Classification of Brainstem Gliomas Based on Tumor Microenvironment Status
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Inclusion and RNA Sequencing
2.2. TME Status Estimation and Clustering
2.3. Estimation of Tumor Biological Features and Radiosensitivity
2.4. Identification of Key Genes Determining TME Clusters
2.5. Multiplex Immunofluorescence Staining
2.6. Generation of Radiomics Models for Identifying TME Clusters
2.7. Statistical Analysis
3. Results
3.1. BSGs Are Grouped into Four Clusters Based on TME Status
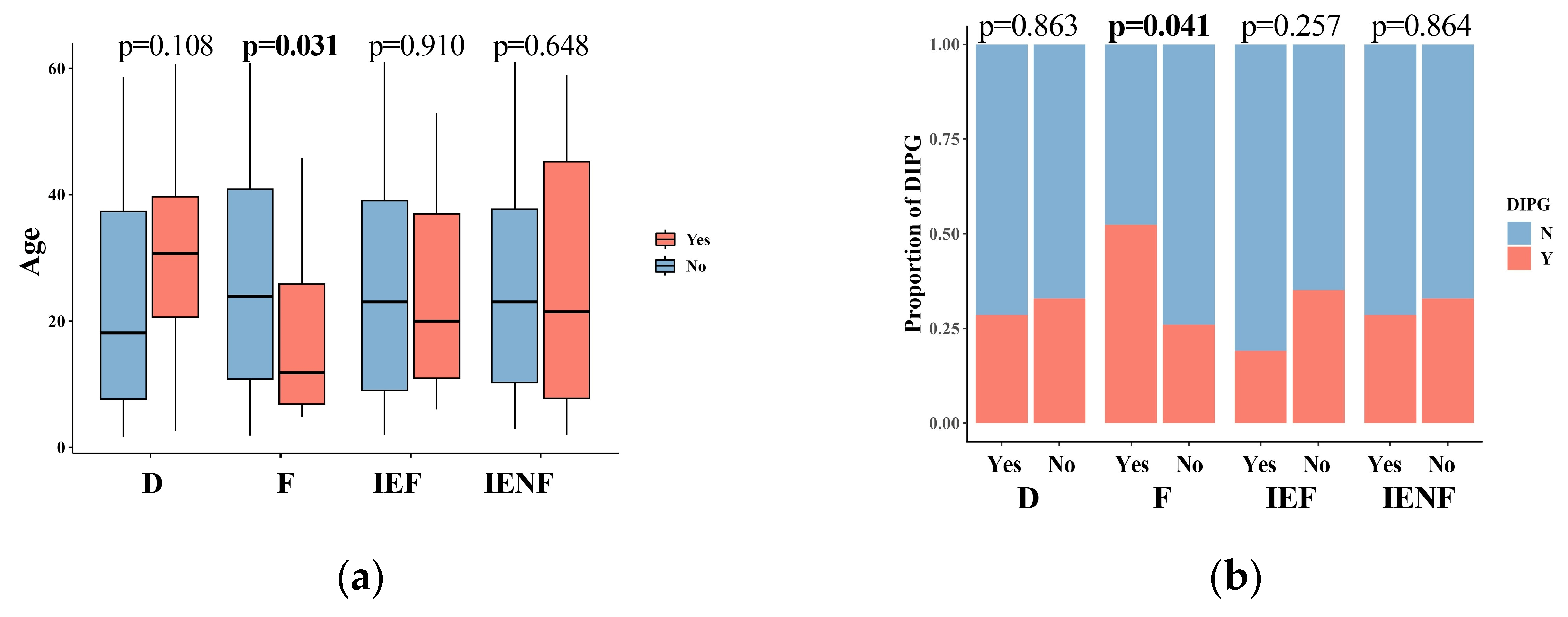
3.2. The TME Clusters Are Correlative with BSG Clinical and Molecular Features
3.3. The TME Clusters Predict Outcomes in BSG Patients
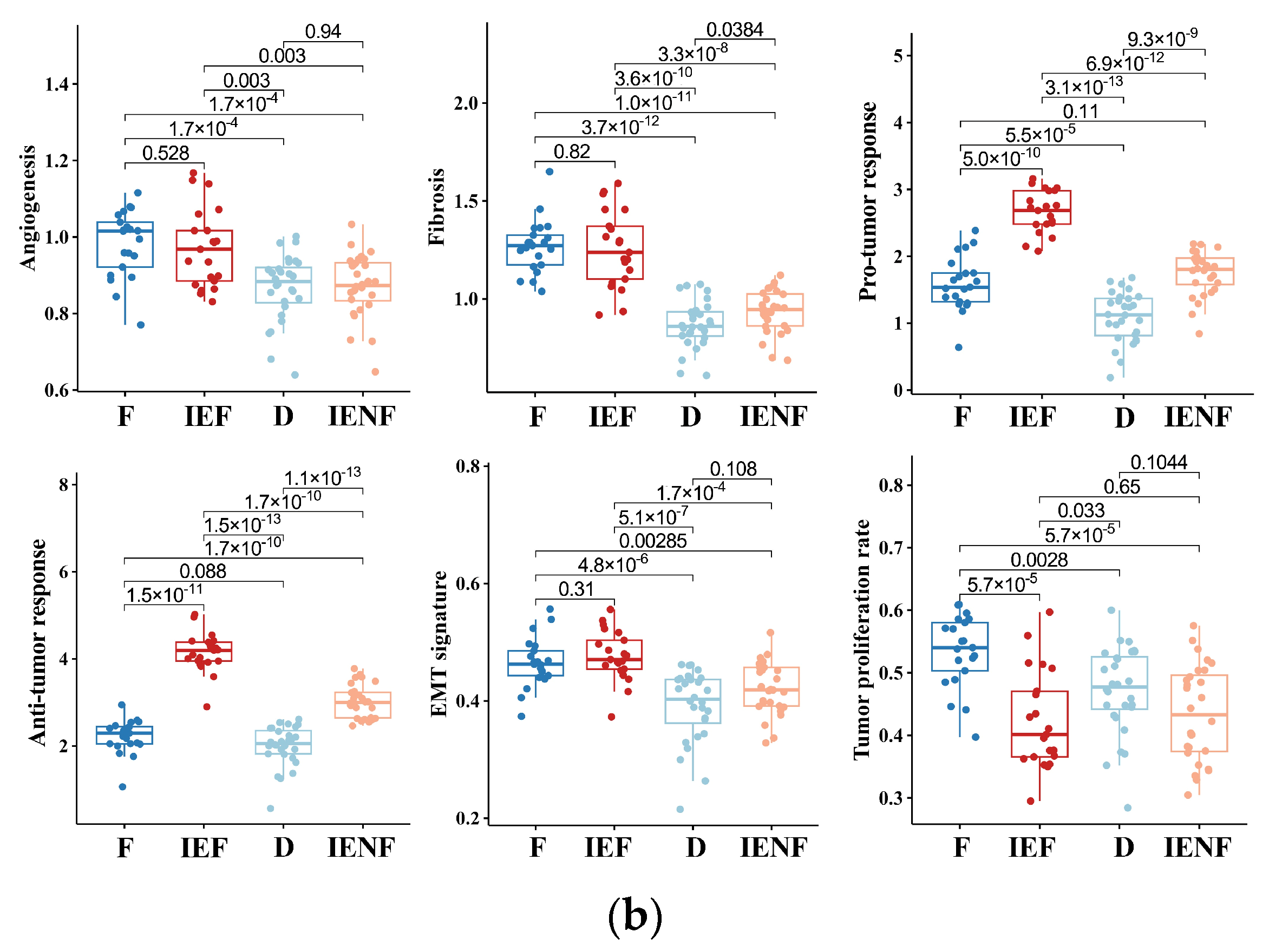
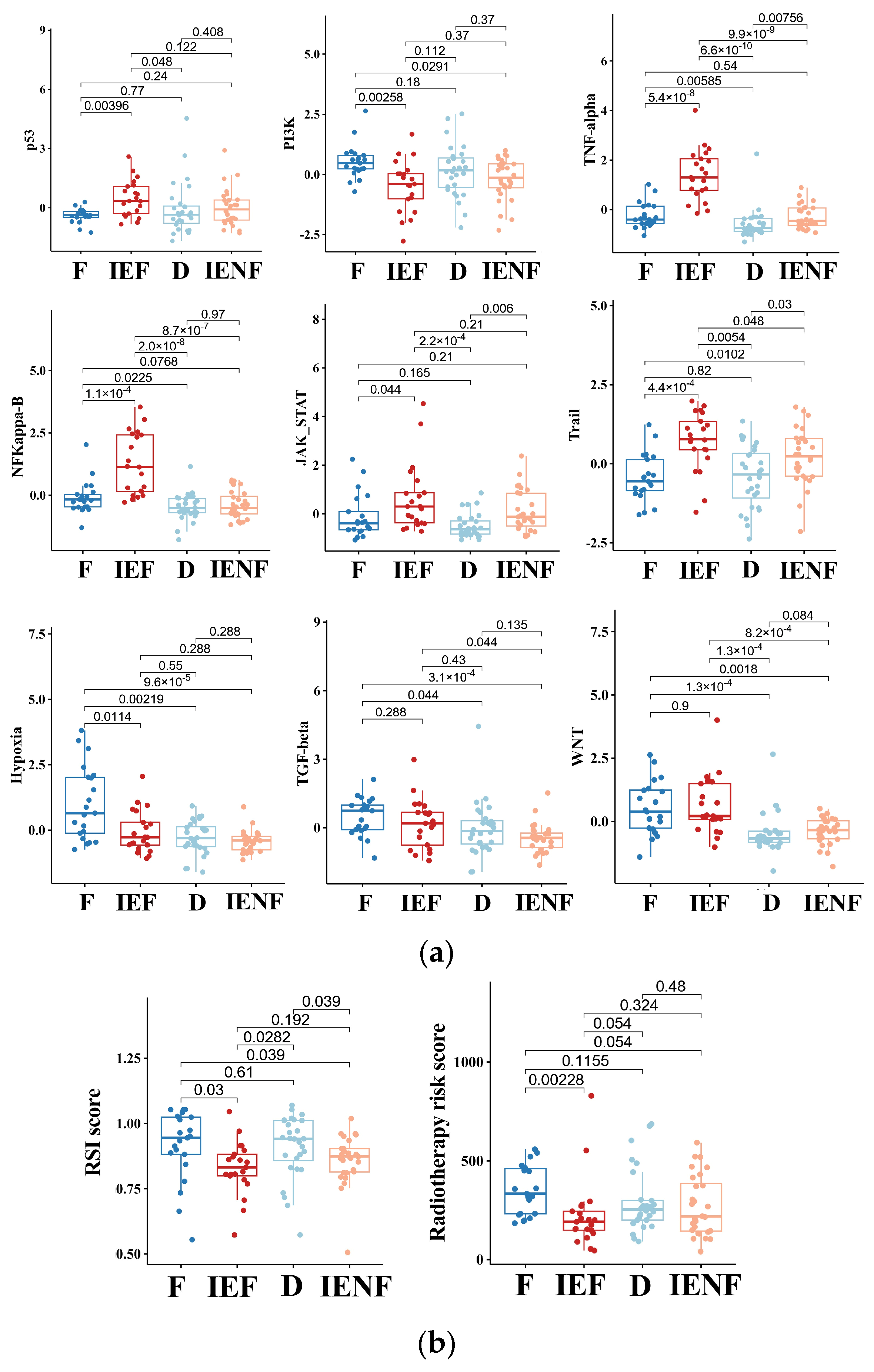
3.4. The TME Clusters Exhibit Distinct Molecular Pathway Activity and Radiation Sensitivity
3.5. A Four-Gene Panel Determines the TME Classification
3.6. Radiomics Features Related to TME Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grimm, S.A.; Chamberlain, M.C. Brainstem glioma: A review. Curr. Neurol. Neurosci. Rep. 2013, 13, 346. [Google Scholar] [CrossRef]
- Chen, L.H.; Pan, C.; Diplas, B.H.; Xu, C.; Hansen, L.J.; Wu, Y.; Chen, X.; Geng, Y.; Sun, T.; Sun, Y.; et al. The integrated genomic and epigenomic landscape of brainstem glioma. Nat. Commun. 2020, 11, 3077. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.G.; Ruggiero, A.; Centonze, A.; Carrieri, A.; Ferorelli, S.; Scilimati, A. Diffuse Intrinsic Pontine Glioma (DIPG): Breakthrough and Clinical Perspective. Curr. Med. Chem. 2021, 28, 3287–3317. [Google Scholar] [CrossRef] [PubMed]
- Cooney, T.M.; Lubanszky, E.; Prasad, R.; Hawkins, C.; Mueller, S. Diffuse midline glioma: Review of epigenetics. J. Neurooncol. 2020, 150, 27–34. [Google Scholar] [CrossRef]
- Cucu, A.I.; Turliuc, S.; Costea, C.F.; Perciaccante, A.; Bianucci, R.; Donell, S.; Scripcariu, D.V.; Turliuc, M.D. The brainstem and its neurosurgical history. Neurosurg. Rev. 2021, 44, 3001–3022. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Fang, Y.; Hui, X.; Jv, Y.; You, C. Radiotherapy for diffuse brainstem glioma in children and young adults. Cochrane Database Syst. Rev. 2016, 2016, CD010439. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Suh, C.O. Radiotherapy for Diffuse Intrinsic Pontine Glioma: Insufficient but Indispensable. Brain Tumor Res. Treat 2023, 11, 79–85. [Google Scholar] [CrossRef]
- Heravi Shargh, V.; Luckett, J.; Bouzinab, K.; Paisey, S.; Turyanska, L.; Singleton, W.G.B.; Lowis, S.; Gershkovich, P.; Bradshaw, T.D.; Stevens, M.F.G.; et al. Chemosensitization of Temozolomide-Resistant Pediatric Diffuse Midline Glioma Using Potent Nanoencapsulated Forms of a N(3)-Propargyl Analogue. ACS Appl. Mater. Interfaces 2021, 13, 35266–35280. [Google Scholar] [CrossRef]
- Majzner, R.G.; Ramakrishna, S.; Yeom, K.W.; Patel, S.; Chinnasamy, H.; Schultz, L.M.; Richards, R.M.; Jiang, L.; Barsan, V.; Mancusi, R.; et al. GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature 2022, 603, 934–941. [Google Scholar] [CrossRef]
- Bernstock, J.D.; Hoffman, S.E.; Kappel, A.D.; Valdes, P.A.; Essayed, W.; Klinger, N.V.; Kang, K.D.; Totsch, S.K.; Olsen, H.E.; Schlappi, C.W.; et al. Immunotherapy approaches for the treatment of diffuse midline gliomas. Oncoimmunology 2022, 11, 2124058. [Google Scholar] [CrossRef]
- Dunn-Pirio, A.M.; Vlahovic, G. Immunotherapy approaches in the treatment of malignant brain tumors. Cancer 2017, 123, 734–750. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, C.; Li, S.; Wang, J.; Zhang, H. Immune Microenvironment and Immunotherapies for Diffuse Intrinsic Pontine Glioma. Cancers 2023, 15, 602. [Google Scholar] [CrossRef]
- Lieberman, N.A.P.; DeGolier, K.; Kovar, H.M.; Davis, A.; Hoglund, V.; Stevens, J.; Winter, C.; Deutsch, G.; Furlan, S.N.; Vitanza, N.A.; et al. Characterization of the immune microenvironment of diffuse intrinsic pontine glioma: Implications for development of immunotherapy. Neuro Oncol. 2019, 21, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Bagaev, A.; Kotlov, N.; Nomie, K.; Svekolkin, V.; Gafurov, A.; Isaeva, O.; Osokin, N.; Kozlov, I.; Frenkel, F.; Gancharova, O.; et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021, 39, 845–865.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zou, C.; Zhang, S.; Chu, T.S.M.; Zhang, Y.; Chen, W.; Zhao, C.; Yang, L.; Xu, Z.; Dong, S.; et al. Reshaping the systemic tumor immune environment (STIE) and tumor immune microenvironment (TIME) to enhance immunotherapy efficacy in solid tumors. J. Hematol. Oncol. 2022, 15, 87. [Google Scholar] [CrossRef]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef]
- Job, S.; Rapoud, D.; Dos Santos, A.; Gonzalez, P.; Desterke, C.; Pascal, G.; Elarouci, N.; Ayadi, M.; Adam, R.; Azoulay, D.; et al. Identification of Four Immune Subtypes Characterized by Distinct Composition and Functions of Tumor Microenvironment in Intrahepatic Cholangiocarcinoma. Hepatology 2020, 72, 965–981. [Google Scholar] [CrossRef]
- Ye, L.; Wang, L.; Yang, J.; Hu, P.; Zhang, C.; Tong, S.; Liu, Z.; Tian, D. Identification of tumor antigens and immune subtypes in lower grade gliomas for mRNA vaccine development. J. Transl. Med. 2021, 19, 352. [Google Scholar] [CrossRef]
- Wang, F. Identification of tumor antigens and immune subtypes of acute myeloid leukemia for mRNA vaccine development. Clin. Transl. Oncol. 2023, 25, 2204–2223. [Google Scholar] [CrossRef]
- Recinos, P.F.; Sciubba, D.M.; Jallo, G.I. Brainstem tumors: Where are we today? Pediatr. Neurosurg. 2007, 43, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Price, G.; Bouras, A.; Hambardzumyan, D.; Hadjipanayis, C.G. Current knowledge on the immune microenvironment and emerging immunotherapies in diffuse midline glioma. eBioMedicine 2021, 69, 103453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.H.; Wan, H.; Yang, R.; Wang, Z.; Feng, J.; Yang, S.; Jones, S.; Wang, S.; Zhou, W.; et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat. Genet 2014, 46, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Zhang, M.; Xiao, X.; Kong, L.; Wu, Y.; Zhao, X.; Sun, T.; Zhang, P.; Geng, Y.; Zuo, P.; et al. A multimodal imaging-based classification for pediatric diffuse intrinsic pontine gliomas. Neurosurg. Rev. 2023, 46, 151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform 2020, 2, lqaa078. [Google Scholar] [CrossRef]
- Schubert, M.; Klinger, B.; Klunemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Bluthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef]
- Grass, G.D.; Alfonso, J.C.L.; Welsh, E.; Ahmed, K.A.; Teer, J.K.; Pilon-Thomas, S.; Harrison, L.B.; Cleveland, J.L.; Mule, J.J.; Eschrich, S.A.; et al. The Radiosensitivity Index Gene Signature Identifies Distinct Tumor Immune Microenvironment Characteristics Associated with Susceptibility to Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 635–647. [Google Scholar] [CrossRef]
- Yan, D.; Zhao, Q.; Du, Z.; Li, H.; Geng, R.; Yang, W.; Zhang, X.; Cao, J.; Yi, N.; Zhou, J.; et al. Development and validation of an immune-related gene signature for predicting the radiosensitivity of lower-grade gliomas. Sci. Rep. 2022, 12, 6698. [Google Scholar] [CrossRef]
- Pan, C.C.; Liu, J.; Tang, J.; Chen, X.; Chen, F.; Wu, Y.L.; Geng, Y.B.; Xu, C.; Zhang, X.; Wu, Z.; et al. A machine learning-based prediction model of H3K27M mutations in brainstem gliomas using conventional MRI and clinical features. Radiother. Oncol. 2019, 130, 172–179. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Beeraka, N.M.; Guo, W.; Lei, Y.; Hu, Q.; Guo, L.; Fan, R.; Liu, J.; Sui, A. Prognosis of Patients with Brainstem Glioblastoma Based on “age, surgery and radiotherapy”: A SEER Database Analysis. Technol. Cancer Res. Treat 2022, 21, 15330338221082760. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, B.; Kocyk, M.; Kijewska, M. TGF beta signaling and its role in glioma pathogenesis. Adv. Exp. Med. Biol. 2013, 986, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.; Kornblum, H.I. Molecular markers in glioma. J. Neurooncol. 2017, 134, 505–512. [Google Scholar] [CrossRef]
- He, L.; Zhou, H.; Zeng, Z.; Yao, H.; Jiang, W.; Qu, H. Wnt/beta-catenin signaling cascade: A promising target for glioma therapy. J. Cell Physiol. 2019, 234, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef]
- Vallee, A.; Guillevin, R.; Vallee, J.N. Vasculogenesis and angiogenesis initiation under normoxic conditions through Wnt/beta-catenin pathway in gliomas. Rev. Neurosci. 2018, 29, 71–91. [Google Scholar] [CrossRef]
- Lang, F.F.; Yung, W.K.; Raju, U.; Libunao, F.; Terry, N.H.; Tofilon, P.J. Enhancement of radiosensitivity of wild-type p53 human glioma cells by adenovirus-mediated delivery of the p53 gene. J. Neurosurg. 1998, 89, 125–132. [Google Scholar] [CrossRef]
- Chedeville, A.L.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Radiotherapy Resistance. Cancers 2021, 13, 542. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, N.; Hu, X. Wnt/beta-catenin signaling inhibitors. Curr. Top Med. Chem. 2023, 23, 880–896. [Google Scholar]
- Persson, M.L.; Douglas, A.M.; Alvaro, F.; Faridi, P.; Larsen, M.R.; Alonso, M.M.; Vitanza, N.A.; Dun, M.D. The intrinsic and microenvironmental features of diffuse midline glioma: Implications for the development of effective immunotherapeutic treatment strategies. Neuro Oncol. 2022, 24, 1408–1422. [Google Scholar] [CrossRef] [PubMed]
- Nepal, C.; Zhu, B.; O’Rourke, C.J.; Bhatt, D.K.; Lee, D.; Song, L.; Wang, D.; Van Dyke, A.L.; Choo-Wosoba, H.; Liu, Z.; et al. Integrative molecular characterisation of gallbladder cancer reveals micro-environment-associated subtypes. J. Hepatol. 2021, 74, 1132–1144. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.W.; Bao, Z.S.; Jiang, T.; Zhu, Y.J. Tumor microenvironment is associated with clinical and genetic properties of diffuse gliomas and predicts overall survival. Cancer Immunol. Immunother. 2022, 71, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, D.; Saw, P.E.; Song, E. Turning cold tumors hot: From molecular mechanisms to clinical applications. Trends Immunol. 2022, 43, 523–545. [Google Scholar] [CrossRef] [PubMed]
- Ho, I.A.; Chan, K.Y.; Ng, W.H.; Guo, C.M.; Hui, K.M.; Cheang, P.; Lam, P.Y. Matrix metalloproteinase 1 is necessary for the migration of human bone marrow-derived mesenchymal stem cells toward human glioma. Stem Cells 2009, 27, 1366–1375. [Google Scholar] [CrossRef]
- Yin, W.; Zhu, H.; Tan, J.; Xin, Z.; Zhou, Q.; Cao, Y.; Wu, Z.; Wang, L.; Zhao, M.; Jiang, X.; et al. Identification of collagen genes related to immune infiltration and epithelial-mesenchymal transition in glioma. Cancer Cell. Int. 2021, 21, 276. [Google Scholar] [CrossRef]
- Khalili, N.; Kazerooni, A.F.; Familiar, A.; Haldar, D.; Kraya, A.; Foster, J.; Koptyra, M.; Storm, P.B.; Resnick, A.C.; Nabavizadeh, A. Radiomics for characterization of the glioma immune microenvironment. NPJ Precis. Oncol. 2023, 7, 59. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Li, X.; Wang, Y.; Pan, C.; Zhang, P.; Gu, G.; Li, T.; Jiang, Z.; Zhang, Y.; Zhang, L. Classification of Brainstem Gliomas Based on Tumor Microenvironment Status. Cancers 2023, 15, 4224. https://doi.org/10.3390/cancers15174224
Xiao X, Li X, Wang Y, Pan C, Zhang P, Gu G, Li T, Jiang Z, Zhang Y, Zhang L. Classification of Brainstem Gliomas Based on Tumor Microenvironment Status. Cancers. 2023; 15(17):4224. https://doi.org/10.3390/cancers15174224
Chicago/Turabian StyleXiao, Xiong, Xiaoou Li, Yi Wang, Changcun Pan, Peng Zhang, Guocan Gu, Tian Li, Zhuang Jiang, Yang Zhang, and Liwei Zhang. 2023. "Classification of Brainstem Gliomas Based on Tumor Microenvironment Status" Cancers 15, no. 17: 4224. https://doi.org/10.3390/cancers15174224
APA StyleXiao, X., Li, X., Wang, Y., Pan, C., Zhang, P., Gu, G., Li, T., Jiang, Z., Zhang, Y., & Zhang, L. (2023). Classification of Brainstem Gliomas Based on Tumor Microenvironment Status. Cancers, 15(17), 4224. https://doi.org/10.3390/cancers15174224





