MYCN Amplifications and Metabolic Rewiring in Neuroblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomics Analyses
2.2. ChIP-Seq Data Analyses
3. Results
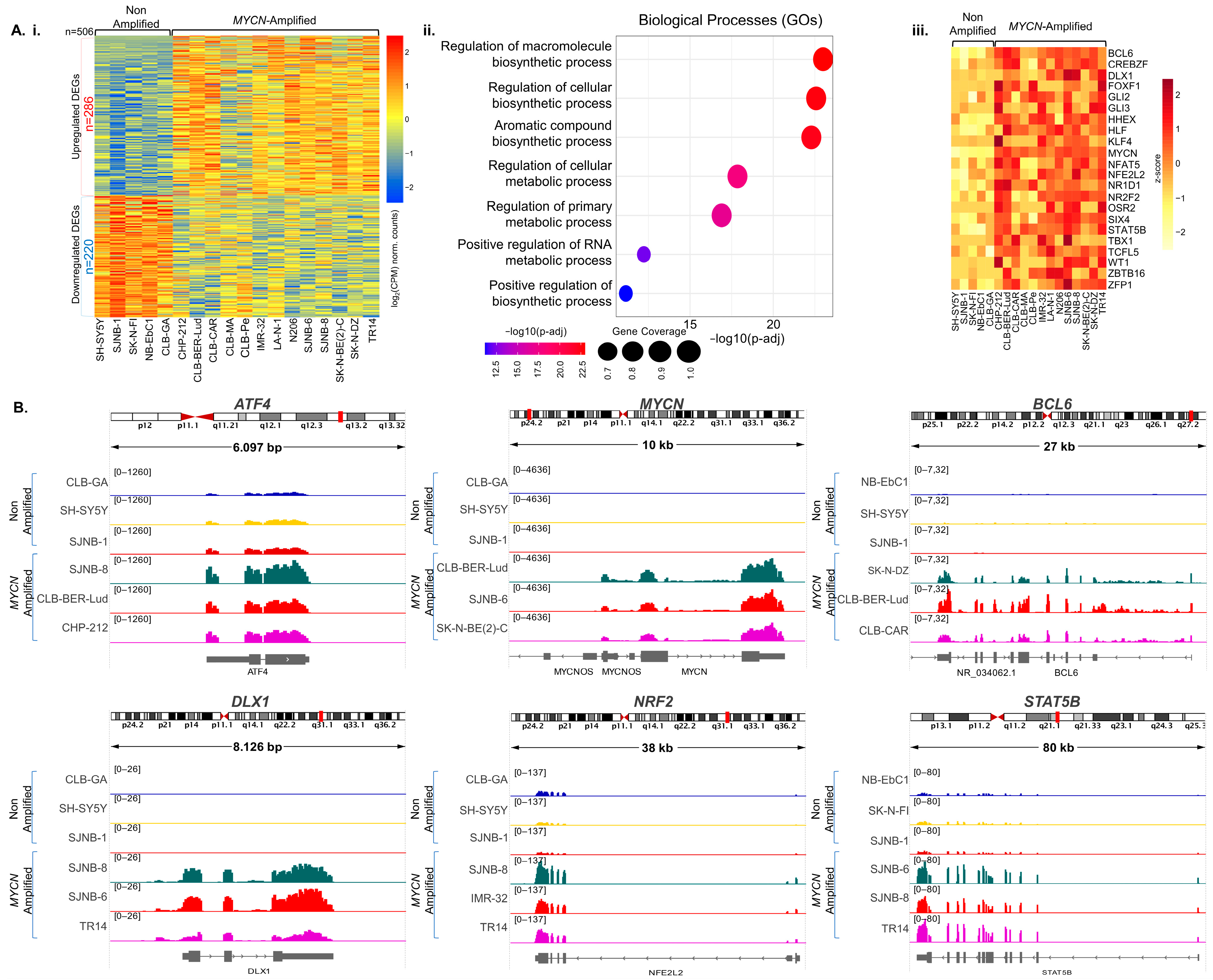
3.1. Gene Expression Rewiring and Metabolic Signatures in MYCN-Amplified Neuroblastoma
3.1.1. MYCN-Binding Landscape in Neuroblastoma Illuminating Novel TF-Encoding Target-Genes
3.1.2. Computational Validation in Neuroblastoma Patient-Derived Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koutsi, M.A.; Pouliou, M.; Champezou, L.; Vatsellas, G.; Giannopoulou, A.I.; Piperi, C.; Agelopoulos, M. Typical Enhancers, Super-Enhancers, and Cancers. Cancers 2022, 14, 4375. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Kim, J.; DeBerardinis, R.J. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019, 30, 434–446. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Peng, X.; Chen, Z.; Farshidfar, F.; Xu, X.; Lorenzi, P.L.; Wang, Y.; Cheng, F.; Tan, L.; Mojumdar, K.; Du, D.; et al. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018, 23, 255–269.e4. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in The Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Yoshida, G.J. Beyond the Warburg Effect: N-Myc Contributes to Metabolic Reprogramming in Cancer Cells. Front. Oncol. 2020, 10, 791. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Ramsey, M.R.; Hayes, D.N.; Fan, C.; McNamara, K.; Kozlowski, P.; Torrice, C.; Wu, M.C.; Shimamura, T.; Perera, S.A.; et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007, 448, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef]
- Wang, X.; Liu, R.; Zhu, W.; Chu, H.; Yu, H.; Wei, P.; Wu, X.; Zhu, H.; Gao, H.; Liang, J.; et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature 2019, 571, 127–131. [Google Scholar] [CrossRef]
- Bradner, J.E.; Hnisz, D.; Young, R.A. Transcriptional Addiction in Cancer. Cell 2017, 168, 629–643. [Google Scholar] [CrossRef]
- Agelopoulos, M.; Foutadakis, S.; Thanos, D. The Causes and Consequences of Spatial Organization of the Genome in Regulation of Gene Expression. Front. Immunol. 2021, 12, 682397. [Google Scholar] [CrossRef]
- Papathanasiou, M.; Tsiftsoglou, S.A.; Polyzos, A.P.; Papadopoulou, D.; Valakos, D.; Klagkou, E.; Karagianni, P.; Pliatska, M.; Talianidis, I.; Agelopoulos, M.; et al. Identification of a dynamic gene regulatory network required for pluripotency factor-induced reprogramming of mouse fibroblasts and hepatocytes. EMBO J. 2021, 40, e102236. [Google Scholar] [CrossRef]
- Bal, E.; Kumar, R.; Hadigol, M.; Holmes, A.B.; Hilton, L.K.; Loh, J.W.; Dreval, K.; Wong, J.; Vlasevska, S.; Corinaldesi, C.; et al. Super-enhancer hypermutation alters oncogene expression in B cell lymphoma. Nature 2022, 607, 808–815. [Google Scholar] [CrossRef]
- Schaub, F.X.; Dhankani, V.; Berger, A.C.; Trivedi, M.; Richardson, A.B.; Shaw, R.; Zhao, W.; Zhang, X.; Ventura, A.; Liu, Y.; et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018, 6, 282–300.e2. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Kohl, N.E.; Kanda, N.; Schreck, R.R.; Bruns, G.; Latt, S.A.; Gilbert, F.; Alt, F.W. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell 1983, 35, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Shi, P.; Wang, Z.; Yuan, C.; Cui, H. Molecular Mechanisms of MYCN Dysregulation in Cancers. Front. Oncol. 2021, 10, 625332. [Google Scholar] [CrossRef] [PubMed]
- Valentijn, L.J.; Koster, J.; Haneveld, F.; Aissa, R.A.; van Sluis, P.; Broekmans, M.E.; Molenaar, J.J.; van Nes, J.; Versteeg, R. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. USA 2012, 109, 19190–19195. [Google Scholar] [CrossRef]
- Schwab, M.; Alitalo, K.; Klempnauer, K.H.; Varmus, H.E.; Bishop, J.M.; Gilbert, F.; Brodeur, G.; Goldstein, M.; Trent, J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 1983, 305, 245–248. [Google Scholar] [CrossRef]
- Wang, D.; Yin, Z.; Wang, H.; Wang, L.; Li, T.; Xiao, R.; Xie, T.; Han, R.; Dong, R.; Liu, H.; et al. The super elongation complex drives transcriptional addiction in MYCN-amplified neuroblastoma. Sci. Adv. 2023, 9, eadf0005. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Bansal, M.; Gupta, A.; Ding, H.F. MYCN and Metabolic Reprogramming in Neuroblastoma. Cancers 2022, 14, 4113. [Google Scholar] [CrossRef]
- Upton, K.; Modi, A.; Patel, K.; Kendsersky, N.M.; Conkrite, K.L.; Sussman, R.T.; Way, G.P.; Adams, R.N.; Sacks, G.I.; Fortina, P.; et al. Epigenomic profiling of neuroblastoma cell lines. Sci. Data 2020, 7, 116. [Google Scholar] [CrossRef]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef]
- Irwin, M.S.; Naranjo, A.; Zhang, F.F.; Cohn, S.L.; London, W.B.; Gastier-Foster, J.M.; Ramirez, N.C.; Pfau, R.; Reshmi, S.; Wagner, E.; et al. Revised Neuroblastoma Risk Classification System: A Report From the Children’s Oncology Group. J. Clin. Oncol. 2021, 39, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Chamdin, A.; Jarzembowski, J.A.; Subramanian, C.; Kuick, R.; Lee, J.S.; Kwok, R.P.; Castle, V.P.; Opipari, A.W. Bcl6 is expressed in neuroblastoma: Tumor cell type-specific expression predicts outcome. Transl. Oncol. 2009, 2, 128–137. [Google Scholar] [CrossRef][Green Version]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Qing, G.; Li, B.; Vu, A.; Skuli, N.; Walton, Z.E.; Liu, X.; Mayes, P.A.; Wise, D.R.; Thompson, C.B.; Maris, J.M.; et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 2012, 22, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Mosseri, V.; London, W.B.; Maris, J.M.; Brodeur, G.M.; Attiyeh, E.; Haber, M.; Khan, J.; Nakagawara, A.; Speleman, F.; et al. Segmental chromosomal alterations have prognostic impact in neuroblastoma: A report from the INRG project. Br. J. Cancer 2012, 107, 1418–1422. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Danßmann, C.; Klironomos, F.; Winkler, A.; Fallmann, J.; Kruetzfeldt, L.M.; Szymansky, A.; Naderi, J.; Bernhart, S.H.; Grunewald, L.; et al. Defining the landscape of circular RNAs in neuroblastoma unveils a global suppressive function of MYCN. Nat. Commun. 2023, 14, 3936. [Google Scholar] [CrossRef]
- Fransson, S.; Martinez-Monleon, A.; Johansson, M.; Sjöberg, R.M.; Björklund, C.; Ljungman, G.; Ek, T.; Kogner, P.; Martinsson, T. Whole-genome sequencing of recurrent neuroblastoma reveals somatic mutations that affect key players in cancer progression and telomere maintenance. Sci. Rep. 2020, 10, 22432. [Google Scholar] [CrossRef]
- van Groningen, T.; Akogul, N.; Westerhout, E.M.; Chan, A.; Hasselt, N.E.; Zwijnenburg, D.A.; Broekmans, M.; Stroeken, P.; Haneveld, F.; Hooijer, G.K.J.; et al. A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat. Commun. 2019, 10, 1530. [Google Scholar] [CrossRef]
- Helmsauer, K.; Valieva, M.E.; Ali, S.; Chamorro González, R.; Schöpflin, R.; Röefzaad, C.; Bei, Y.; Dorado Garcia, H.; Rodriguez-Fos, E.; Puiggròs, M.; et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat. Commun. 2020, 11, 5823. [Google Scholar] [CrossRef]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef]
- Koneru, B.; Lopez, G.; Farooqi, A.; Conkrite, K.L.; Nguyen, T.H.; Macha, S.J.; Modi, A.; Rokita, J.L.; Urias, E.; Hindle, A.; et al. Telomere Maintenance Mechanisms Define Clinical Outcome in High-Risk Neuroblastoma. Cancer Res. 2020, 80, 2663–2675. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Takita, J.; Sanada, M.; Hayashi, Y. Oncogenic mutations of ALK in neuroblastoma. Cancer Sci. 2011, 102, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Trochet, D.; Bourdeaut, F.; Janoueix-Lerosey, I.; Deville, A.; de Pontual, L.; Schleiermacher, G.; Coze, C.; Philip, N.; Frébourg, T.; Munnich, A.; et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am. J. Hum. Genet. 2004, 74, 761–764. [Google Scholar] [CrossRef]
- Xia, Y.; Ye, B.; Ding, J.; Yu, Y.; Alptekin, A.; Thangaraju, M.; Prasad, P.D.; Ding, Z.C.; Park, E.J.; Choi, J.H.; et al. Metabolic Reprogramming by MYCN Confers Dependence on the Serine-Glycine-One-Carbon Biosynthetic Pathway. Cancer Res. 2019, 79, 3837–3850. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Mohammad, M.A.; Milazzo, G.; Moreno-Smith, M.; Patel, T.D.; Zorman, B.; Badachhape, A.; Hernandez, B.E.; Wolf, A.B.; Zeng, Z.; et al. MYCN-driven fatty acid uptake is a metabolic vulnerability in neuroblastoma. Nat. Commun. 2022, 13, 3728. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.F.; Duan, J.J.; Wang, J.; Li, L.; Wang, D.; Liu, X.Z.; Yang, J.; Zhang, H.R.; Lv, J.; Yang, Y.J.; et al. Inhibition of the ALDH18A1-MYCN positive feedback loop attenuates MYCN-amplified neuroblastoma growth. Sci. Transl. Med. 2020, 12, eaax8694. [Google Scholar] [CrossRef]
- Boeva, V.; Louis-Brennetot, C.; Peltier, A.; Durand, S.; Pierre-Eugène, C.; Raynal, V.; Etchevers, H.C.; Thomas, S.; Lermine, A.; Daudigeos-Dubus, E.; et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 2017, 49, 1408–1413. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Rifatbegovic, F.; Frech, C.; Abbasi, M.R.; Taschner-Mandl, S.; Weiss, T.; Schmidt, W.M.; Schmidt, I.; Ladenstein, R.; Ambros, I.M.; Ambros, P.F. Neuroblastoma cells undergo transcriptomic alterations upon dissemination into the bone marrow and subsequent tumor progression. Int. J. Cancer 2018, 142, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Monaco, G.; Lee, B.; Xu, W.; Mustafah, S.; Hwang, Y.Y.; Carré, C.; Burdin, N.; Visan, L.; Ceccarelli, M.; Poidinger, M.; et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep. 2019, 26, 1627–1640.e7. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.S.; Kuznetsov, I.B.; Zhang, S.; Song, Y.K.; Asgharzadeh, S.; Sindiri, S.; Wen, X.; Patidar, R.; Najaraj, S.; Walton, A.; et al. Clinically Relevant Cytotoxic Immune Cell Signatures and Clonal Expansion of T-Cell Receptors in High-Risk MYCN-Not-Amplified Human Neuroblastoma. Clin. Cancer Res. 2018, 24, 5673–5684. [Google Scholar] [CrossRef]
- Wang, L.; Tan, T.K.; Durbin, A.D.; Zimmerman, M.W.; Abraham, B.J.; Tan, S.H.; Ngoc, P.C.T.; Weichert-Leahey, N.; Akahane, K.; Lawton, L.N.; et al. ASCL1 is a MYCN- and LMO1-dependent member of the adrenergic neuroblastoma core regulatory circuitry. Nat. Commun. 2019, 10, 5622. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 9354. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- McLeay, R.C.; Bailey, T.L. Motif Enrichment Analysis: A unified framework and an evaluation on ChIP data. BMC Bioinform. 2010, 11, 165. [Google Scholar] [CrossRef]
- Bailey, T.L.; Gribskov, M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef]
- Michael, A.K.; Stoos, L.; Crosby, P.; Eggers, N.; Nie, X.Y.; Makasheva, K.; Minnich, M.; Healy, K.L.; Weiss, J.; Kempf, G.; et al. Cooperation between bHLH transcription factors and histones for DNA access. Nature 2023, 619, 385–393. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Tong, A.J.; Liu, X.; Thomas, B.J.; Lissner, M.M.; Baker, M.R.; Senagolage, M.D.; Allred, A.L.; Barish, G.D.; Smale, S.T. A Stringent Systems Approach Uncovers Gene-Specific Mechanisms Regulating Inflammation. Cell 2016, 165, 165–179. [Google Scholar] [CrossRef]
- Ren, P.; Yue, M.; Xiao, D.; Xiu, R.; Gan, L.; Liu, H.; Qing, G. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J. Pathol. 2015, 235, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef] [PubMed]
- Westermann, F.; Muth, D.; Benner, A.; Bauer, T.; Henrich, K.O.; Oberthuer, A.; Brors, B.; Beissbarth, T.; Vandesompele, J.; Pattyn, F.; et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008, 9, R150. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.W.; Hui, W.W.; Wang, J.J.; Yung, M.M.; Hui, L.M.; Qin, Y.; Liang, R.R.; Leung, T.H.; Xu, D.; Chan, K.K.; et al. DLX1 acts as a crucial target of FOXM1 to promote ovarian cancer aggressiveness by enhancing TGF-β/SMAD4 signaling. Oncogene 2017, 36, 1404–1416. [Google Scholar] [CrossRef]
- Suenaga, Y.; Kaneko, Y.; Matsumoto, D.; Hossain, M.S.; Ozaki, T.; Nakagawara, A. Positive auto-regulation of MYCN in human neuroblastoma. Biochem. Biophys. Res. Commun. 2009, 390, 21–26. [Google Scholar] [CrossRef]
- Sivak, L.E.; Tai, K.F.; Smith, R.S.; Dillon, P.A.; Brodeur, G.M.; Carroll, W.L. Autoregulation of the human N-myc oncogene is disrupted in amplified but not single-copy neuroblastoma cell lines. Oncogene 1997, 15, 1937–1946. [Google Scholar] [CrossRef][Green Version]
- Reiter, J.L.; Brodeur, G.M. High-resolution mapping of a 130-kb core region of the MYCN amplicon in neuroblastomas. Genomics 1996, 32, 97–103. [Google Scholar] [CrossRef]
- Schwab, M. MYCN in neuronal tumours. Cancer Lett. 2004, 204, 179–187. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, C.; Zwahlen, M.; von Feilitzen, K.; Karlsson, M.; Shi, M.; Yuan, M.; Song, X.; Li, X.; Yang, H.; et al. Systematic transcriptional analysis of human cell lines for gene expression landscape and tumor representation. Nat. Commun. 2023, 14, 5417. [Google Scholar] [CrossRef]
- Ye, B.H.; Lista, F.; Lo Coco, F.; Knowles, D.M.; Offit, K.; Chaganti, R.S.; Dalla-Favera, R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 1993, 262, 747–750. [Google Scholar] [CrossRef]
- Yang, H.; Green, M.R. Epigenetic Programing of B-Cell Lymphoma by BCL6 and Its Genetic Deregulation. Front. Cell Dev. Biol. 2019, 7, 272. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Cermak, L.; Pagan, J.K.; Rossi, M.; Martinengo, C.; di Celle, P.F.; Chapuy, B.; Shipp, M.; Chiarle, R.; Pagano, M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 2012, 481, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Batmanov, K.; Wang, W.; Bjørås, M.; Delabie, J.; Wang, J. Integrative whole-genome sequence analysis reveals roles of regulatory mutations in BCL6 and BCL2 in follicular lymphoma. Sci. Rep. 2017, 7, 7040. [Google Scholar] [CrossRef] [PubMed]
- Brescia, P.; Schneider, C.; Holmes, A.B.; Shen, Q.; Hussein, S.; Pasqualucci, L.; Basso, K.; Dalla-Favera, R. MEF2B Instructs Germinal Center Development and Acts as an Oncogene in B Cell Lymphomagenesis. Cancer Cell 2018, 34, 453–465.e9. [Google Scholar] [CrossRef]
- Cardenas, M.G.; Oswald, E.; Yu, W.; Xue, F.; MacKerell, A.D., Jr.; Melnick, A.M. The Expanding Role of the BCL6 Oncoprotein as a Cancer Therapeutic Target. Clin. Cancer Res. 2017, 23, 885–893. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Migliazza, A.; Basso, K.; Houldsworth, J.; Chaganti, R.S.; Dalla-Favera, R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood 2003, 101, 2914–2923. [Google Scholar] [CrossRef]
- Cattoretti, G.; Pasqualucci, L.; Ballon, G.; Tam, W.; Nandula, S.V.; Shen, Q.; Mo, T.; Murty, V.V.; Dalla-Favera, R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 2005, 7, 445–455. [Google Scholar] [CrossRef]
- Haniuda, K.; Fukao, S.; Kitamura, D. Metabolic Reprogramming Induces Germinal Center B Cell Differentiation through Bcl6 Locus Remodeling. Cell Rep. 2020, 33, 108333. [Google Scholar] [CrossRef]
- Geng, H.; Brennan, S.; Milne, T.A.; Chen, W.Y.; Li, Y.; Hurtz, C.; Kweon, S.M.; Zickl, L.; Shojaee, S.; Neuberg, D.; et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012, 2, 1004–1023. [Google Scholar] [CrossRef]
- Hurtz, C.; Hatzi, K.; Cerchietti, L.; Braig, M.; Park, E.; Kim, Y.M.; Herzog, S.; Ramezani-Rad, P.; Jumaa, H.; Müller, M.C.; et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J. Exp. Med. 2011, 208, 2163–2174. [Google Scholar] [CrossRef]
- Walker, S.R.; Liu, S.; Xiang, M.; Nicolais, M.; Hatzi, K.; Giannopoulou, E.; Elemento, O.; Cerchietti, L.; Melnick, A.; Frank, D.A. The transcriptional modulator BCL6 as a molecular target for breast cancer therapy. Oncogene 2015, 34, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Kume, K.; Chen, Z.; Chan, L.N.; Robinson, M.E.; Leveille, E.; Lee, J.; Cosgun, K.N.; Arce, D.F.; Khanduja, D.; Klemm, L.; et al. Divergent MYC- and BCL6-Driven Metabolic Programs Enable Dynamic Regulation of Cell Biomass in B-Cell Malignancies. Blood 2022, 140, 5900–5901. [Google Scholar] [CrossRef]
- Sommars, M.A.; Ramachandran, K.; Senagolage, M.D.; Futtner, C.R.; Germain, D.M.; Allred, A.L.; Omura, Y.; Bederman, I.R.; Barish, G.D. Dynamic repression by BCL6 controls the genome-wide liver response to fasting and steatosis. Elife 2019, 8, e43922. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Shields, J.M.; Yang, V.W. Identification of the DNA sequence that interacts with the gut-enriched Krüppel-like factor. Nucleic Acids Res. 1998, 26, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Choi, K.J.; Quan, M.D.; Sharma, S.; Sankaran, B.; Park, H.; LaGrone, A.; Kim, J.J.; MacKenzie, K.R.; Ferreon, A.C.M.; et al. Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization. Nat. Commun. 2021, 12, 5579. [Google Scholar] [CrossRef] [PubMed]
- Rowland, B.D.; Peeper, D.S. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 2006, 6, 11–23. [Google Scholar] [CrossRef]
- Gamper, A.M.; Qiao, X.; Kim, J.; Zhang, L.; DeSimone, M.C.; Rathmell, W.K.; Wan, Y. Regulation of KLF4 turnover reveals an unexpected tissue-specific role of pVHL in tumorigenesis. Mol. Cell 2012, 45, 233–243. [Google Scholar] [CrossRef]
- Moon, J.S.; Kim, H.E.; Koh, E.; Park, S.H.; Jin, W.J.; Park, B.W.; Park, S.W.; Kim, K.S. Krüppel-like factor 4 (KLF4) activates the transcription of the gene for the platelet isoform of phosphofructokinase (PFKP) in breast cancer. J. Biol. Chem. 2011, 286, 23808–23816. [Google Scholar] [CrossRef]
- Shi, M.; Cui, J.; Du, J.; Wei, D.; Jia, Z.; Zhang, J.; Zhu, Z.; Gao, Y.; Xie, K. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin. Cancer Res. 2014, 20, 4370–4380. [Google Scholar] [CrossRef]
- Chen, X.; Ding, X.; Wu, Q.; Qi, J.; Zhu, M.; Miao, C. Monomethyltransferase SET8 facilitates hepatocellular carcinoma growth by enhancing aerobic glycolysis. Cell Death Dis. 2019, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- Oyinlade, O.; Wei, S.; Lal, B.; Laterra, J.; Zhu, H.; Goodwin, C.R.; Wang, S.; Ma, D.; Wan, J.; Xia, S. Targeting UDP-α-D-glucose 6-dehydrogenase inhibits glioblastoma growth and migration. Oncogene 2018, 37, 2615–2629. [Google Scholar] [CrossRef] [PubMed]
- Cotterman, R.; Knoepfler, P.S. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS ONE 2009, 4, e5799. [Google Scholar] [CrossRef] [PubMed]
- Shum, C.K.; Lau, S.T.; Tsoi, L.L.; Chan, L.K.; Yam, J.W.; Ohira, M.; Nakagawara, A.; Tam, P.K.; Ngan, E.S. Krüppel-like factor 4 (KLF4) suppresses neuroblastoma cell growth and determines non-tumorigenic lineage differentiation. Oncogene 2013, 32, 4086–4099. [Google Scholar] [CrossRef] [PubMed]
- Panganiban, G.; Rubenstein, J.L. Developmental functions of the Distal-less/Dlx homeobox genes. Development 2002, 129, 4371–4386. [Google Scholar] [CrossRef]
- Giorgianni, M.W.; Mann, R.S. Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless. Dev. Cell 2011, 20, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Gebelein, B.; McKay, D.J.; Mann, R.S. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 2004, 431, 653–659. [Google Scholar] [CrossRef]
- Agelopoulos, M.; McKay, D.J.; Mann, R.S. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012, 1, 350–359. [Google Scholar] [CrossRef]
- Agelopoulos, M.; McKay, D.J.; Mann, R.S. cgChIP: A cell type- and gene-specific method for chromatin analysis. Methods Mol. Biol. 2014, 1196, 291–306. [Google Scholar] [CrossRef]
- Tan, Y.; Testa, J.R. DLX Genes: Roles in Development and Cancer. Cancers 2021, 13, 3005. [Google Scholar] [CrossRef]
- Goel, S.; Bhatia, V.; Kundu, S.; Biswas, T.; Carskadon, S.; Gupta, N.; Asim, M.; Morrissey, C.; Palanisamy, N.; Ateeq, B. Transcriptional network involving ERG and AR orchestrates Distal-less homeobox-1 mediated prostate cancer progression. Nat. Commun. 2021, 12, 5325. [Google Scholar] [CrossRef]
- Liang, M.; Sun, Y.; Yang, H.L.; Zhang, B.; Wen, J.; Shi, B.K. DLX1, a binding protein of beta-catenin, promoted the growth and migration of prostate cancer cells. Exp. Cell Res. 2018, 363, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Takeshita, K.; Imai, Y.; Kumano, K.; Kurokawa, M.; Masuda, S.; Shimizu, K.; Nakamura, S.; Ruddle, F.H.; Hirai, H. Homeoprotein DLX-1 interacts with Smad4 and blocks a signaling pathway from activin A in hematopoietic cells. Proc. Natl. Acad. Sci. USA 2003, 100, 15577–15582. [Google Scholar] [CrossRef] [PubMed]
- Starkova, J.; Gadgil, S.; Qiu, Y.H.; Zhang, N.; Hermanova, I.; Kornblau, S.M.; Drabkin, H.A. Up-regulation of homeodomain genes, DLX1 and DLX2, by FLT3 signaling. Haematologica 2011, 96, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Stirewalt, D.L.; Radich, J.P. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer 2003, 3, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Mimura, S.; Suga, M.; Okada, K.; Kinehara, M.; Nikawa, H.; Furue, M.K. Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells. Int. J. Dev. Biol. 2016, 60, 21–28. [Google Scholar] [CrossRef]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Hybertson, B.M.; Gao, B.; Bose, S.K.; McCord, J.M. Oxidative stress in health and disease: The therapeutic potential of Nrf2 activation. Mol. Asp. Med. 2011, 32, 234–246. [Google Scholar] [CrossRef]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef]
- Panieri, E.; Saso, L. Potential Applications of NRF2 Inhibitors in Cancer Therapy. Oxid. Med. Cell Longev. 2019, 2019, 8592348. [Google Scholar] [CrossRef]
- Weiss-Sadan, T.; Ge, M.; Hayashi, M.; Gohar, M.; Yao, C.H.; de Groot, A.; Harry, S.; Carlin, A.; Fischer, H.; Shi, L.; et al. NRF2 activation induces NADH-reductive stress, providing a metabolic vulnerability in lung cancer. Cell Metab. 2023, 35, 487–503.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Fahl, W.E. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem. Biophys. Res. Commun. 2001, 289, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Pouremamali, F.; Pouremamali, A.; Dadashpour, M.; Soozangar, N.; Jeddi, F. An update of Nrf2 activators and inhibitors in cancer prevention/promotion. Cell Commun. Signal. 2022, 20, 100. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Sun, Z.; Villeneuve, N.F.; Zhang, S.; Zhao, F.; Li, Y.; Chen, W.; Yi, X.; Zheng, W.; Wondrak, G.T.; et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 2008, 29, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- de Miranda Ramos, V.; Zanotto-Filho, A.; de Bittencourt Pasquali, M.A.; Klafke, K.; Gasparotto, J.; Dunkley, P.; Gelain, D.P.; Moreira, J.C.F. NRF2 Mediates Neuroblastoma Proliferation and Resistance to Retinoic Acid Cytotoxicity in a Model of In Vitro Neuronal Differentiation. Mol. Neurobiol. 2016, 53, 6124–6135. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, A.L.; Piras, S.; Passalacqua, M.; Domenicotti, C.; Parodi, A.; Fenoglio, D.; Pronzato, M.A.; Marinari, U.M.; Moretta, L.; Traverso, N.; et al. HO-1 up-regulation: A key point in high-risk neuroblastoma resistance to bortezomib. Biochim. Biophys. Acta 2014, 1842, 613–622. [Google Scholar] [CrossRef]
- Soldaini, E.; John, S.; Moro, S.; Bollenbacher, J.; Schindler, U.; Leonard, W.J. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell Biol. 2000, 20, 389–401. [Google Scholar] [CrossRef]
- Lin, J.X.; Li, P.; Liu, D.; Jin, H.T.; He, J.; Ata Ur Rasheed, M.; Rochman, Y.; Wang, L.; Cui, K.; Liu, C.; et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 2012, 36, 586–599. [Google Scholar] [CrossRef]
- Xiong, H.; Su, W.Y.; Liang, Q.C.; Zhang, Z.G.; Chen, H.M.; Du, W.; Chen, Y.X.; Fang, J.Y. Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Lab. Investig. 2009, 89, 717–725. [Google Scholar] [CrossRef]
- Coffer, P.J.; Koenderman, L.; de Groot, R.P. The role of STATs in myeloid differentiation and leukemia. Oncogene 2000, 19, 2511–2522. [Google Scholar] [CrossRef]
- Cholez, E.; Debuysscher, V.; Bourgeais, J.; Boudot, C.; Leprince, J.; Tron, F.; Brassart, B.; Regnier, A.; Bissac, E.; Pecnard, E.; et al. Evidence for a protective role of the STAT5 transcription factor against oxidative stress in human leukemic pre-B cells. Leukemia 2012, 26, 2390–2397. [Google Scholar] [CrossRef]
- Chueh, F.Y.; Leong, K.F.; Yu, C.L. Mitochondrial translocation of signal transducer and activator of transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated cells. Biochem. Biophys. Res. Commun. 2010, 402, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Li, Z.; Xiao, L.; Hu, W.; Zhang, L.; Xie, B.; Zhou, Q.; He, J.; Qiu, Y.; Wen, M.; et al. Glutamine Synthetase Promotes Radiation Resistance via Facilitating Nucleotide Metabolism and Subsequent DNA Damage Repair. Cell Rep. 2019, 28, 1136–1143.e4. [Google Scholar] [CrossRef] [PubMed]
- Georganta, E.M.; Tsoutsi, L.; Gaitanou, M.; Georgoussi, Z. δ-opioid receptor activation leads to neurite outgrowth and neuronal differentiation via a STAT5B-Gαi/o pathway. J. Neurochem. 2013, 127, 329–341. [Google Scholar] [CrossRef]
- Pallaki, P.; Georganta, E.M.; Serafimidis, I.; Papakonstantinou, M.P.; Papanikolaou, V.; Koutloglou, S.; Papadimitriou, E.; Agalou, A.; Tserga, A.; Simeonof, A.; et al. A novel regulatory role of RGS4 in STAT5B activation, neurite outgrowth and neuronal differentiation. Neuropharmacology 2017, 117, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Sanaei, M.; Kavoosi, F. The Effect of 5-aza,2’-deoxyCytidine (5 AZA CdR or Decitabine) on Extrinsic, Intrinsic, and JAK/STAT Pathways in Neuroblastoma and Glioblastoma Cells Lines. Asian Pac. J. Cancer Prev. 2023, 24, 1841–1854. [Google Scholar] [CrossRef] [PubMed]
- Agelopoulos, M.; Thanos, D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006, 25, 4843–4853. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G. A homoeotic mutation transforming leg to antenna in Drosophila. Nature 1981, 292, 635–638. [Google Scholar] [CrossRef]
- Thng, D.K.H.; Toh, T.B.; Chow, E.K. Capitalizing on Synthetic Lethality of MYC to Treat Cancer in the Digital Age. Trends Pharmacol. Sci. 2021, 42, 166–182. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pouliou, M.; Koutsi, M.A.; Champezou, L.; Giannopoulou, A.-I.; Vatsellas, G.; Piperi, C.; Agelopoulos, M. MYCN Amplifications and Metabolic Rewiring in Neuroblastoma. Cancers 2023, 15, 4803. https://doi.org/10.3390/cancers15194803
Pouliou M, Koutsi MA, Champezou L, Giannopoulou A-I, Vatsellas G, Piperi C, Agelopoulos M. MYCN Amplifications and Metabolic Rewiring in Neuroblastoma. Cancers. 2023; 15(19):4803. https://doi.org/10.3390/cancers15194803
Chicago/Turabian StylePouliou, Marialena, Marianna A. Koutsi, Lydia Champezou, Angeliki-Ioanna Giannopoulou, Giannis Vatsellas, Christina Piperi, and Marios Agelopoulos. 2023. "MYCN Amplifications and Metabolic Rewiring in Neuroblastoma" Cancers 15, no. 19: 4803. https://doi.org/10.3390/cancers15194803
APA StylePouliou, M., Koutsi, M. A., Champezou, L., Giannopoulou, A.-I., Vatsellas, G., Piperi, C., & Agelopoulos, M. (2023). MYCN Amplifications and Metabolic Rewiring in Neuroblastoma. Cancers, 15(19), 4803. https://doi.org/10.3390/cancers15194803








