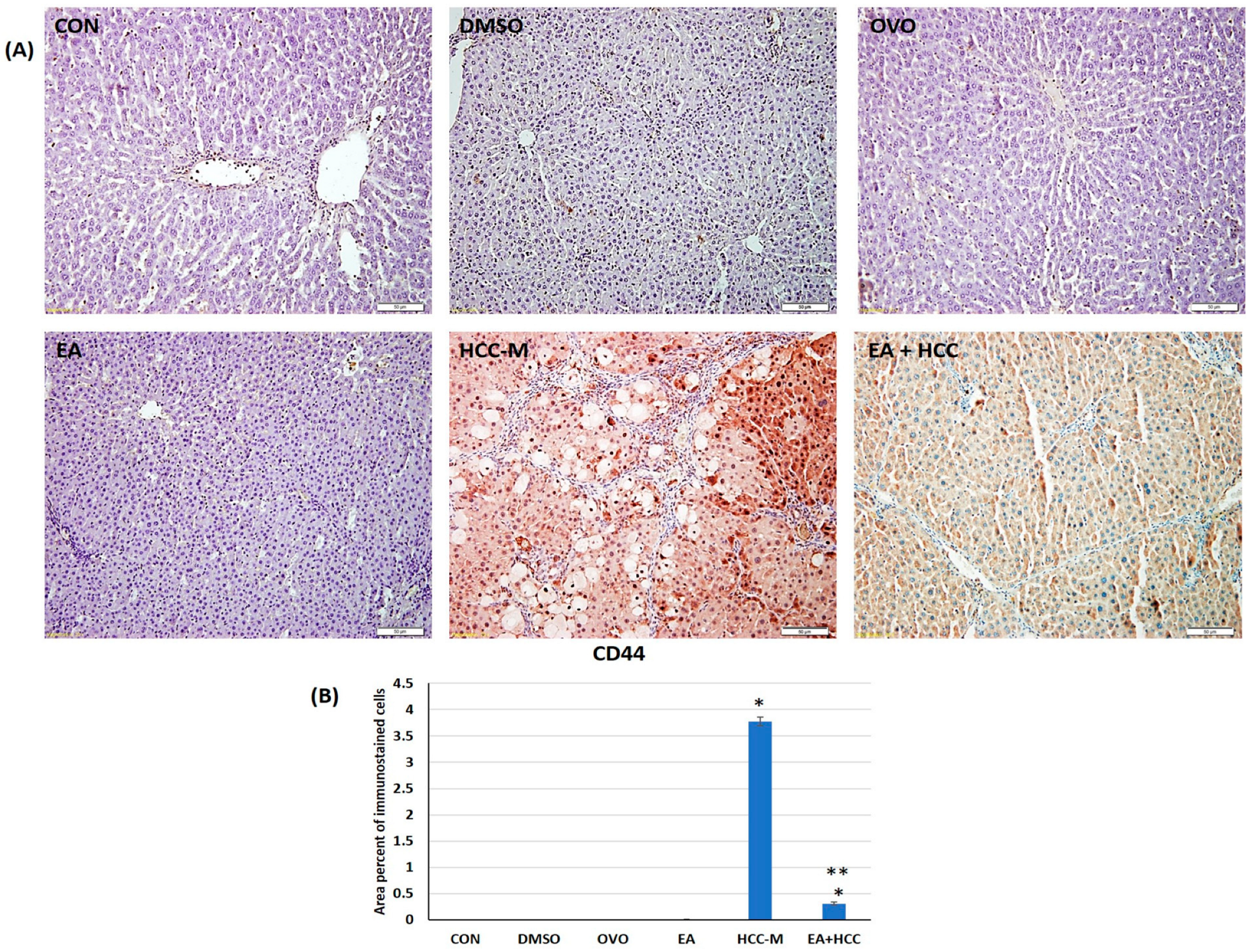
2.2.6. Immunohistochemical Study
Image Analysis
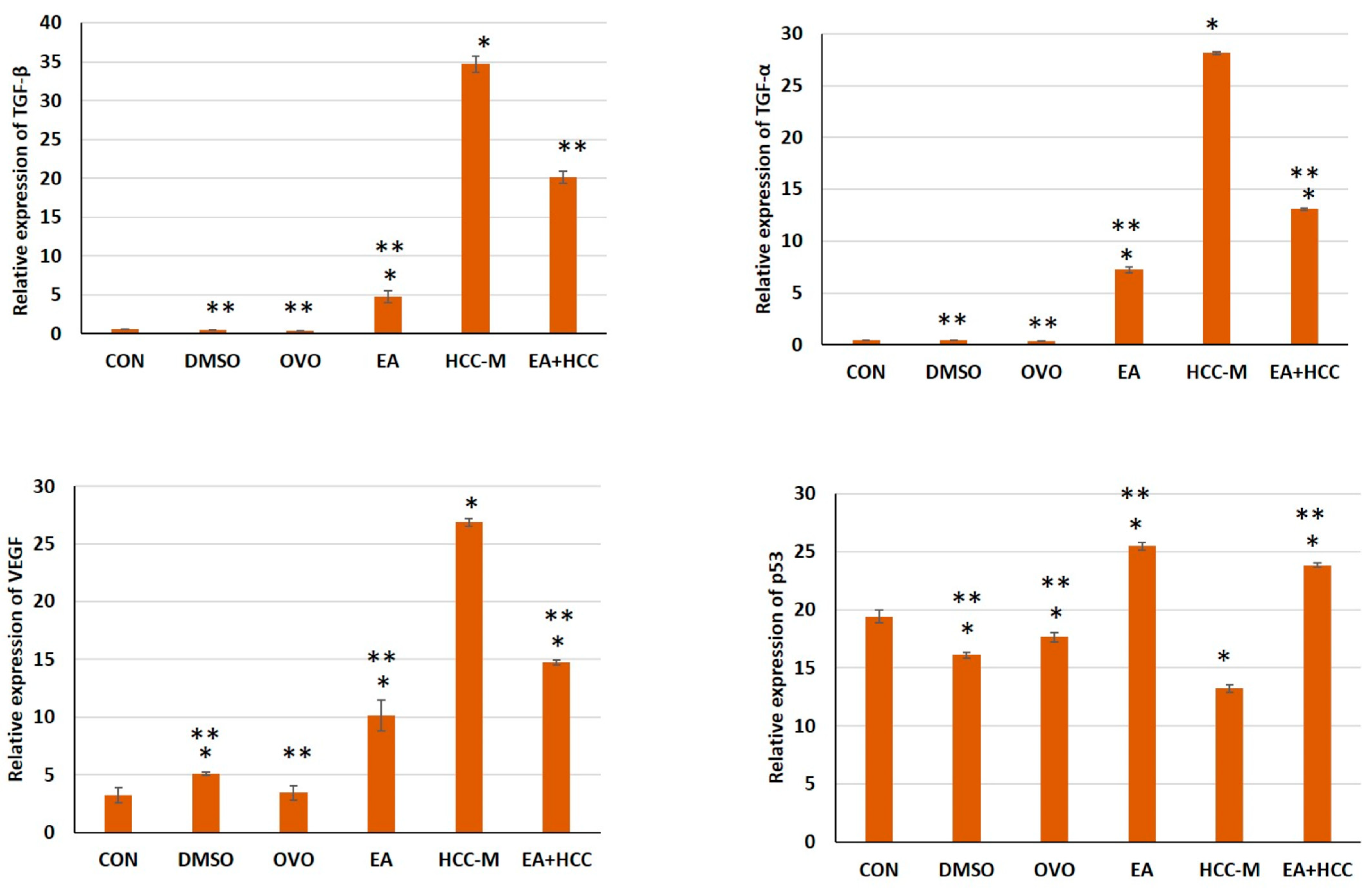
2.2.7. Quantitative Real-Time PCR (qRT-PCR)
Statistical Study
3. Results
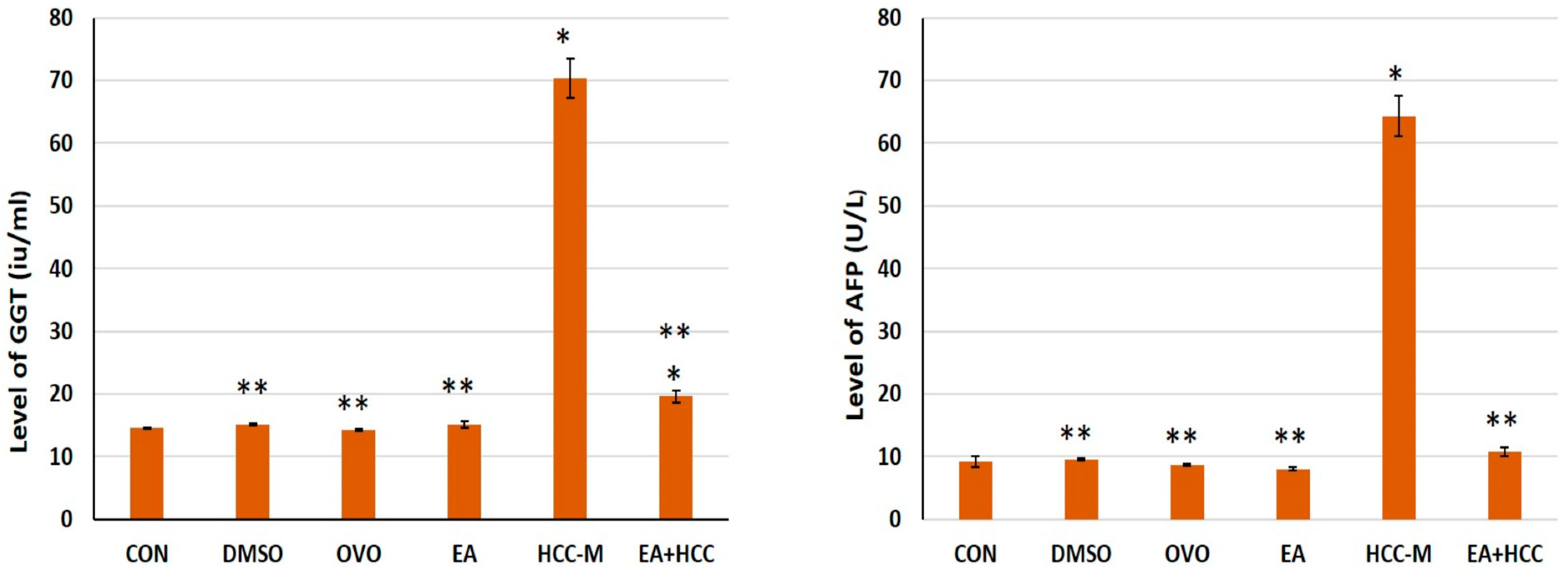
3.1. Biochemical Analysis
3.1.1. Morphological Study
3.1.2. Histopathology
3.1.3. Immunohistochemical Study
3.1.4. qRT-PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Schulte, L.A.; López-Gil, J.C.; Sainz, B., Jr.; Hermann, P.C. The cancer stem cell in hepatocellular carcinoma. Cancers 2020, 12, 684. [Google Scholar]
- Papaccio, F.; Paino, F.; Regad, T.; Papaccio, G.; Desiderio, V.; Tirino, V. Concise review: Cancer cells, cancer stem cells, and mesenchymal stem cells: Influence in cancer development. Stem. Cells Transl. Med. 2017, 6, 2115–2125. [Google Scholar]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer stem cells: The architects of the tumor ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar]
- Afify, S.M.; Seno, M. Conversion of stem cells to cancer stem cells: Undercurrent of cancer initiation. Cancers 2019, 11, 345. [Google Scholar]
- Nguyen, L.T.; Tretiakova, M.S.; Silvis, M.R.; Lucas, J.; Klezovitch, O.; Coleman, I.; Bolouri, H.; Kutyavin, V.I.; Morrissey, C.; True, L.D.; et al. ERG activates the YAP1 transcriptional program and induces the development of age-related prostate tumors. Cancer Cell 2015, 27, 797–808. [Google Scholar]
- Zhang, L.; Yang, S.; Chen, X.; Stauffer, S.; Yu, F.; Lele, S.M.; Fu, K.; Datta, K.; Palermo, N.; Chen, Y.; et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol. Cell Biol. 2015, 35, 1350–1362. [Google Scholar]
- Bourgeais, J.; Gouilleux-Gruart, V.; Gouilleux, F. Oxidative metabolism in cancer: A STAT affair? Jak-Stat 2013, 2, e25764. [Google Scholar]
- Yu, L.-X.; Ling, Y.; Wang, H.-Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis. Oncol. 2018, 2, 6. [Google Scholar] [PubMed]
- Cicalese, A.; Bonizzi, G.; Pasi, C.E.; Faretta, M.; Ronzoni, S.; Giulini, B.; Brisken, C.; Minucci, S.; Di Fiore, P.P.; Pelicci, P.G. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 2009, 138, 1083–1095. [Google Scholar] [PubMed]
- Gao, B.; Bataller, R. Alcoholic liver disease: Pathogenesis and new therapeutic targets. Gastroenterology 2011, 141, 1572–1585. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Adams, L.S.; Henning, S.M.; Niu, Y.; Zhang, Y.; Nair, M.G.; Heber, D. In vitro antiproliferative, apoptotic and antioxidant activities of punicalagin, ellagic acid and a total pomegranate tannin extract are enhanced in combination with other polyphenols as found in pomegranate juice. J. Nutr. Biochem. 2005, 16, 360–367. [Google Scholar]
- Wang, L.; Ho, J.; Glackin, C.; Martins-Green, M. Specific pomegranate juice components as potential inhibitors of prostate cancer metastasis. Translational. Oncol. 2012, 5, 344–355. [Google Scholar]
- Vicinanza, R.; Zhang, Y.; Henning, S.M.; Heber, D. Pomegranate Juice Metabolites, Ellagic Acid and Urolithin A, Synergistically Inhibit Androgen-Independent Prostate Cancer Cell Growth via Distinct Effects on Cell Cycle Control and Apoptosis. Evid.-Based Complement. Altern. Med. 2013, 2013, 247504. [Google Scholar]
- Li, L.W.; Na, C.; Tian, S.Y.; Chen, J.; Ma, R.; Gao, Y.; Lou, G. Ellagic acid induces HeLa cell apoptosis via regulating signal transducer and activator of transcription 3 signaling. Exp. Ther. Med. 2018, 16, 29–36. [Google Scholar]
- Umesalma, S.; Sudhandiran, G. Ellagic acid prevents rat colon carcinogenesis induced by 1, 2 dimethyl hydrazine through inhibition of AKT-phosphoinositide-3 kinase pathway. Eur. J. Pharmacol. 2011, 660, 249–258. [Google Scholar]
- Aslan, A.; Hussein, Y.T.; Gok, O.; Beyaz, S.; Erman, O.; Baspinar, S. Ellagic acid ameliorates lung damage in rats via modulating antioxidant activities, inhibitory effects on inflammatory mediators and apoptosis-inducing activities. Environ. Sci. Pollut. Res. 2020, 27, 7526–7537. [Google Scholar]
- Promsong, A.; Chung, W.O.; Satthakarn, S.; Nittayananta, W. Ellagic acid modulates the expression of oral innate immune mediators: Potential role in mucosal protection. J. Oral Pathol. Med. 2015, 44, 214–221. [Google Scholar]
- Zhou, Z.; Liu, Y.; Song, W.; Jiang, X.; Deng, Z.; Xiong, W.; Shen, J. Metabolic reprogramming mediated PD-L1 depression and hypoxia reversion to reactivate tumor therapy. J. Control Release 2022, 352, 793–812. [Google Scholar] [PubMed]
- Su, Y.C.; Davuluri, G.V.; Chen, C.H.; Shiau, D.C.; Chen, C.C.; Chen, C.L.; Lin, Y.S.; Chang, C.P. Galectin-1-induced autophagy facilitates cisplatin resistance of hepatocellular carcinoma. PLoS ONE 2016, 11, e0148408. [Google Scholar]
- Uehara, T.; Pogribny, I.P.; Rusyn, I. The DEN and CCl4-Induced Mouse Model of Fibrosis and Inflammation-Associated Hepatocellular Carcinoma. Curr. Protoc. Pharmacol. 2014, 66, 14–30. [Google Scholar] [CrossRef]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [PubMed]
- Wang, Z.; Li, Q.; Xiang, M.; Zhang, F.; Wei, D.; Wen, Z.; Zhou, Y. Astragaloside alleviates hepatic fibrosis function via PAR2 signaling pathway in diabetic rats. Cell. Physiol. Biochem. 2017, 4, 1156–1166. [Google Scholar] [CrossRef]
- Drury, R.A.; Walington, E.A. Carlton’s Histological Techniques, 5th ed.; Oxford University Press: Oxford, NY, USA, 1983. [Google Scholar]
- Zhu, X.D.; Zhang, J.B.; Zhuang, P.Y.; Zhu, H.G.; Zhang, W.; Xiong, Y.Q.; Wu, W.Z.; Wang, L.; Tang, Z.Y.; Sun, H.C. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J. Clin. Oncol. 2008, 26, 2707–2716. [Google Scholar]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Hu, X.Q.; Song, Y.N.; Wu, R.; Cai, F.F.; Zhang, Y.; Peng, J.H.; Su, S.B. Metabolic mechanisms of Fuzheng-Huayu formula against liver fibrosis in rats. J. Ethnopharmacol. 2019, 238, 111888–111897. [Google Scholar] [CrossRef]
- Huo, X.; Meng, X.; Zhang, J.; Zhao, Y. Hepatoprotective effect of different combinations of 18α-and 18β-Glycyrrhizic acid against CCl4-induced liver injury in rats. Biomed. Pharmacother. 2020, 122, 109354. [Google Scholar]
- Ahn, M.; Kim, J.; Bang, H.; Moon, J.; Kim, G.O.; Shin, T. Hepatoprotective effects of allyl isothiocyanateagainst carbon tetrachloride-induced hepatotoxicity in rat. Chem. Biol. Interact. 2016, 254, 102–108. [Google Scholar]
- Torres, L.R.; Santana, F.C.; Torres-Leal, F.L.; Melo, L.L.; Yoshime, L.T.; Matos-Neto, E.M.; Seelaender, M.C.; Araújo, C.M.; Cogliati, B.; Mancini-Filho, J. Pequi (Caryocar brasiliense Camb.) almond oil attenuates carbon tetrachloride-induced acute hepatic injury in rats: Antioxidant and anti-inflammatory effects. Food Chem. Toxicol. 2016, 97, 205–216. [Google Scholar] [PubMed]
- Gur, E.; Waner, T. The variability of organ weight background data in rats. Lab. Anim. 1993, 27, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Carr, B.; Guerra, V.; Giannini, E.G.; Farinati, F.; Ciccarese, F.; Rapaccini, G.L.; Di Marco, M.; Benvegnù, L.; Zoli, M.; Borzio, F.; et al. A Liver Index and its Relationship to Indices of HCC Aggressiveness. J. Integr. Oncol. 2016, 5, 178. [Google Scholar] [CrossRef]
- Greaves, P. Liver and Pancreas. In Histopathology of Preclinical Toxicity Studies, 4th ed.; Academic Press: Cambridge, MA, USA; Elsevier: London, UK, 2012; pp. 433–535. ISBN 9780444538567. [Google Scholar]
- Leist, M.; Gantner, F.; Bohlinger, I.; Tiegs, G.; Germann, P.G.; Wendel, A. Tumor necrosis factor-induced hepatocyte apoptosis precedes liver failure in experiment murine shock models. Am. J. Pathol. 1995, 146, 1220–1234. [Google Scholar]
- Aubert, J.; Begriche, K.; Delannoy, M.; Morel, I.; Pajaud, J.; Ribault, C.; Lepage, S.; McGill, M.R.; Lucas-Clerc, C.; Turlin, B.; et al. Differences in early acetaminophen hepatotoxicity between obese ob/ob and db/db mice. J. Pharmacol. Exp. Ther. 2012, 342, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Nair, K.; Deepadevi, K.; Arun, P.; Kumar, V.; Santhosh, A.; Lekshmi, L.; Kurup, P.A. Toxic effect of systemic administration of low doses of the plasticizer di-(2-ethyl hexyl) phthalate [DEHP] in rats. Indian J. Exp. Biol. 1998, 36, 264–272. [Google Scholar]
- Lu, Y.; Zhu, M.; Li, W.; Lin, B.; Dong, X.; Chen, Y.; Xie, X.; Guo, J.; Li, M. Alpha fetoprotein plays a critical role in promoting metastasis of hepatocellular carcinoma cells. J. Cell. Mol. Med. 2016, 20, 549–558. [Google Scholar] [CrossRef]
- Carr, B.I.; Guerra, V. Low alpha-fetoprotein levels are associated with improved survival in hepatocellular carcinoma patients with portal vein thrombosis. Dig. Dis. Sci. 2016, 61, 937–947. [Google Scholar] [CrossRef]
- Giannini, E.G.; Marenco, S.; Borgonovo, G.; Savarino, V.; Farinati, F.; Del Poggio, P.; Rapaccini, G.L.; Anna Di Nolfo, M.; Benvegnu, L.; Zoli, M.; et al. Alpha-fetoprotein has no prognostic role in small hepatocellular carcinoma identified during surveillance in compensated cirrhosis. Hepatology 2012, 56, 1371–1379. [Google Scholar] [CrossRef]
- Yamamoto, K.; Imamura, H.; Matsuyama, Y.; Kume, Y.; Ikeda, H.; Norman, G.L.; Shums, Z.; Aoki, T.; Hasegawa, K.; Beck, Y. AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of clinic pathological variables of HCC. J. Gastroenterol. 2010, 45, 1272–1282. [Google Scholar] [CrossRef]
- Bignold, L.P. Pathogenetic mechanisms of nuclear pleomorphism of tumour cells based on the mutator phenotype theory of carcinogenesis. Histol. Histopathol. 2003, 18, 657–664. [Google Scholar]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar]
- Lowes, K.N.; Brennan, B.A.; Yeoh, G.C.; Olynyk, J.K. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am. J. Pathol. 1999, 154, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Mikhail, S.; He, A.R. Liver cancer stem cells. Int. J. Hepatol. 2011, 2011, 486954. [Google Scholar] [CrossRef] [PubMed]
- Szakacs, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [PubMed]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA damage in stem cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [PubMed]
- Dzobo, K.; Senthebane, D.A.; Rowe, A.; Thomford, N.E.; Mwapagha, L.M.; Al-Awwad, N.; Dandara, C.; Parker, M.I. Cancer Stem Cell Hypothesis for Therapeutic Innovation in Clinical Oncology? Taking the Root Out, Not Chopping the Leaf. OMICS J. Integr. Biol. 2016, 20, 681–691. [Google Scholar]
- Yang, X.R.; Xu, Y.; Yu, B.; Zhou, J.; Qiu, S.J.; Shi, G.M.; Zhang, B.H.; Wu, W.Z.; Shi, Y.; Wu, B.; et al. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut 2010, 59, 953–962. [Google Scholar] [CrossRef]
- Wan, S.; Zhao, E.; Kryczek, I.; Vatan, L.; Sadovskaya, A.; Ludema, G.; Ludema, G.; Simeone, D.M.; Zou, W.; Welling, T.H. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology 2014, 147, 1393–1404. [Google Scholar]
- Fan, Q.M.; Jing, Y.Y.; Yu, G.F.; Kou, X.R.; Ye, F.; Gao, L.; Li, R.; Zhao, Q.; Yang, Y.; Lu, Z.; et al. Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2014, 352, 160–168. [Google Scholar]
- Soares, R.; Pereira, M.B.; Silva, C.; Amendoeira, I.; Wagner, R.; Ferro, J.; Schmitt, F.C. Expression of TGF-alpha and EGFR in breast cancer and its relation to angiogenesis. Breast J. 2002, 6, 171–177. [Google Scholar]
- Ciardiello, F.; Kim, N.; McGeady, M.L.; Liscia, D.S.; Saeki, T.; Bianco, C.; Salomon, D.S. Expression of transforming growth factor alpha (TGFα) in breast cancer. Ann. Oncol. 1991, 2, 169–182. [Google Scholar] [PubMed]
- CavigLia, J.M.; Schwabe, R.F. Experimental Hepatocarcinogenesis. In Pathobiology of Human Disease: A Dynamic Encyclopedia of Disease Mechanisms; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Laouar, Y.; Sutterwala, F.; Gorelik, L.; Flavell, R. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 2005, 6, 600–607. [Google Scholar] [CrossRef]
- Korn, T.; Bettelli, E.; Oukka, M.; Kuchroo, V.K. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009, 27, 485–517. [Google Scholar] [CrossRef]
- Lemarie, C.A.; Tharaux, P.L.; Esposito, B.; Tedgui, A.; Lehoux, S. TGF-α mediates NF-kB activation in strained arteries. Circ. Res. 2006, 99, 434–441. [Google Scholar] [CrossRef]
- Lawrence, T. The nucear factor NF_KappaB pathway in inflammation. Cold Spring Harbor Perspect. Biol. 2009, 1, a00165. [Google Scholar] [CrossRef] [PubMed]
- Tak, P.P.; Firestein, G.S. NF-kappaB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef]
- Sutterwala, F.S.; Haasken, S.; Cassel, S.L. Mechanism of NLRP3 inflammasome activation. Ann. N. Y. Acad. Sci. 2014, 1319, 82–95. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Z.; Hou, J.; Xiong, W.; Kim, H.; Chen, J.; Zheng, C.; Jian, X.; Yoon, J.; Shen, J. Tumor Selective Metabolic Reprogramming as a Prospective PD-L1 Depression Strategy to Reactivate Immunotherapy. Adv. Mater. 2022, 34, 2206121. [Google Scholar] [CrossRef]
- Deng, H.A.; Chen, Y.; Yuan, Y.; Zhang, H.Y. Regulatory effect of ellagic acid on immune function in burned rats. J. Burn. Care Res. 2023, 18, irad041. [Google Scholar]
- Kang, E.H.; Kown, T.Y.; Oh, G.T.; Park, W.F.; Park, S.I.; Park, S.K.; Lee, Y.I. The flavonoid ellagic acid from a medicinal herb inhibits host immune tolerance induced by the hepatitis B virus-e antigen. Antivir. Res. 2006, 72, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Min, J.K.; Han, K.Y.; Kim, E.C.; Kim, Y.M.; Lee, S.W.; Kim, O.H.; Kim, K.W.; Gho, Y.S.; Kwon, Y.G. Capsaicin inhibits in vitro and in vivo angiogenesis. Cancer Res. 2004, 64, 644–651. [Google Scholar] [PubMed]
- Tang, K.H.; Ma, S.; Lee, T.K.; Chan, Y.P.; Kwan, P.S.; Tong, C.M.; Ng, I.O.; Man, K.; To, K.F.; Lai, P.B.; et al. CD133+ liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology 2012, 55, 807–820. [Google Scholar] [CrossRef]
- Sakr, M.A.; Al-Azzawi, M.A.; Anis, A.; Abd El-Aziz, A.A.; Ebeid, M.E.; Shokeer, M.A.; Fayed, A. The correlation between P53 and COX-2 expression and the pathological alteration in hepatocellular carcinoma. Egypt J. Med. Hum. Genet. 2022, 23, 14. [Google Scholar] [CrossRef]
- Zheng, P.; Lv, S.; Wang, Q.; Cai, B.; Zhang, F.; Huo, F. LncRNA PLAC2 upregulates p53 to induce hepatocellular carcinoma cell apoptosis. Gene 2019, 712, 143944. [Google Scholar] [PubMed]
- You, J.; Yang, H.; Lai, Y.; Simon, L.; Au, J.; Burkart, A.L. Erratum to ARID2, p110α, p53, and β-catenin protein expression in hepatocellular carcinoma and clinicopathologic implications. Hum. Pathol. 2015, 46, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Jiang, H.; Liu, Y.; Chen, J.; Zhou, X.; Zhao, C.; Chen, X.; Lin, M. Rhein induces liver cancer cells apoptosis via activating ROS-dependent JNK/Jun/caspase-3 signaling pathway. J. Cancer 2020, 11, 500–507. [Google Scholar] [CrossRef]
- Gonzalez-Sarrias, A.; Azorin-Ortuno, M.; Yanez-Gascon, M.J.; Tomas-Barberan, F.A.; Garcia-Conesa, M.T.; Espin, J.C. Dissimilar in vitro and in vivo effects of ellagic acid and its microbiota-derived metabolites, urolithins, on the cytochrome P450 1A1. J. Agric. Food Chem. 2009, 57, 5623–5632. [Google Scholar]
- Ramos, S. Cancer chemoprevention and chemotherapy: Dietary polyphenols and signalling pathways. Mol. Nutr. Food Res. 2008, 52, 507–526. [Google Scholar]



| Forward | Reverse | |
|---|---|---|
| P53 | 5′GTT CCGAGA GCT GAA TGA GG 3′ | 5′ TTT TAT GGCGGG ACG TAG AC3′ |
| TGF alpha (TGF-α) | 5′AAACACACGAGACGCTGAAG3′ | 5′ATCCAGTGAGTTCCGAAAGC3′ |
| TGF beta TGF-β | 5′TACCTGAACCCGTGTTGCTCTC3′ | 5′GTTGCTGAGGTATCGCCAGGAA3′ |
| VEGF | 5′CAG CTA TTG CCG TCC AAT TGA3′ | 5′CCA GGG CTT CAT CAT TGC A 3′ |
| GAPDH | 5′GCT CAC TAA AGG GCA TCC TG 3′ | 5′CCA TAGAGG CCA TGA GAT CC3′ |
| CON | DMSO | OVO | EA | HCC-M | EA + HCC | |
|---|---|---|---|---|---|---|
| ALT (U/L) | 16.66 ± 0.18 | 17.7 ± 0.15 ** | 18.62 ± 0.13 ** | 19.40 ± 0.23 ** | 175 ± 8.39 * | 27.50 ± 3.10 **,* |
| AST (U/L) | 21.80 ± 0.83 | 23.60 ± 0.54 ** | 25.20 ± 0.83 ** | 30.40 ± 2.30 ** | 192.50 ± 9.34 * | 33.60 ± 4.52 **,* |
| ALP (U/L) | 45 ± 1.0 | 49.20 ± 0.83 ** | 65.20 ± 0.83 **,* | 47.40 ± 2.30 ** | 195.10 ± 9.24 * | 57.70 ± 2.71 **,* |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramadan, W.S.; Alkarim, S.; Moulay, M.; Alrefeai, G.; Alkudsy, F.; Hakeem, K.R.; Iskander, A. Modulation of the Tumor Microenvironment by Ellagic Acid in Rat Model for Hepatocellular Carcinoma: A Potential Target against Hepatic Cancer Stem Cells. Cancers 2023, 15, 4891. https://doi.org/10.3390/cancers15194891
Ramadan WS, Alkarim S, Moulay M, Alrefeai G, Alkudsy F, Hakeem KR, Iskander A. Modulation of the Tumor Microenvironment by Ellagic Acid in Rat Model for Hepatocellular Carcinoma: A Potential Target against Hepatic Cancer Stem Cells. Cancers. 2023; 15(19):4891. https://doi.org/10.3390/cancers15194891
Chicago/Turabian StyleRamadan, Wafaa S., Saleh Alkarim, Mohammed Moulay, Ghadeer Alrefeai, Fatma Alkudsy, Khalid Rehman Hakeem, and Ashwaq Iskander. 2023. "Modulation of the Tumor Microenvironment by Ellagic Acid in Rat Model for Hepatocellular Carcinoma: A Potential Target against Hepatic Cancer Stem Cells" Cancers 15, no. 19: 4891. https://doi.org/10.3390/cancers15194891
APA StyleRamadan, W. S., Alkarim, S., Moulay, M., Alrefeai, G., Alkudsy, F., Hakeem, K. R., & Iskander, A. (2023). Modulation of the Tumor Microenvironment by Ellagic Acid in Rat Model for Hepatocellular Carcinoma: A Potential Target against Hepatic Cancer Stem Cells. Cancers, 15(19), 4891. https://doi.org/10.3390/cancers15194891







