Critical Role of Novel O-GlcNAcylation of S550 and S551 on the p65 Subunit of NF-κB in Pancreatic Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Liquid Chromatography-Tandem Mass Spectrometric Analysis
2.2. Cell Lines and Transfections
2.3. Western Blotting and Antibodies
2.4. Immunofluorescence
2.5. Luciferase Assays
2.6. Co-Immunoprecipitations
2.7. Cell Proliferation and 3D Growth Assays
2.8. Migration Assay
2.9. Quantitative PCR (qPCR)
2.10. Ingenuity Pathway Analysis (IPA)
3. Results
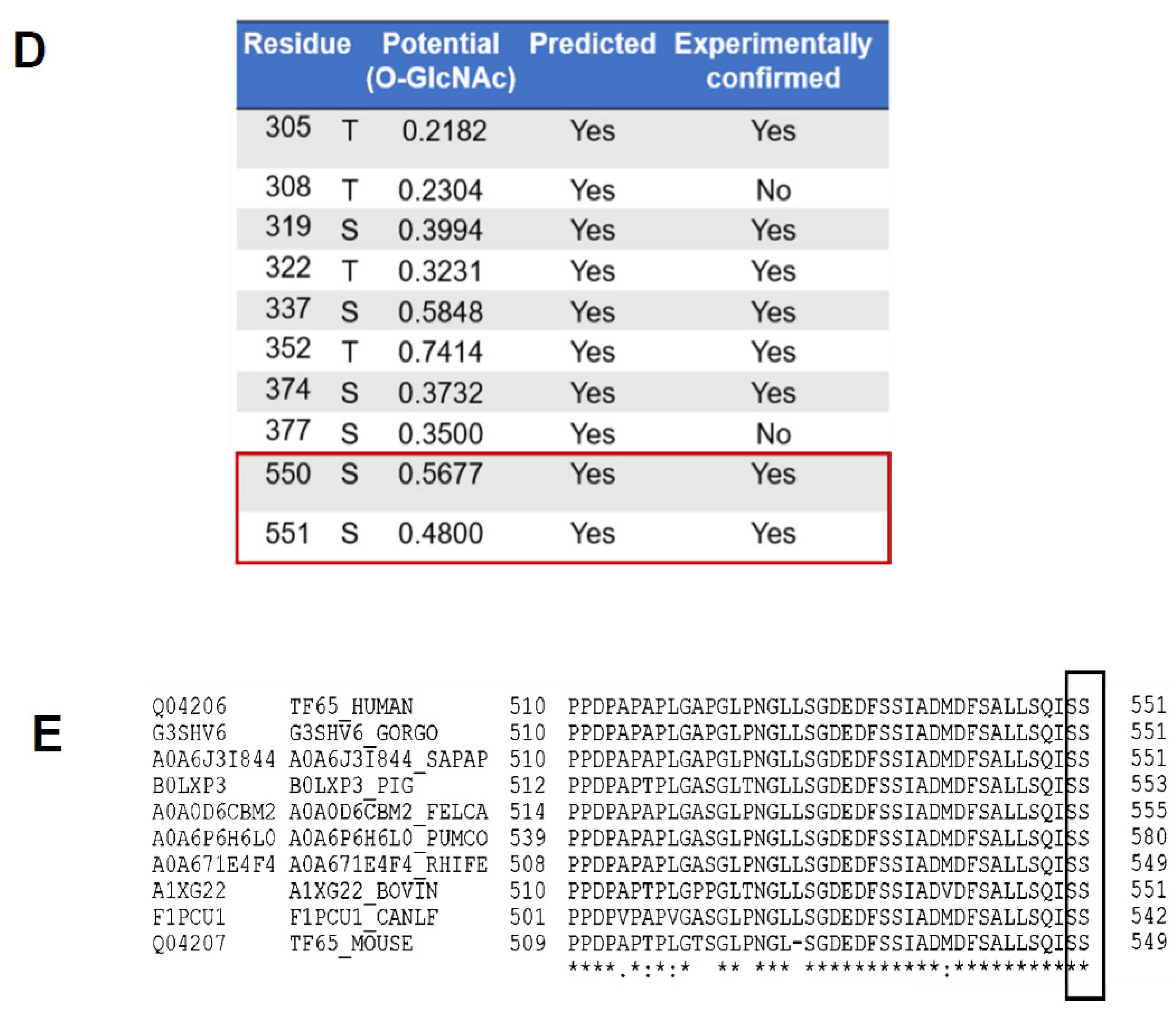
3.1. The p65 Subunit of NF-κB Is O-GlcNAcylated in the Transcriptional Activating Domain at S550 and S551
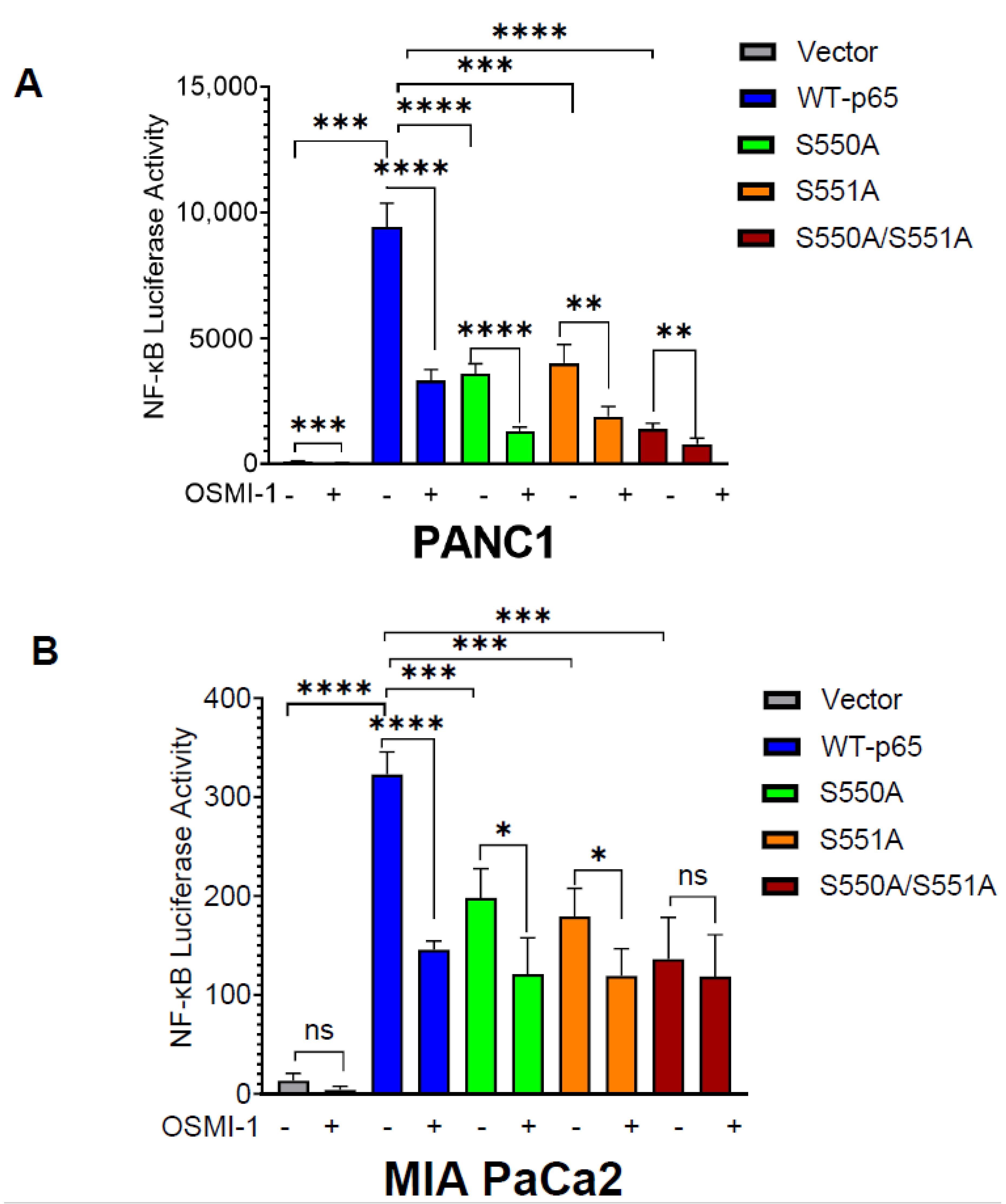
3.2. O-GlcNAcylation of p65 at S550 and S551 Is Important for NF-κB Transcriptional Activity
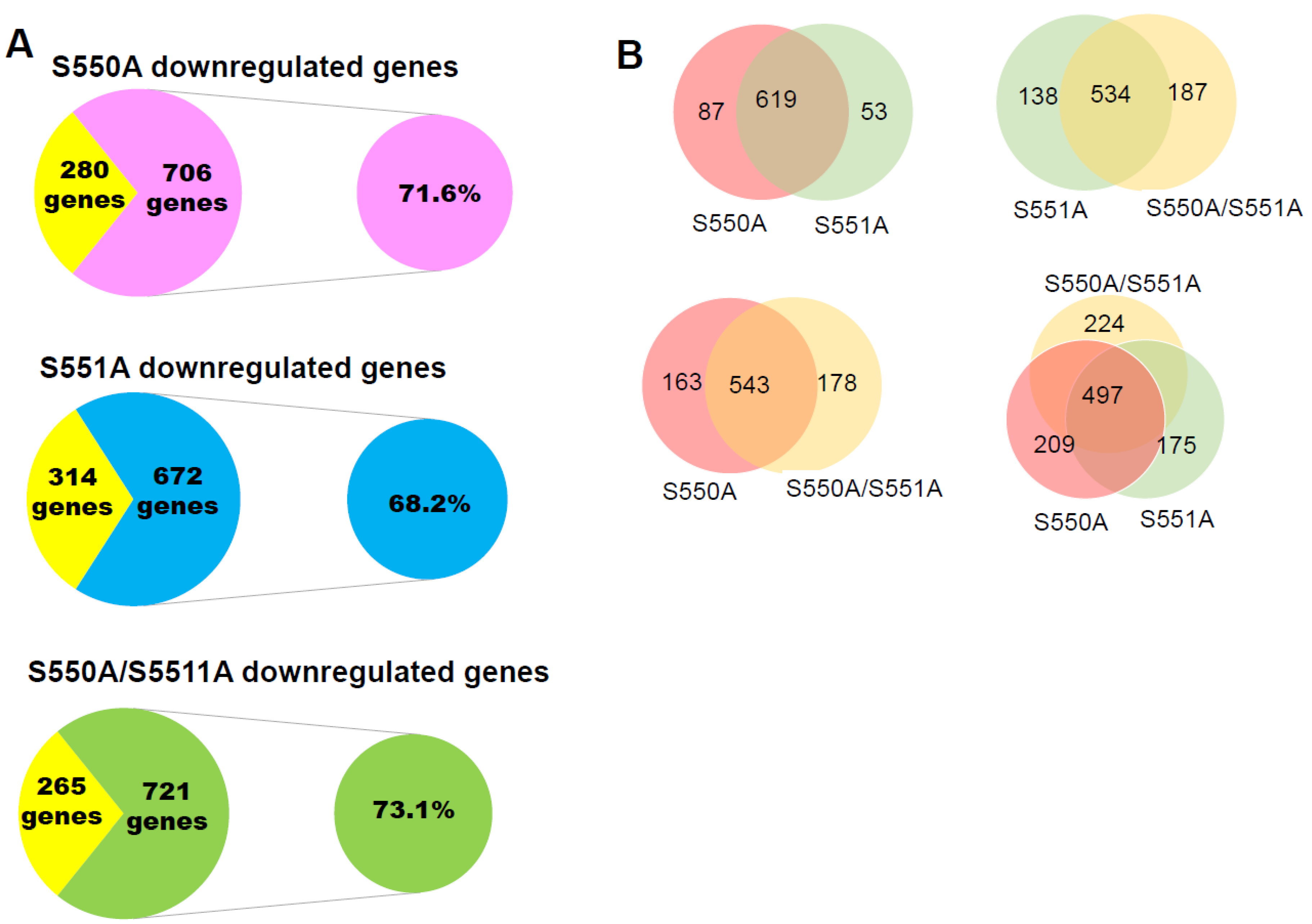
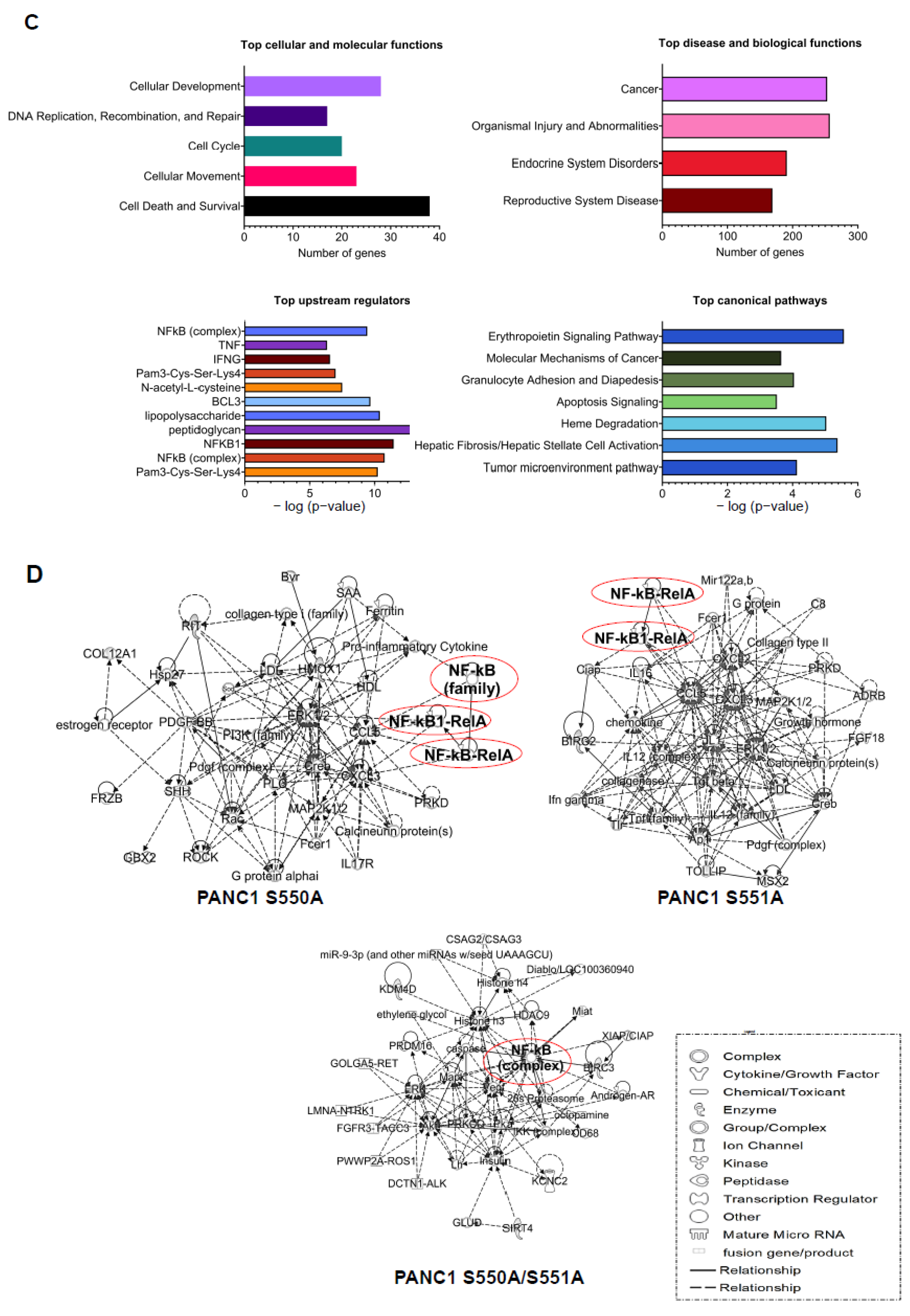
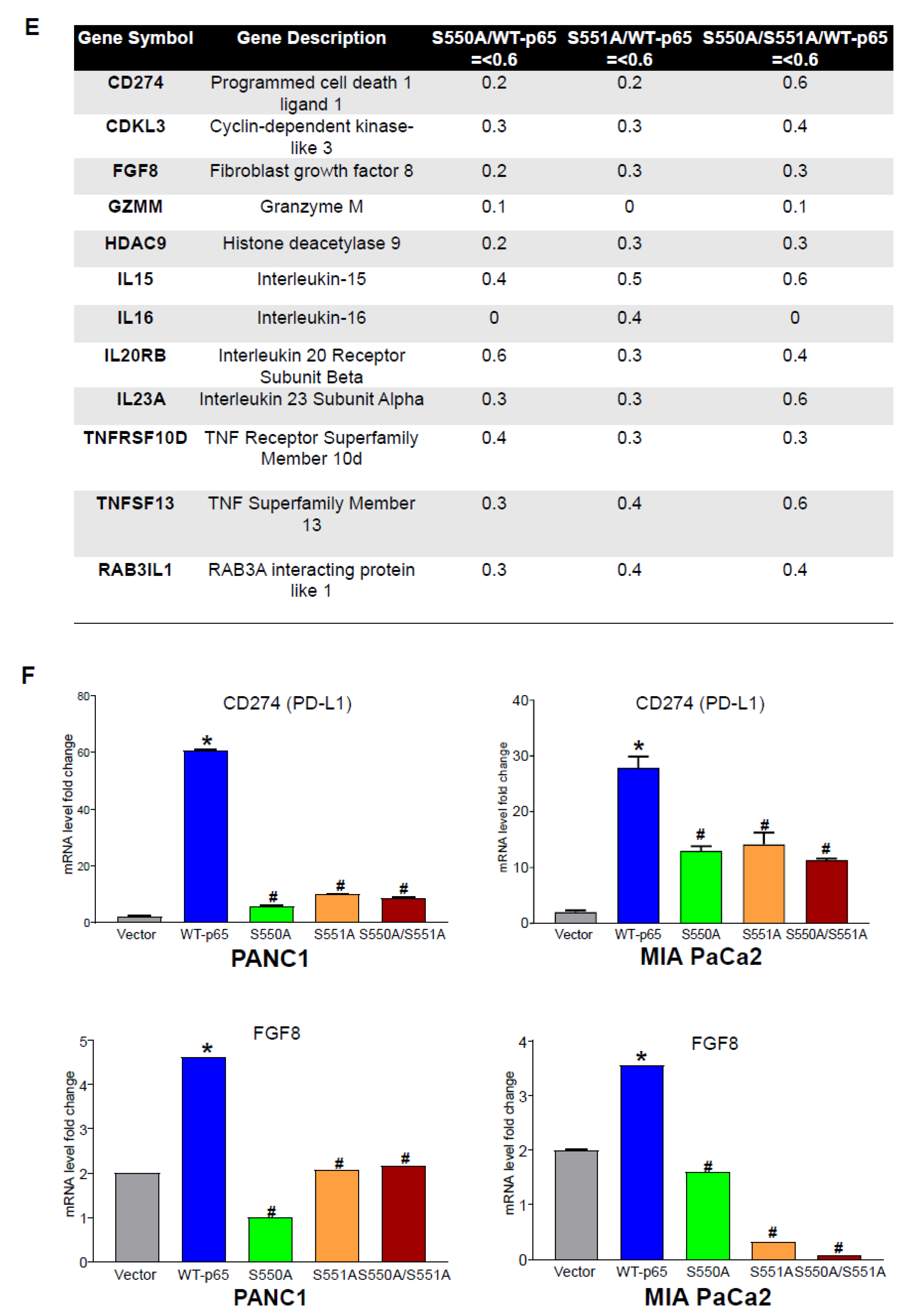
3.3. The S550A and S551A mutations of p65 Downregulate a Subset of NF-κB Target Genes
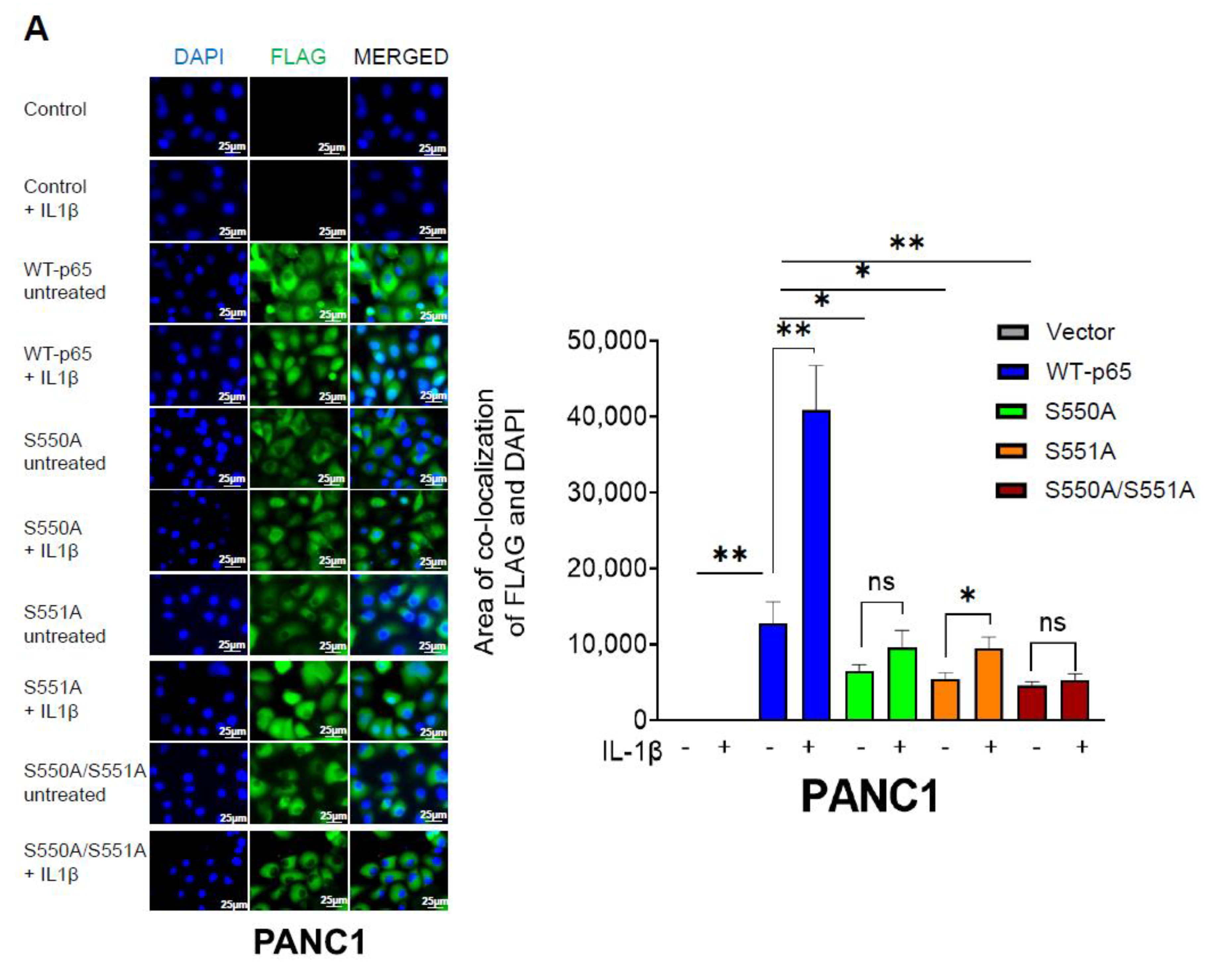
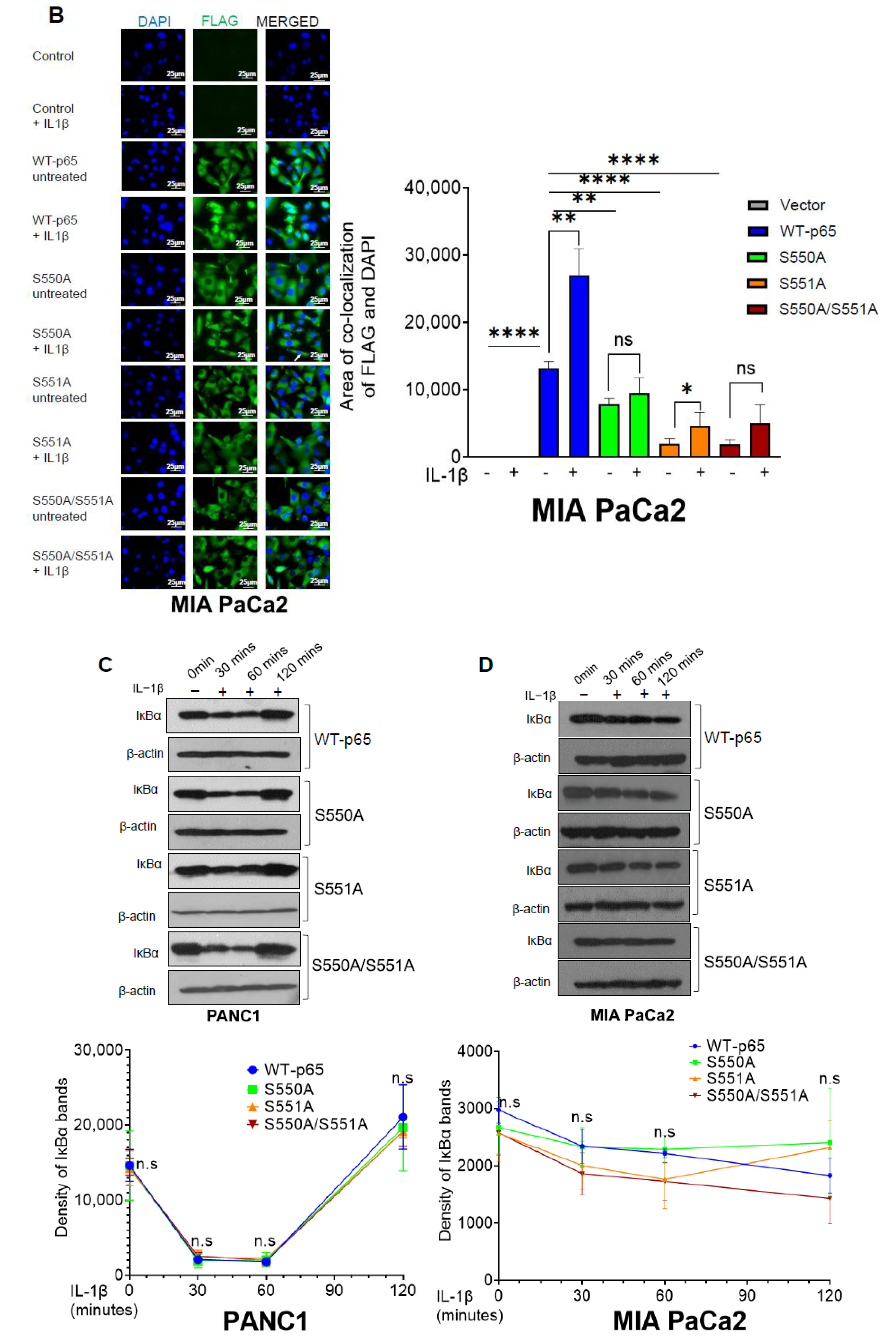
3.4. The S550A and S551A Mutations of p65 Reduce NF-ĸB Nuclear Translocation and Does Not Affect IĸBα Degradation
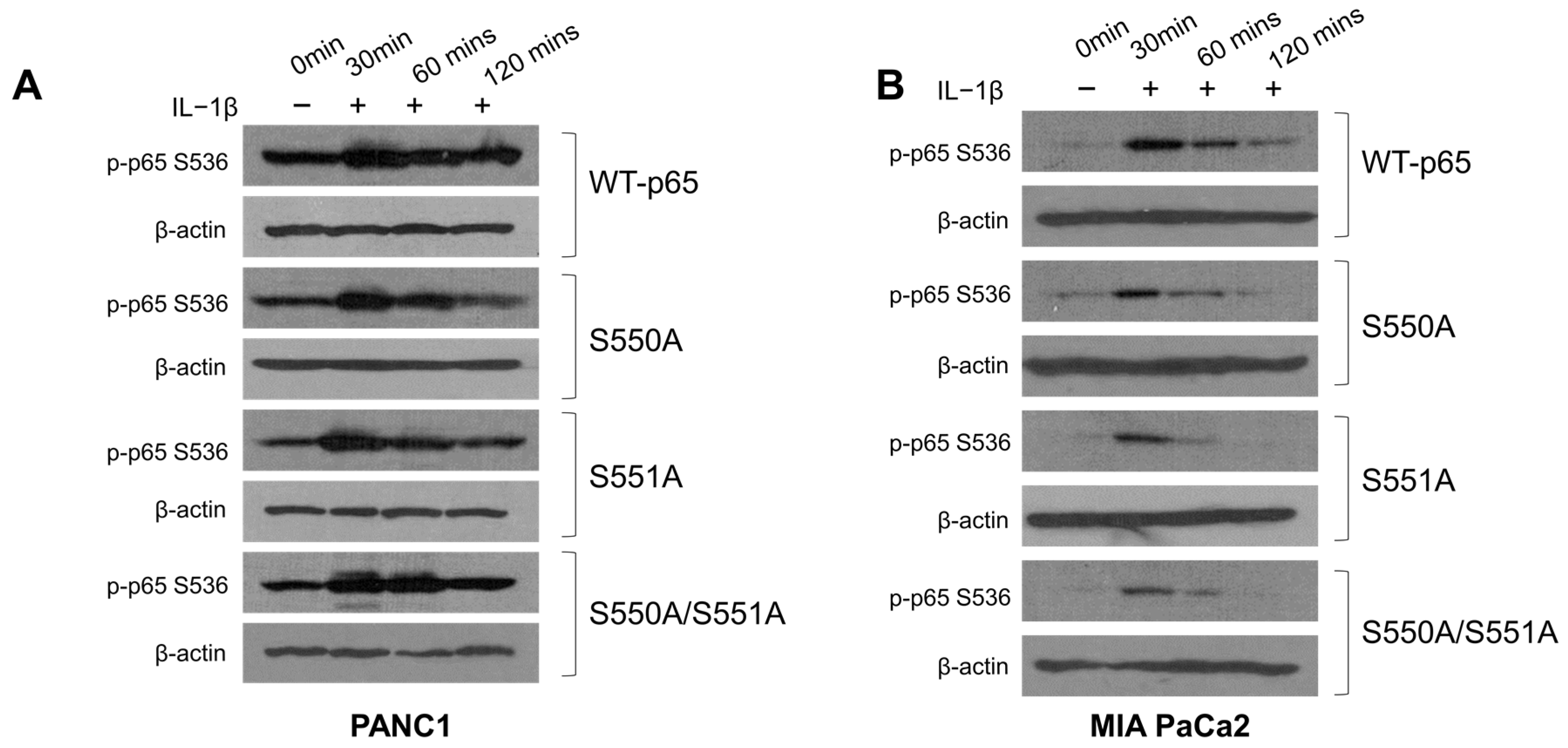
3.5. S550A and S551A Mutations of p65 May Compromise S536 Phosphorylation in PDAC
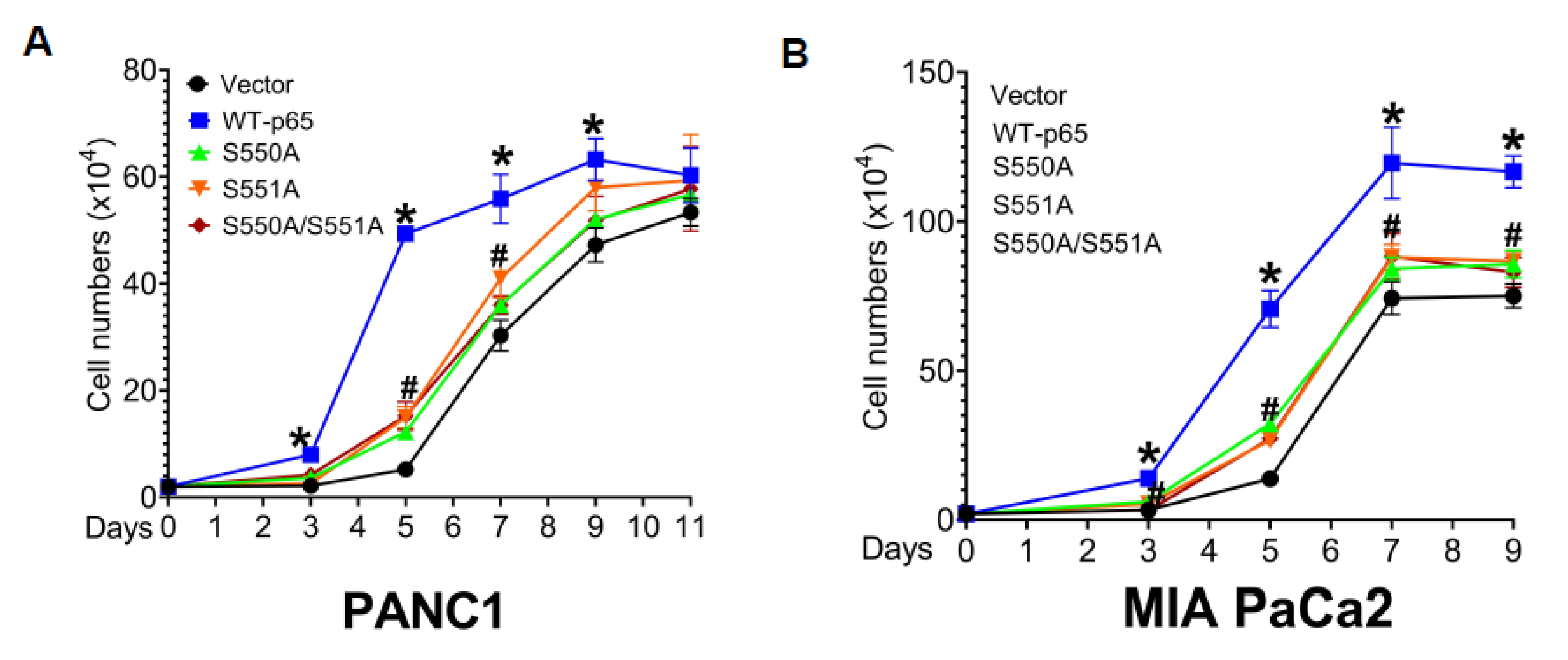
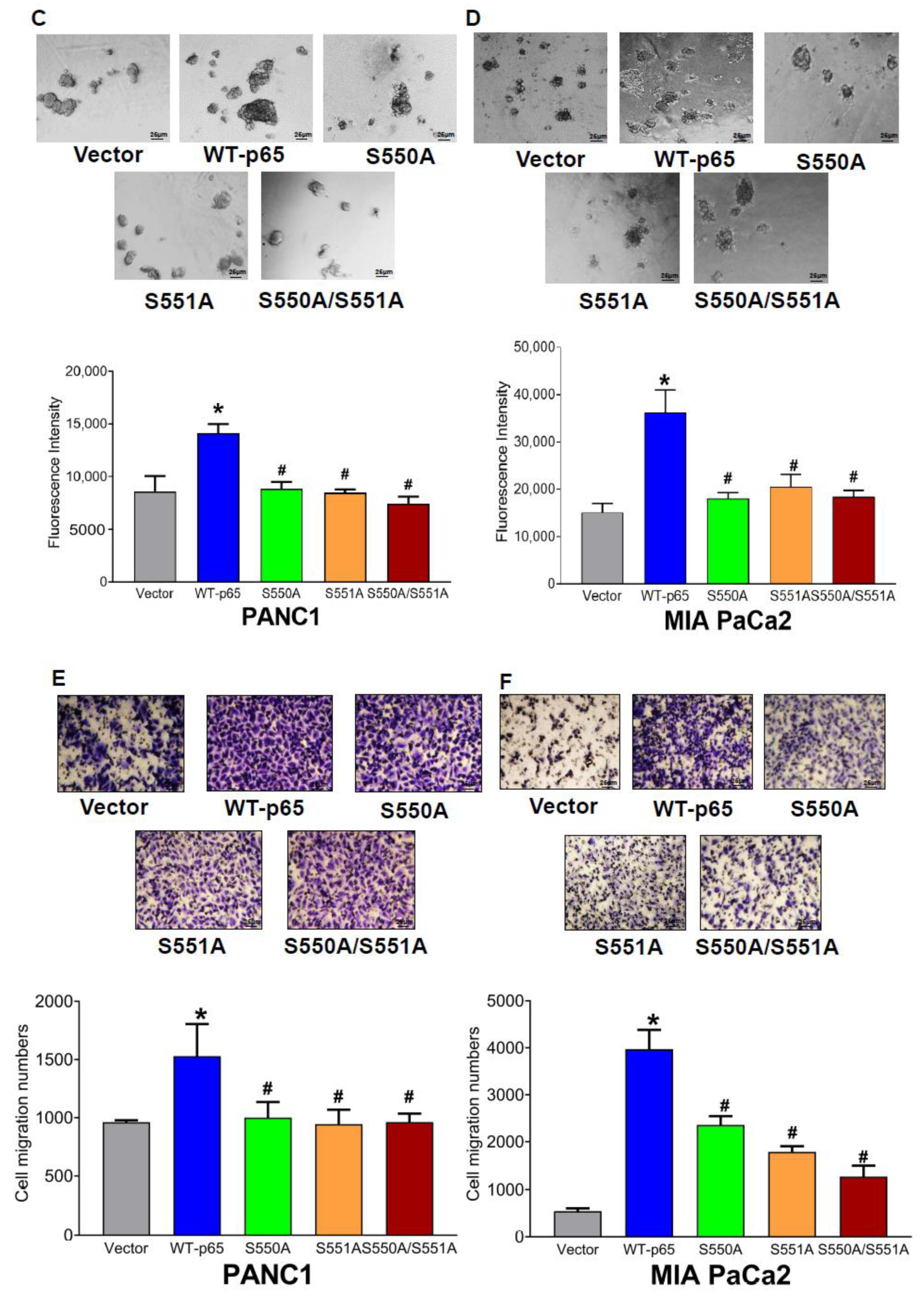
3.6. The S550A and S551A Mutations of p65 Decreases Cell Proliferation, 3D Growth, and Cell Migration in PDAC
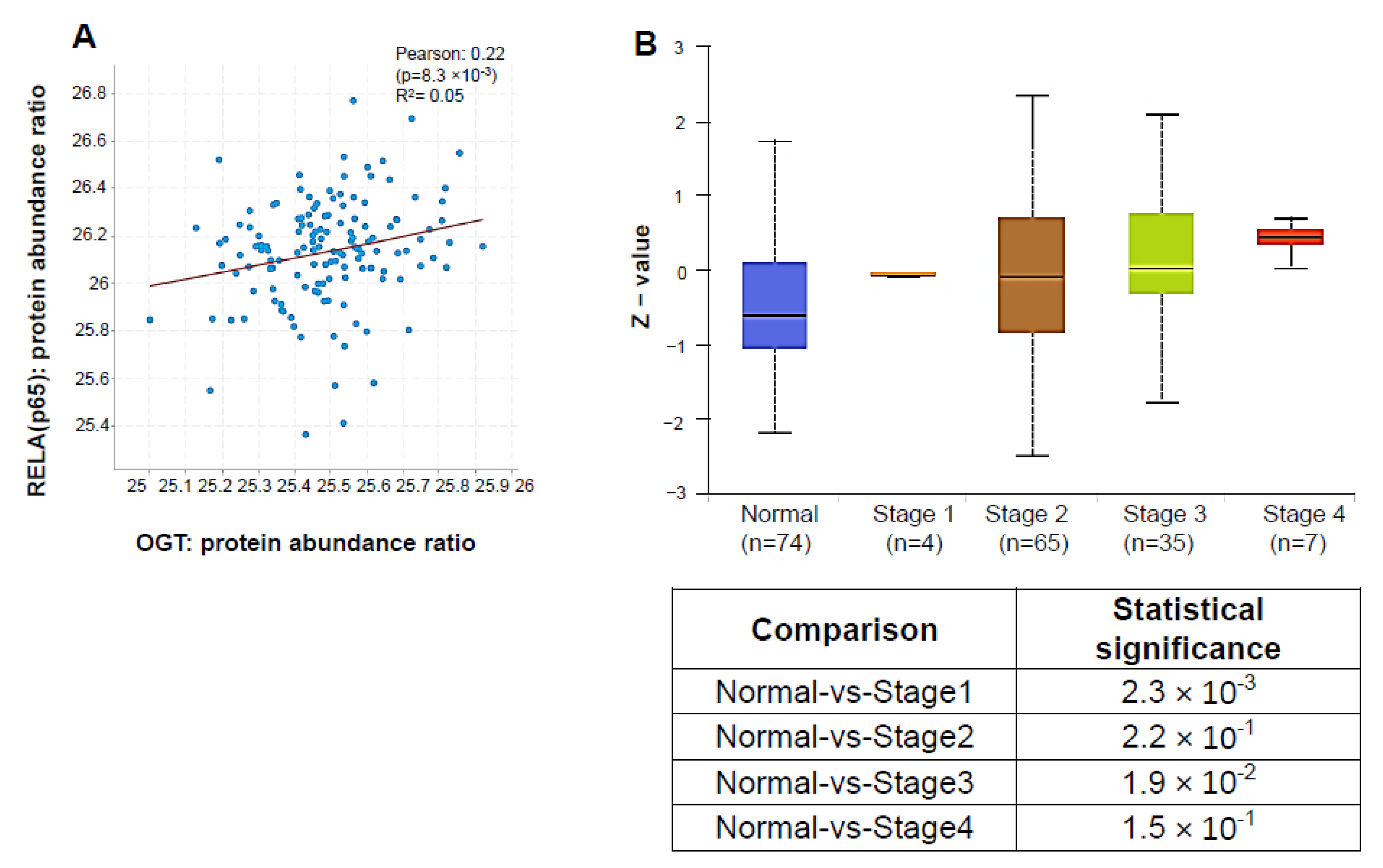
3.7. OGT Protein Abundance Positively Correlates with p65 and Is Elevated in PDAC Patients
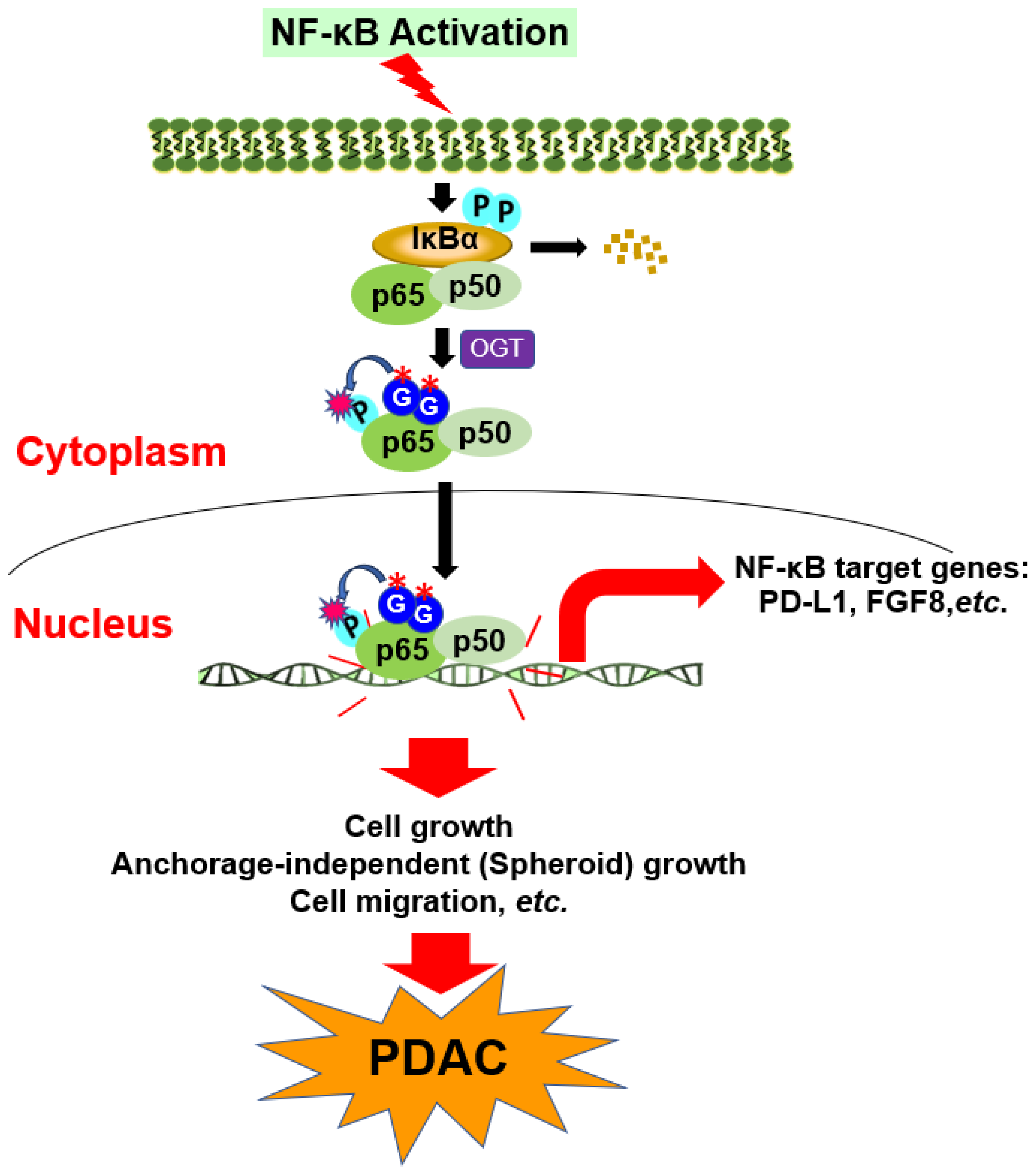
3.8. Hypothetical Model
4. Discussion
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat. Oncol. 2019, 14, 141. [Google Scholar] [CrossRef]
- Roth, M.T.; Cardin, D.B.; Berlin, J.D. Recent advances in the treatment of pancreatic cancer. F1000Research 2020, 9, F1000 Faculty Rev-131. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, L.; Mundade, R.; Korc, M.; Loehrer, P.J.; Lu, T. Critical role of NF-κB in pancreatic cancer. Oncotarget 2014, 5, 10969–10975. [Google Scholar] [CrossRef]
- Motolani, A.A.; Martin, M.; Sun, M.; Lu, T. Phosphorylation of the Regulators, a Complex Facet of NF-κB Signaling in Cancer. Biomolecules 2021, 11, 15. [Google Scholar] [CrossRef]
- Motolani, A.; Martin, M.; Sun, M.; Lu, T. NF-κB and Cancer Therapy Drugs. In Reference Module in Biomedical Sciences; Elsevier: Oxford, UK, 2021; pp. 351–363. [Google Scholar] [CrossRef]
- Perkins, N.D. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene 2006, 25, 51. [Google Scholar] [CrossRef]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wei, H.; Prabhu, L.; Zhao, W.; Martin, M.; Hartley, A.-V.; Lu, T. Role of Novel Serine 316 Phosphorylation of the p65 Subunit of NF-κB in Differential Gene Regulation. J. Biol. Chem. 2015, 290, 20336–20347. [Google Scholar] [CrossRef]
- Wei, H.; Wang, B.; Miyagi, M.; She, Y.; Gopalan, B.; Huang, D.-B.; Ghosh, G.; Stark, G.R.; Lu, T. PRMT5 dimethylates R30 of the p65 subunit to activate NF- B. Proc. Natl. Acad. Sci. USA 2013, 110, 13516–13521. [Google Scholar] [CrossRef]
- Hart, G.W.; Akimoto, Y. The O-GlcNAc Modification. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. Available online: http://www.ncbi.nlm.nih.gov/books/NBK1954/ (accessed on 7 September 2022).
- Ferrer, C.M.; Sodi, V.L.; Reginato, M.J. O-GlcNAcylation in Cancer Biology: Linking Metabolism and Signaling. J. Mol. Biol. 2016, 428, 3282–3294. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Vocadlo, D.J.; Vosseller, K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem. 2013, 288, 15121–15130. [Google Scholar] [CrossRef]
- O’Shea, J.M.; Perkins, N.D. Regulation of the RelA (p65) transactivation domain. Biochem. Soc. Trans. 2008, 36 Pt 4, 603–608. [Google Scholar] [CrossRef]
- Hartley, A.-V.; Wang, B.; Jiang, G.; Wei, H.; Sun, M.; Prabhu, L.; Martin, M.; Safa, A.; Sun, S.; Liu, Y.; et al. Regulation of a PRMT5/NF-κB Axis by Phosphorylation of PRMT5 at Serine 15 in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 3684. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Sathe, S.S.; Sizemore, N.; Li, X.; Vithalani, K.; Commane, M.; Swiatkowski, S.M.; Stark, G.R. Mutant human cells with constitutive activation of NF-κB. Proc. Natl. Acad. Sci. USA 2004, 101, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2022, 2002, 310–322. [Google Scholar]
- Allison, D.F.; Wamsley, J.J.; Kumar, M.; Li, D.; Gray, L.G.; Hart, G.W.; Jones, D.R.; Mayo, M.W. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl. Acad. Sci. USA 2012, 109, 16888–16893. [Google Scholar] [CrossRef]
- Yang, W.H.; Park, S.Y.; Nam, H.W.; Kim, D.H.; Kang, J.G.; Kang, E.S.; Kim, Y.S.; Lee, H.C.; Kim, K.S.; Cho, J.W. NFκB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 17345–17350. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nature Reviews. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Müerköster, S.; Arlt, A.; Gehrz, A.; Vorndamm, J.; Witt, M.; Grohmann, F.; Fölsch, U.R.; Schäfer, H. Autokrine IL-1β-Sekretion führt zuerhöhter NF-κB-Aktivität und zu Chemoresistenz in Pankreaskarzinomzellen in vivo. Med. Klin. 2004, 99, 185–190. [Google Scholar] [CrossRef]
- Ortiz-Meoz, R.F.; Jiang, J.; Lazarus, M.B.; Orman, M.; Janetzko, J.; Fan, C.; Duveau, D.Y.; Tan, Z.-W.; Thomas, C.J.; Walker, S. A Small Molecule That Inhibits OGT Activity in Cells. ACS Chem. Biol. 2015, 10, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Shimoji, S.; Hart, G.W. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010, 584, 2526–2538. [Google Scholar] [CrossRef] [PubMed]
- Christian, F.; Smith, E.L.; Carmody, R.J. The Regulation of NF-κB Subunits by Phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef]
- Mori, S.; Chang, J.T.; Andrechek, E.R.; Matsumura, N.; Baba, T.; Yao, G.; Kim, J.W.; Gatza, M.; Murphy, S.; Nevins, J.R. An Anchorage-Independent Cell Growth Signature Identifies Tumors with Metastatic Potential. Oncogene 2009, 28, 2796–2805. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-H.; Weng, C.-L.; Lin, K.-I. O-GlcNAcylation and its role in the immune system. J. Biomed. Sci. 2020, 27, 57. [Google Scholar] [CrossRef]
- Sharma, N.S.; Gupta, V.K.; Dauer, P.; Kesh, K.; Hadad, R.; Giri, B.; Chandra, A.; Dudeja, V.; Slawson, C.; Banerjee, S.; et al. O-GlcNAc modification of Sox2 regulates self-renewal in pancreatic cancer by promoting its stability. Theranostics 2019, 9, 3410–3424. [Google Scholar] [CrossRef]
- Huang, B.; Yang, X.-D.; Lamb, A.; Chen, L.-F. Posttranslational modifications of NF-κB: Another layer of regulation for NF-κB signaling pathway. Cell. Signal. 2010, 22, 1282–1290. [Google Scholar] [CrossRef]
- Li, Y.; Xie, M.; Men, L.; Du, J. O-GlcNAcylation in immunity and inflammation: An intricate system (Review). Int. J. Mol. Med. 2019, 44, 363–374. [Google Scholar] [CrossRef]
- Torres, C.R.; Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar] [CrossRef]
- Zhang, D.; Cai, Y.; Chen, M.; Gao, L.; Shen, Y.; Huang, Z. OGT-mediated O-GlcNAcylation promotes NF-κB activation and inflammation in acute pancreatitis. Inflamm. Res. 2015, 64, 943–952. [Google Scholar] [CrossRef]
- Ali, A.; Kim, S.H.; Kim, M.J.; Choi, M.Y.; Kang, S.S.; Cho, G.J.; Kim, Y.S.; Choi, J.-Y.; Choi, W.S. O-GlcNAcylation of NF-κB Promotes Lung Metastasis of Cervical Cancer Cells via Upregulation of CXCR4 Expression. Mol. Cells 2017, 40, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Phoomak, C.; Vaeteewoottacharn, K.; Sawanyawisuth, K.; Seubwai, W.; Wongkham, C.; Silsirivanit, A.; Wongkham, S. Mechanistic insights of O-GlcNAcylation that promote progression of cholangiocarcinoma cells via nuclear translocation of NF-κB. Sci. Rep. 2016, 6, 27853. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, P.; Clark, P.M.; Mason, D.E.; Peters, E.C.; Hsieh-Wilson, L.C.; Baltimore, D. Activation of the Transcriptional Function of the NF-κB Protein c-Rel by O-GlcNAc Glycosylation. Sci. Signal. 2013, 6, ra75. [Google Scholar] [CrossRef]
- Ma, Z.; Chalkley, R.J.; Vosseller, K. Hyper-O-GlcNAcylation activates nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) signaling through interplay with phosphorylation and acetylation. J. Biol. Chem. 2017, 292, 9150–9163. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Ding, D.; Yan, Y.; Li, H.; Wang, B.; Ma, L.; Ye, Z.; Ma, T.; Wu, Q.; Rodrigues, D.N.; et al. Phosphorylated RB Promotes Cancer Immunity by Inhibiting NF-κB Activation and PD-L1 Expression. Mol. Cell 2019, 73, 22–35.e6. [Google Scholar] [CrossRef]
- Jomrich, G.; Wilfing, L.; Radosavljevic, S.; Parak, A.; Winkler, D.; Timelthaler, G.; Schindl, M.; Schoppmann, S.F.; Klösch, B. Fibroblast growth factor 8 overexpression is predictive of poor prognosis in pancreatic ductal adenocarcinoma. Eur. Surg. 2020, 52, 282–289. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motolani, A.; Martin, M.; Wang, B.; Jiang, G.; Alipourgivi, F.; Huang, X.; Safa, A.; Liu, Y.; Lu, T. Critical Role of Novel O-GlcNAcylation of S550 and S551 on the p65 Subunit of NF-κB in Pancreatic Cancer. Cancers 2023, 15, 4742. https://doi.org/10.3390/cancers15194742
Motolani A, Martin M, Wang B, Jiang G, Alipourgivi F, Huang X, Safa A, Liu Y, Lu T. Critical Role of Novel O-GlcNAcylation of S550 and S551 on the p65 Subunit of NF-κB in Pancreatic Cancer. Cancers. 2023; 15(19):4742. https://doi.org/10.3390/cancers15194742
Chicago/Turabian StyleMotolani, Aishat, Matthew Martin, Benlian Wang, Guanglong Jiang, Faranak Alipourgivi, Xiumei Huang, Ahmad Safa, Yunlong Liu, and Tao Lu. 2023. "Critical Role of Novel O-GlcNAcylation of S550 and S551 on the p65 Subunit of NF-κB in Pancreatic Cancer" Cancers 15, no. 19: 4742. https://doi.org/10.3390/cancers15194742
APA StyleMotolani, A., Martin, M., Wang, B., Jiang, G., Alipourgivi, F., Huang, X., Safa, A., Liu, Y., & Lu, T. (2023). Critical Role of Novel O-GlcNAcylation of S550 and S551 on the p65 Subunit of NF-κB in Pancreatic Cancer. Cancers, 15(19), 4742. https://doi.org/10.3390/cancers15194742







