Histomorphometric Analysis of 38 Giant Cell Tumors of Bone after Recurrence as Compared to Changes Following Denosumab Treatment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Histology and Immunohistochemistry
2.3. Statistics and Graphs
3. Results
3.1. Clinic
3.2. Histological Findings
3.3. Immunohistochemical Findings
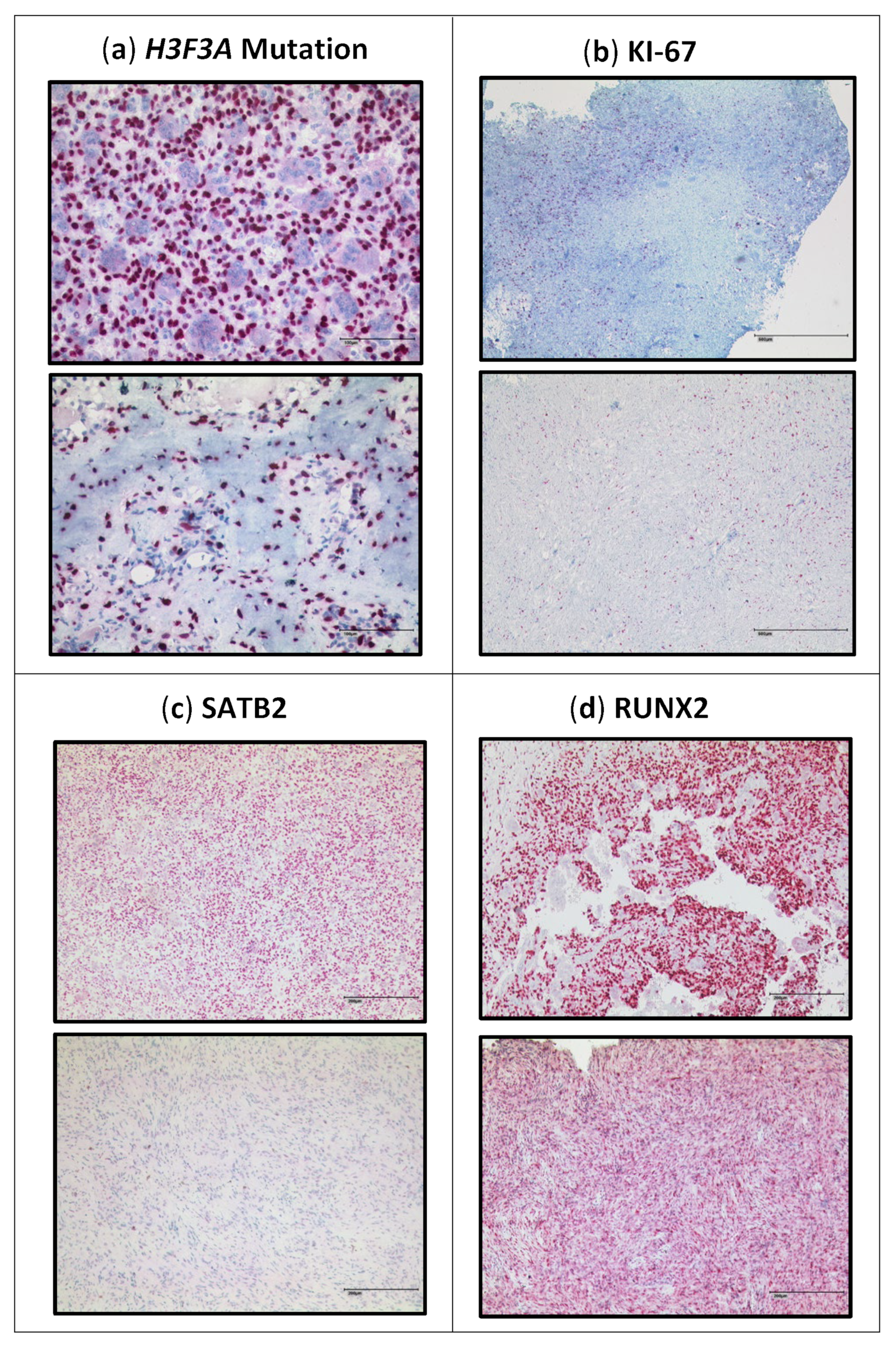
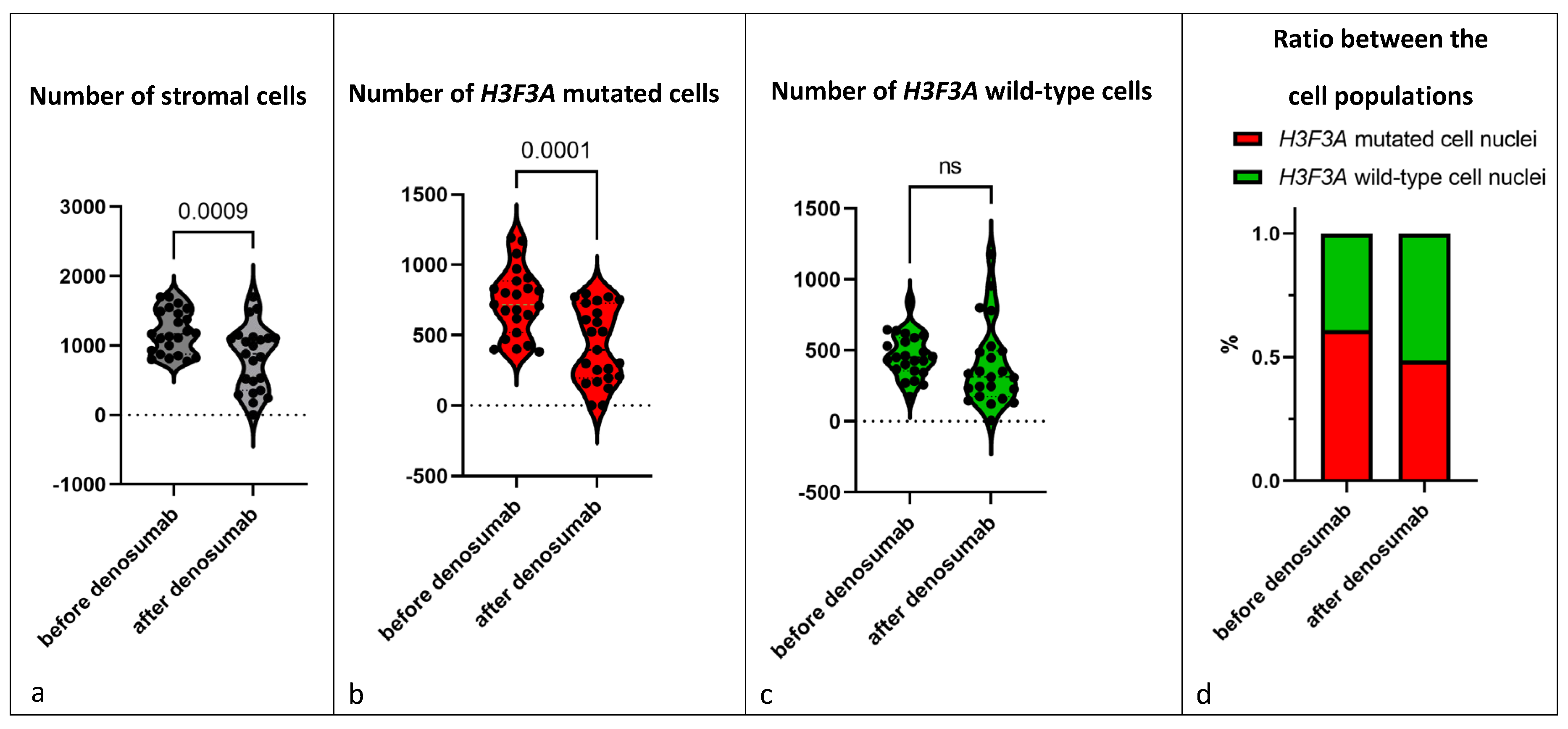
3.3.1. H3F3A G34W Staining
3.3.2. Proliferation Rate
3.3.3. SATB2
3.3.4. RUNX2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Athanasou, N.A.; Bansal, M.; Forsyth, R. WHO Classification of Tumours. Soft Tissue and Bone Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Sobti, A.; Agrawal, P.; Agarwala, S.; Agarwal, M. Giant Cell Tumor of Bone—An Overview. Arch. Bone Jt. Surg. 2016, 4, 2–9. [Google Scholar] [PubMed]
- Chen, C.-C.; Liau, C.-T.; Chang, C.-H.; Hsu, Y.-H.; Shih, H.-N. Giant Cell Tumors of the Bone With Pulmonary Metastasis. Orthopedics 2016, 39, e68–e73. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.T.; Dohle, J.; Bernd, L.; Braun, A.; Cserhati, M.; Enderle, A.; Hovy, L.; Matejovsky, Z.; Szendroi, M.; Trieb, K.; et al. Local recurrence of giant cell tumor of bone after intralesional treatment with and without adjuvant therapy. J. Bone Jt. Surg. Am. 2008, 90, 1060–1067. [Google Scholar] [CrossRef]
- Hild, V.; Mellert, K.; Möller, P.; Barth, T.F.E. Giant Cells of Various Lesions Are Characterised by Different Expression Patterns of HLA-Molecules and Molecules Involved in the Cell Cycle, Bone Metabolism, and Lineage Affiliation: An Immunohistochemical Study with a Review of the Literature. Cancers 2023, 15, 3702. [Google Scholar] [CrossRef]
- Forsyth, R.G.; Krenács, T.; Athanasou, N.; Hogendoorn, P.C.W. Cell Biology of Giant Cell Tumour of Bone: Crosstalk between m/wt Nucleosome H3.3, Telomeres and Osteoclastogenesis. Cancers 2021, 13, 5119. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S.; Presneau, N.; Scheipl, S.; Pillay, N.; van Loo, P.; Wedge, D.C.; Cooke, S.L.; Gundem, G.; Davies, H.; et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 2013, 45, 1479–1482. [Google Scholar] [CrossRef] [PubMed]
- Cleven, A.H.; Höcker, S.; Briaire-de Bruijn, I.; Szuhai, K.; Cleton-Jansen, A.-M.; Bovée, J.V. Mutation Analysis of H3F3A and H3F3B as a Diagnostic Tool for Giant Cell Tumor of Bone and Chondroblastoma. Am. J. Surg. Pathol. 2015, 39, 1576–1583. [Google Scholar] [CrossRef]
- Presneau, N.; Baumhoer, D.; Behjati, S.; Pillay, N.; Tarpey, P.; Campbell, P.J.; Jundt, G.; Hamoudi, R.; Wedge, D.C.; van Loo, P.; et al. Diagnostic value of H3F3A mutations in giant cell tumour of bone compared to osteoclast-rich mimics. J. Pathol. Clin. Res. 2015, 1, 113–123. [Google Scholar] [CrossRef]
- Lüke, J.; von Baer, A.; Schreiber, J.; Lübbehüsen, C.; Breining, T.; Mellert, K.; Marienfeld, R.; Schultheiss, M.; Möller, P.; Barth, T.F.E. H3F3A mutation in giant cell tumour of the bone is detected by immunohistochemistry using a monoclonal antibody against the G34W mutated site of the histone H3.3 variant. Histopathology 2017, 71, 125–133. [Google Scholar] [CrossRef]
- van der Heijden, L.; Dijkstra, P.D.S.; van de Sande, M.A.J.; Kroep, J.R.; Nout, R.A.; van Rijswijk, C.S.P.; Bovée, J.V.M.G.; Hogendoorn, P.C.W.; Gelderblom, H. The clinical approach toward giant cell tumor of bone. Oncologist 2014, 19, 550–561. [Google Scholar] [CrossRef]
- Branstetter, D.G.; Nelson, S.D.; Manivel, J.C.; Blay, J.-Y.; Chawla, S.; Thomas, D.M.; Jun, S.; Jacobs, I. Denosumab induces tumor reduction and bone formation in patients with giant-cell tumor of bone. Clin. Cancer Res. 2012, 18, 4415–4424. [Google Scholar] [CrossRef] [PubMed]
- Arfee, S.; Malik, A.T.; Nehru, A.; Ali, U.; Arfee, A.; Arfee, A.A. Comparison of Local and Intravenous Zoledronic Acid on Histopathology and Recurrence Rate after Extended Curettage in Giant Cell Tumors of Proximal Tibia: A Prospective Study. J. Pharm. Bioallied Sci. 2022, 14, S289–S291. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Henshaw, R.; Skubitz, K.; Chawla, S.; Staddon, A.; Blay, J.-Y.; Roudier, M.; Smith, J.; Ye, Z.; Sohn, W.; et al. Denosumab in patients with giant-cell tumour of bone: An open-label, phase 2 study. Lancet Oncol. 2010, 11, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Chawla, S.; Blay, J.-Y.; Rutkowski, P.; Le Cesne, A.; Reichardt, P.; Gelderblom, H.; Grimer, R.J.; Choy, E.; Skubitz, K.; Seeger, L.; et al. Denosumab in patients with giant-cell tumour of bone: A multicentre, open-label, phase 2 study. Lancet Oncol. 2019, 20, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Chawla, S.; Henshaw, R.; Seeger, L.; Choy, E.; Blay, J.-Y.; Ferrari, S.; Kroep, J.; Grimer, R.; Reichardt, P.; Rutkowski, P.; et al. Safety and efficacy of denosumab for adults and skeletally mature adolescents with giant cell tumour of bone: Interim analysis of an open-label, parallel-group, phase 2 study. Lancet Oncol. 2013, 14, 901–908. [Google Scholar] [CrossRef] [PubMed]
- XGEVA® 120 mg Injektionslösung (Denosumab) und das Risiko Eines Neuen Primären Malignoms. Available online: https://www.pei.de/SharedDocs/Downloads/DE/newsroom/veroeffentlichungen-arzneimittel/anhaenge-am-sik-infos/2018-05-16-informationsbrief-xgeva.pdf?__blob=publicationFile&v=2 (accessed on 14 November 2022).
- Hasenfratz, M.; Mellert, K.; Marienfeld, R.; von Baer, A.; Schultheiss, M.; Roitman, P.D.; Aponte-Tinao, L.A.; Lehner, B.; Möller, P.; Mechtersheimer, G.; et al. Profiling of three H3F3A-mutated and denosumab-treated giant cell tumors of bone points to diverging pathways during progression and malignant transformation. Sci. Rep. 2021, 11, 5709. [Google Scholar] [CrossRef]
- Fittall, M.W.; Lyskjaer, I.; Ellery, P.; Lombard, P.; Ijaz, J.; Strobl, A.-C.; Oukrif, D.; Tarabichi, M.; Sill, M.; Koelsche, C.; et al. Drivers underpinning the malignant transformation of giant cell tumour of bone. J. Pathol. 2020, 252, 433–440. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Aponte-Tinao, L.A.; Piuzzi, N.S.; Roitman, P.; Farfalli, G.L. A High-grade Sarcoma Arising in a Patient With Recurrent Benign Giant Cell Tumor of the Proximal Tibia While Receiving Treatment With Denosumab. Clin. Orthop. Relat. Res. 2015, 473, 3050–3055. [Google Scholar] [CrossRef]
- Ud Din, N.; Umer, M.; Park, Y.-K. Histomorphometric Analysis of Pre- and Post-Denosumab-Treated Giant Cell Tumor of Bone. Int. J. Surg. Pathol. 2020, 28, 859–867. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, H.; Zhang, X.; Tang, Y.; Wu, Z.; Wang, Y.; Huang, H.; Fu, X.; Liu, J.; Hogendoorn, P.C.W.; et al. Clinicopathologic and molecular features of denosumab-treated giant cell tumour of bone (GCTB): Analysis of 21 cases. Ann. Diagn. Pathol. 2022, 57, 151882. [Google Scholar] [CrossRef]
- Treffel, M.; Lardenois, E.; Larousserie, F.; Karanian, M.; Gomez-Brouchet, A.; Bouvier, C.; Le Loarer, F.; Aubert, S.; de Pinieux, G.; Audard, V.; et al. Denosumab-treated Giant Cell Tumors of Bone: A Clinicopathologic Analysis of 35 Cases From the French Group of Bone Pathology. Am. J. Surg. Pathol. 2020, 44, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, K.E.; DevecI, M.A.; Paydas, S.; Gonlusen, G. Morphologic evaluation of the effect of denosumab on giant cell tumors of bone and a new grading scheme. Pol. J. Pathol. 2016, 67, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Girolami, I.; Mancini, I.; Simoni, A.; Baldi, G.G.; Simi, L.; Campanacci, D.; Beltrami, G.; Scoccianti, G.; D’Arienzo, A.; Capanna, R.; et al. Denosumab treated giant cell tumour of bone: A morphological, immunohistochemical and molecular analysis of a series. J. Clin. Pathol. 2016, 69, 240–247. [Google Scholar] [CrossRef]
- Roitman, P.D.; Jauk, F.; Farfalli, G.L.; Albergo, J.I.; Aponte-Tinao, L.A. Denosumab-treated giant cell tumor of bone. Its histologic spectrum and potential diagnostic pitfalls. Hum. Pathol. 2017, 63, 89–97. [Google Scholar] [CrossRef]
- Rekhi, B.; Dave, V. Giant cell tumor of bone: An update, including spectrum of pathological features, pathogenesis, molecular profile and the differential diagnoses. Histol. Histopathol. 2023, 38, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Mallya, V.; Mandal, S.; Tomar, R.; Khurana, N.; Maini, L. Histopathological response to denosumab in giant cell tumours of bone—A review of 11 cases. J. Cancer Res. Ther. 2023, 19, 768–772. [Google Scholar] [CrossRef]
- Kato, I.; Furuya, M.; Matsuo, K.; Kawabata, Y.; Tanaka, R.; Ohashi, K. Giant cell tumours of bone treated with denosumab: Histological, immunohistochemical and H3F3A mutation analyses. Histopathology 2018, 72, 914–922. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Gordon, J.A.R.; Beloti, M.M.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 2010, 107, 19879–19884. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef]
- Palmerini, E.; Seeger, L.L.; Gambarotti, M.; Righi, A.; Reichardt, P.; Bukata, S.; Blay, J.-Y.; Dai, T.; Jandial, D.; Picci, P. Malignancy in giant cell tumor of bone: Analysis of an open-label phase 2 study of denosumab. BMC Cancer 2021, 21, 89. [Google Scholar] [CrossRef] [PubMed]




| Number | Sex | Tumor Localization | Duration of Denosumab Therapy | Age at Diagnosis |
|---|---|---|---|---|
| Patient 1 | m | lateral condyle of left femur | 6 months | 40 |
| Patient 2 | m | left distal tibia | 25 months | 37 |
| Patient 3 | m | distal radius left | 6 months | 45 |
| Patient 4 | f | spina iliaca anterior | 12 months | 33 |
| Patient 5 | f | proximal femur left | 6 months | 20 |
| Patient 6 | f | lateral femoral condyle left | 4 months | 28 |
| Patient 7 | m | proximal femur right | 10 months | 52 |
| Patient 8 | m | proximal tibia left | 20 months | 55 |
| Patient 9 | m | proximal tibia right | 4 months | 48 |
| Patient 10 | m | left radius | 6 months | 39 |
| Patient 11 | m | distal radius right | 8 months | 44 |
| Patient 12 | f | right patella | 9 months | 32 |
| Patient 13 | f | distal femur right | 3 months | 26 |
| Patient 14 | m | proximal femur left | one single dose | 28 |
| Patient 15 | f | femoral neck right | 12 months | 28 |
| Patient 16 | f | right os ischii | 12 months | 20 |
| Patient 17 | f | right tibia | 4 months | 26 |
| Patient 18 | m | left hand, phalanx digitus 3 | 11 months | 44 |
| Patient 19 | f | proximal tibia right | 9 months | 13 |
| Patient 20 | m | distal femur left | 9 months | 26 |
| Patient 21 | m | proximal tibia left | 9 months | 31 |
| Patient 22 | m | proximal tibia right | 2 months | 51 |
| Patient 23 | f | 4th thoracic vertebrae | 5 months | 32 |
| Patient 24 | f | proximal tibia right | 12 months | 15 |
| Patient 25 | m | left femur neck | 6 months | 28 |
| Patient 26 | m | left hand, os metacarpale IV | none | 49 |
| Patient 27 | m | left hand, phalanx digitus1 | none | 41 |
| Patient 28 | m | right tibia | none | 48 |
| Patient 29 | f | left tibia | none | 63 |
| Patient 30 | m | proximal humerus right | none | 24 |
| Patient 31 | f | proximal tibia left | none | 17 |
| Patient 32 | f | proximal tibia | none | 20 |
| Patient 33 | f | proximal tibia right | none | 19 |
| Patient 34 | m | distal radius left | none | 38 |
| Patient 35 | f | distal radius right | none | 27 |
| Patient 36 | m | femur left | none | 38 |
| Patient 37 | f | femur right | none | 27 |
| Patient 38 | m | proximal tibia right | none | 39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arndt, S.; Hartmann, W.; Rókusz, A.; Leinauer, B.; von Baer, A.; Schultheiss, M.; Pablik, J.; Fritzsche, H.; Mogler, C.; Antal, I.; et al. Histomorphometric Analysis of 38 Giant Cell Tumors of Bone after Recurrence as Compared to Changes Following Denosumab Treatment. Cancers 2023, 15, 4249. https://doi.org/10.3390/cancers15174249
Arndt S, Hartmann W, Rókusz A, Leinauer B, von Baer A, Schultheiss M, Pablik J, Fritzsche H, Mogler C, Antal I, et al. Histomorphometric Analysis of 38 Giant Cell Tumors of Bone after Recurrence as Compared to Changes Following Denosumab Treatment. Cancers. 2023; 15(17):4249. https://doi.org/10.3390/cancers15174249
Chicago/Turabian StyleArndt, Sophia, Wolfgang Hartmann, András Rókusz, Benedikt Leinauer, Alexandra von Baer, Markus Schultheiss, Jessica Pablik, Hagen Fritzsche, Carolin Mogler, Imre Antal, and et al. 2023. "Histomorphometric Analysis of 38 Giant Cell Tumors of Bone after Recurrence as Compared to Changes Following Denosumab Treatment" Cancers 15, no. 17: 4249. https://doi.org/10.3390/cancers15174249
APA StyleArndt, S., Hartmann, W., Rókusz, A., Leinauer, B., von Baer, A., Schultheiss, M., Pablik, J., Fritzsche, H., Mogler, C., Antal, I., Baumhoer, D., Mellert, K., Möller, P., Szendrői, M., Jundt, G., & Barth, T. F. E. (2023). Histomorphometric Analysis of 38 Giant Cell Tumors of Bone after Recurrence as Compared to Changes Following Denosumab Treatment. Cancers, 15(17), 4249. https://doi.org/10.3390/cancers15174249






