YY1 Knockdown Relieves the Differentiation Block and Restores Apoptosis in AML Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Ethics Statement
2.3. Human Samples and AML Cell Lines
2.4. Plasmid Constructs, Lentiviral Infection, and Cell Transfection
2.5. Protein Extraction and Western Blot Analysis
2.6. RNA Isolated and Analysis
2.7. Chromatin Immunoprecipitation (ChIp)
2.8. Immunophenotypic Analysis
2.9. Morphological Analysis
2.10. Statistical Analysis
3. Results
3.1. An Inhibitor of NF-kB Activation, Bortezomib, Downregulates YY1 in AML Cells
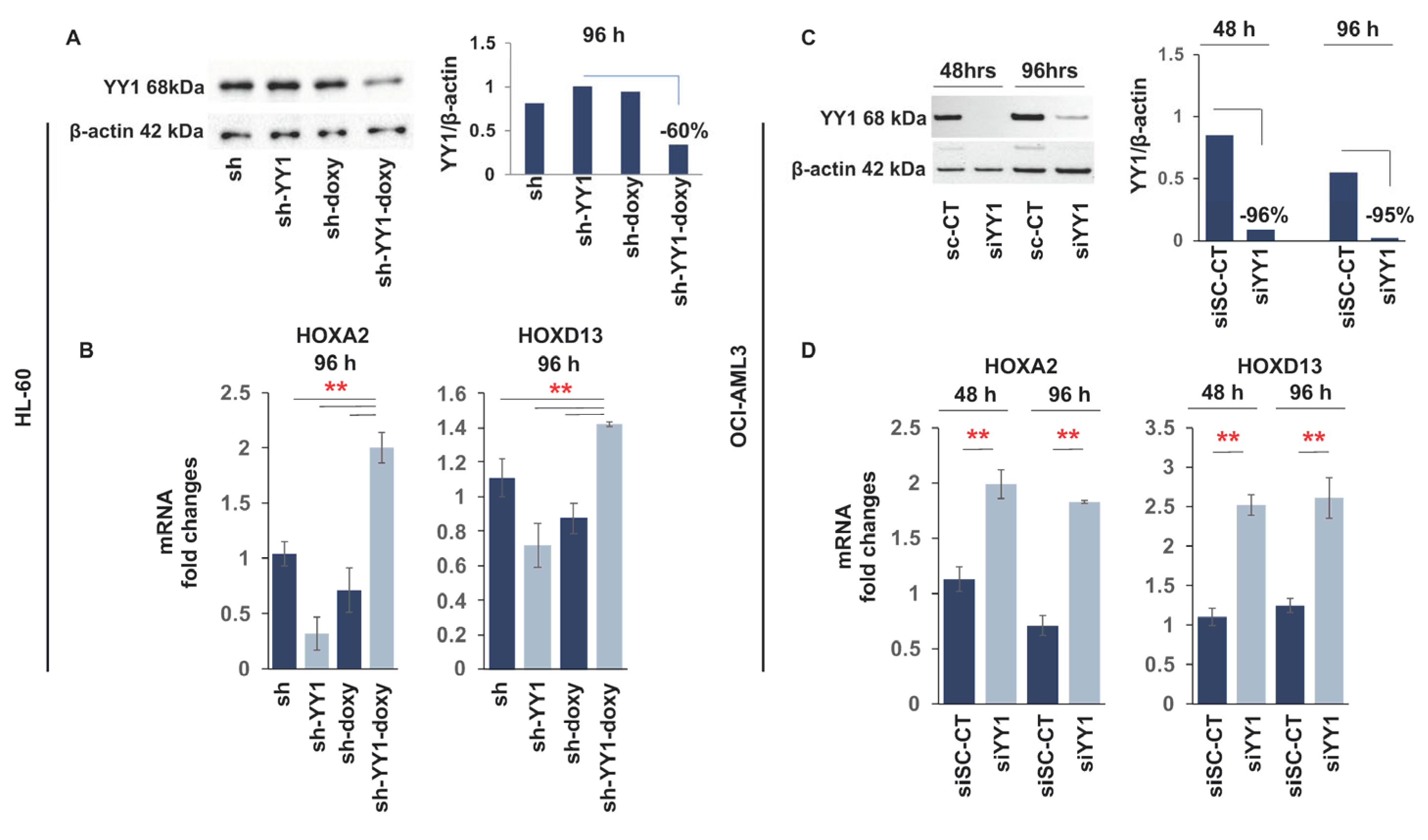
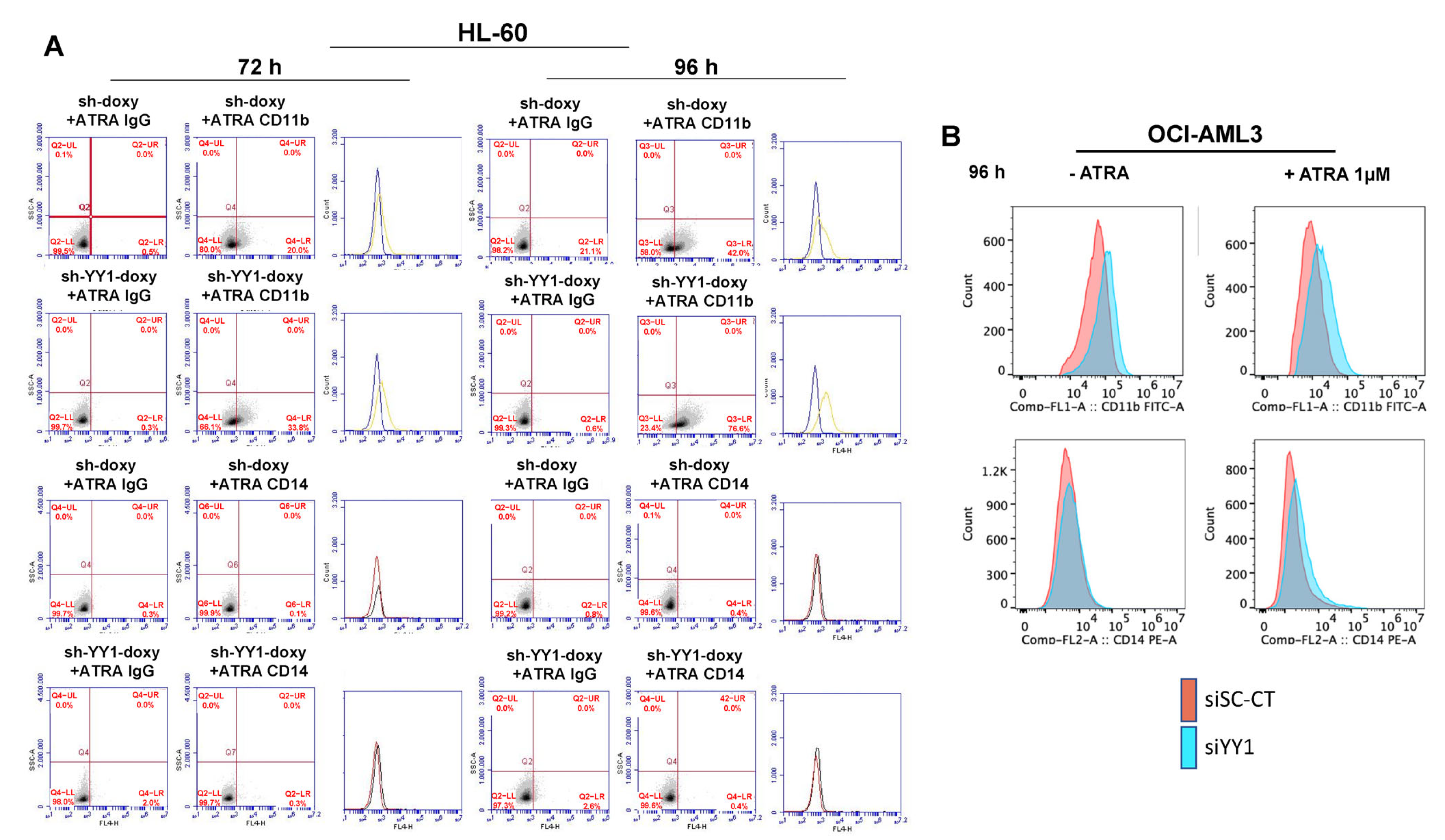
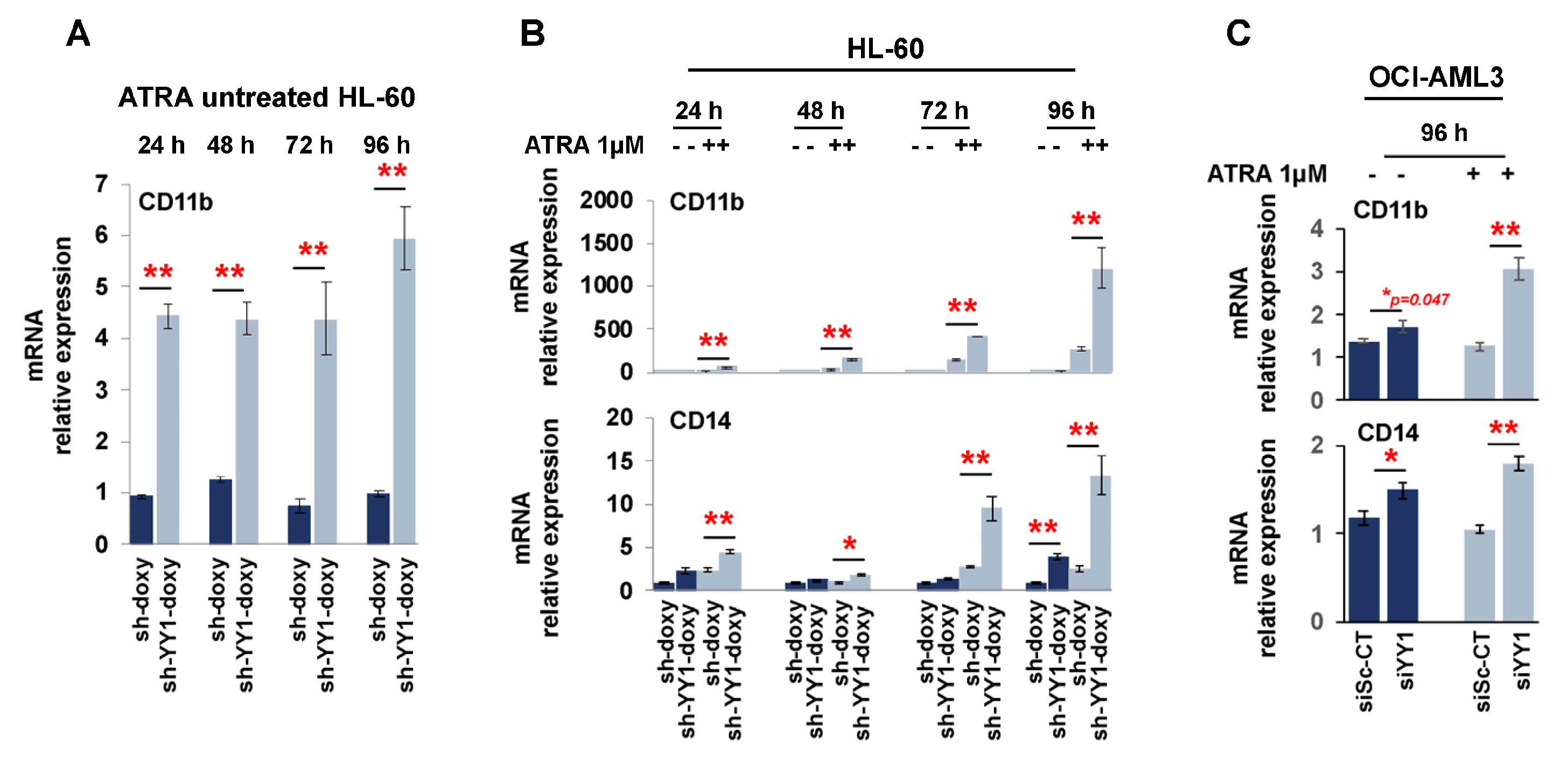
3.2. YY1 Knockdown Induces Expression of Myeloid Differentiation Markers
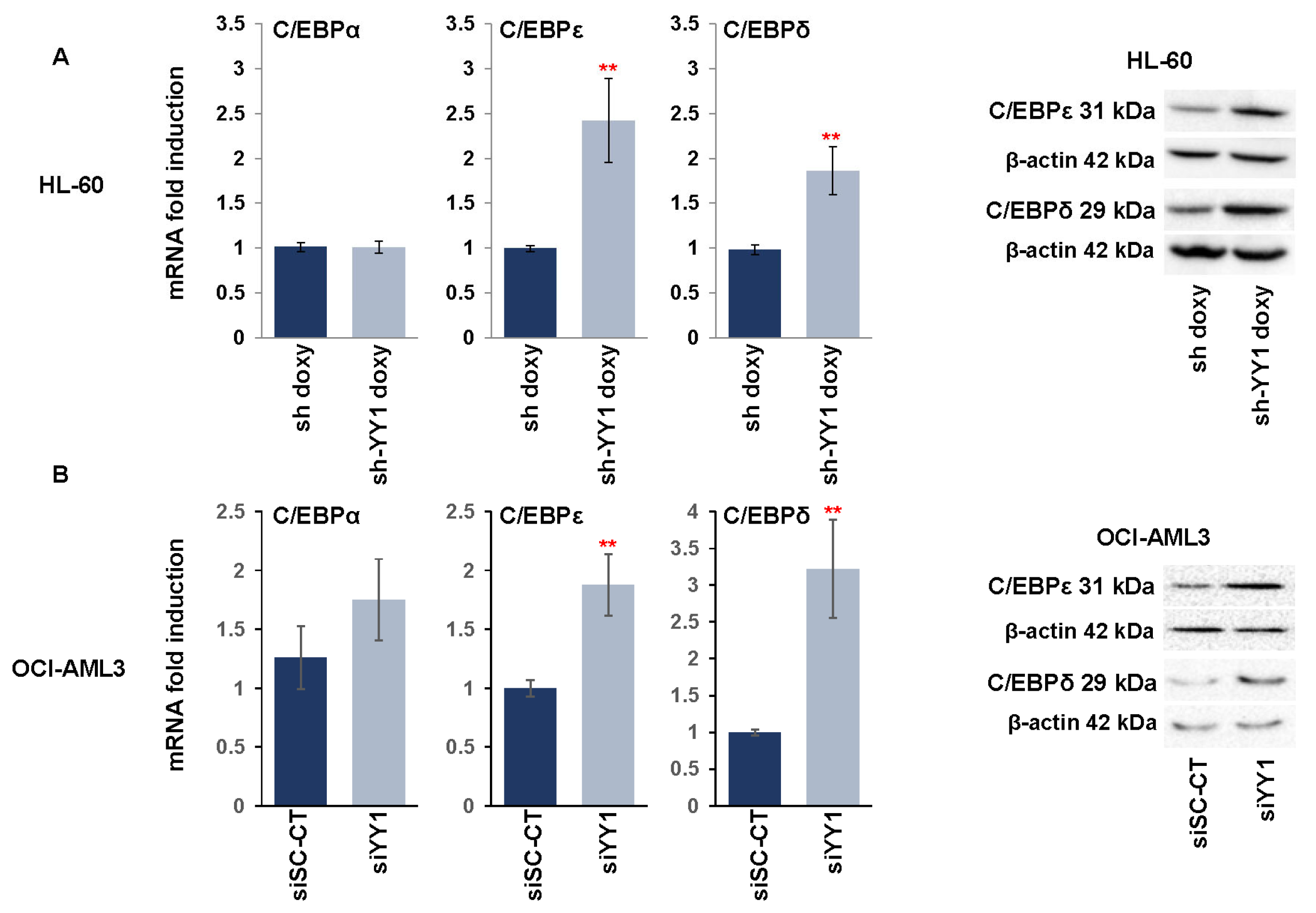
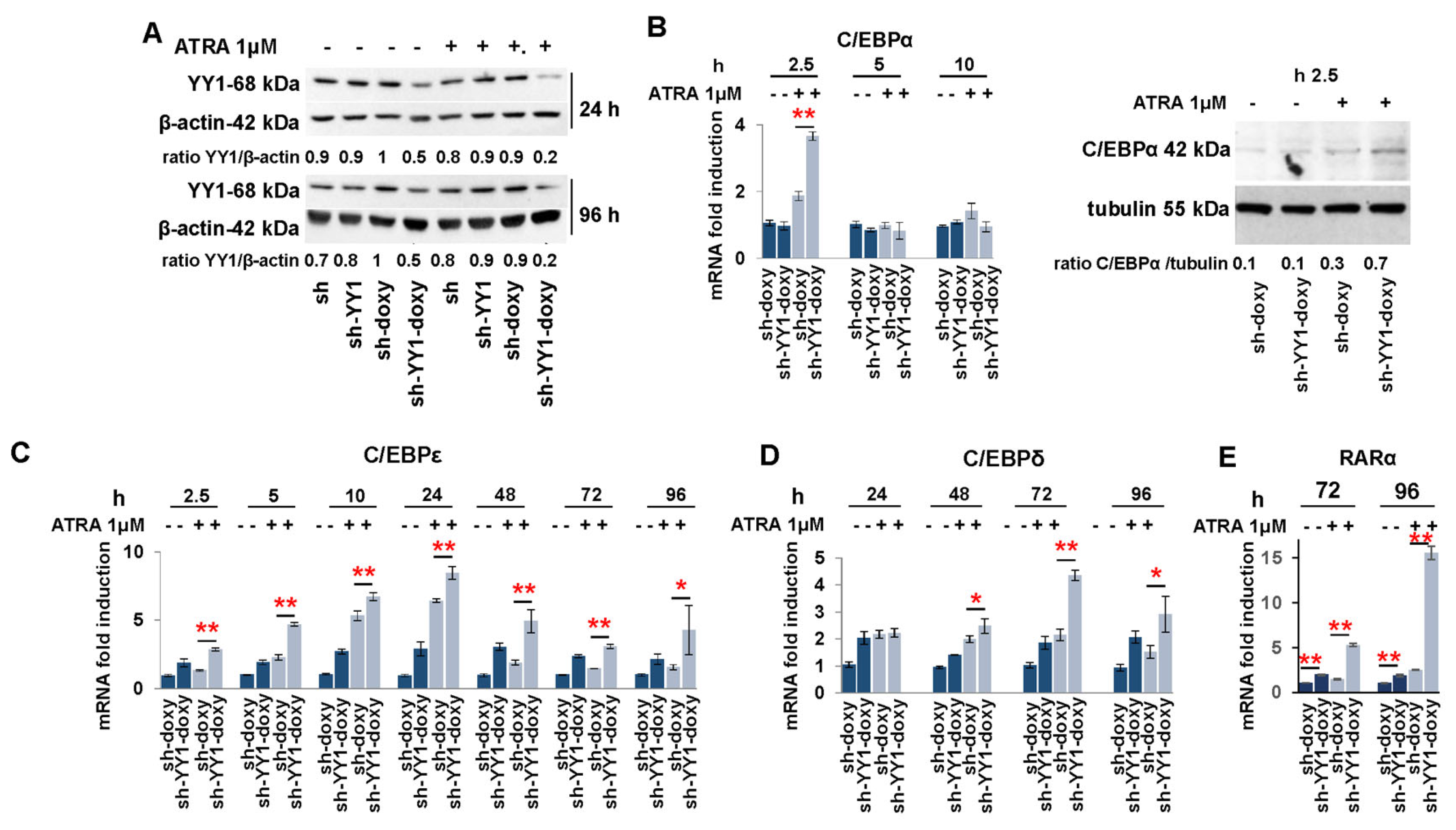
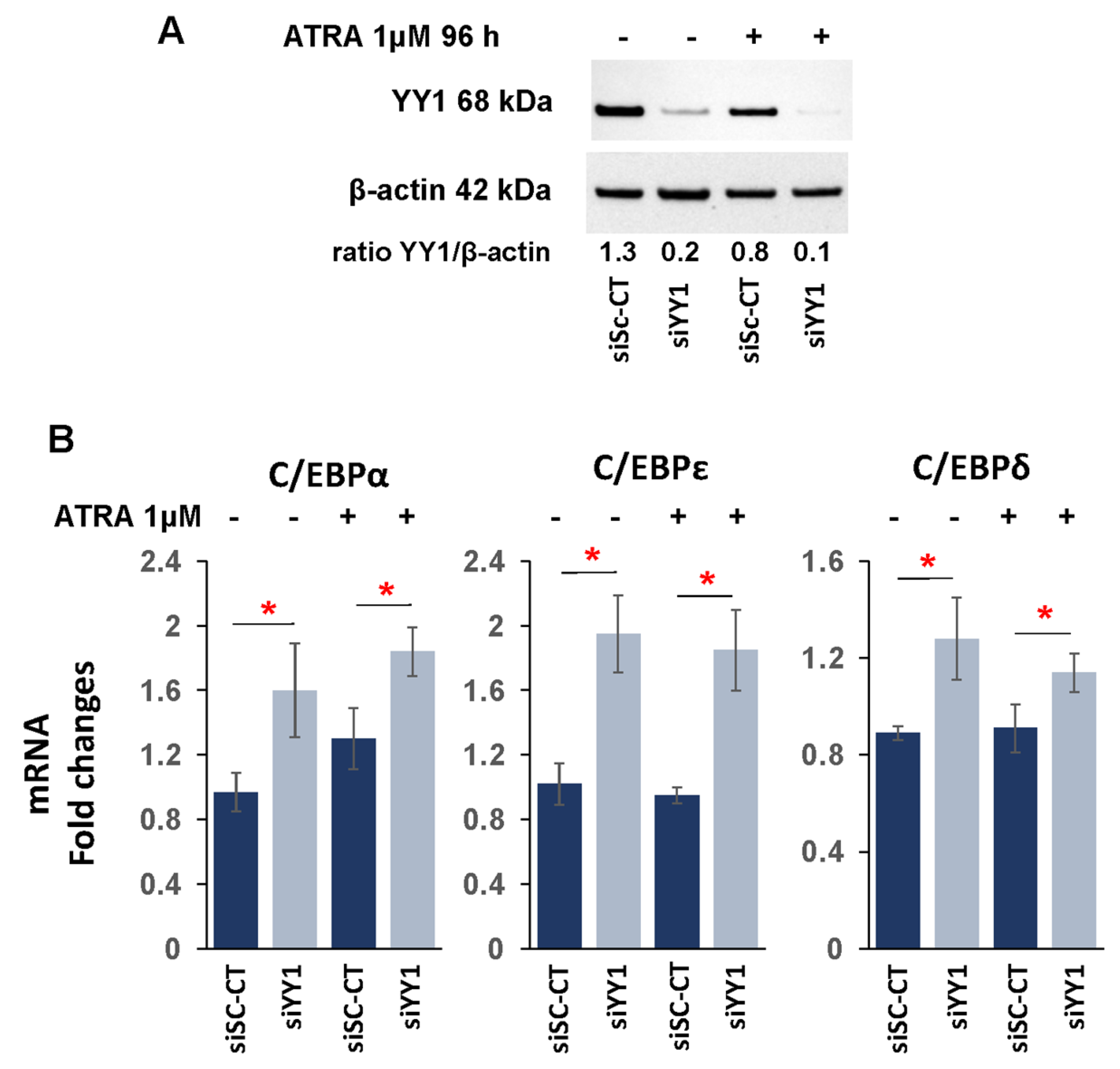
3.3. YY1 Knockdown Increases Expression of C/EBP Family Proteins and Myeloid Growth Factors/Receptors in ATRA-Treated HL-60 and OCI-AML3 Cell Lines
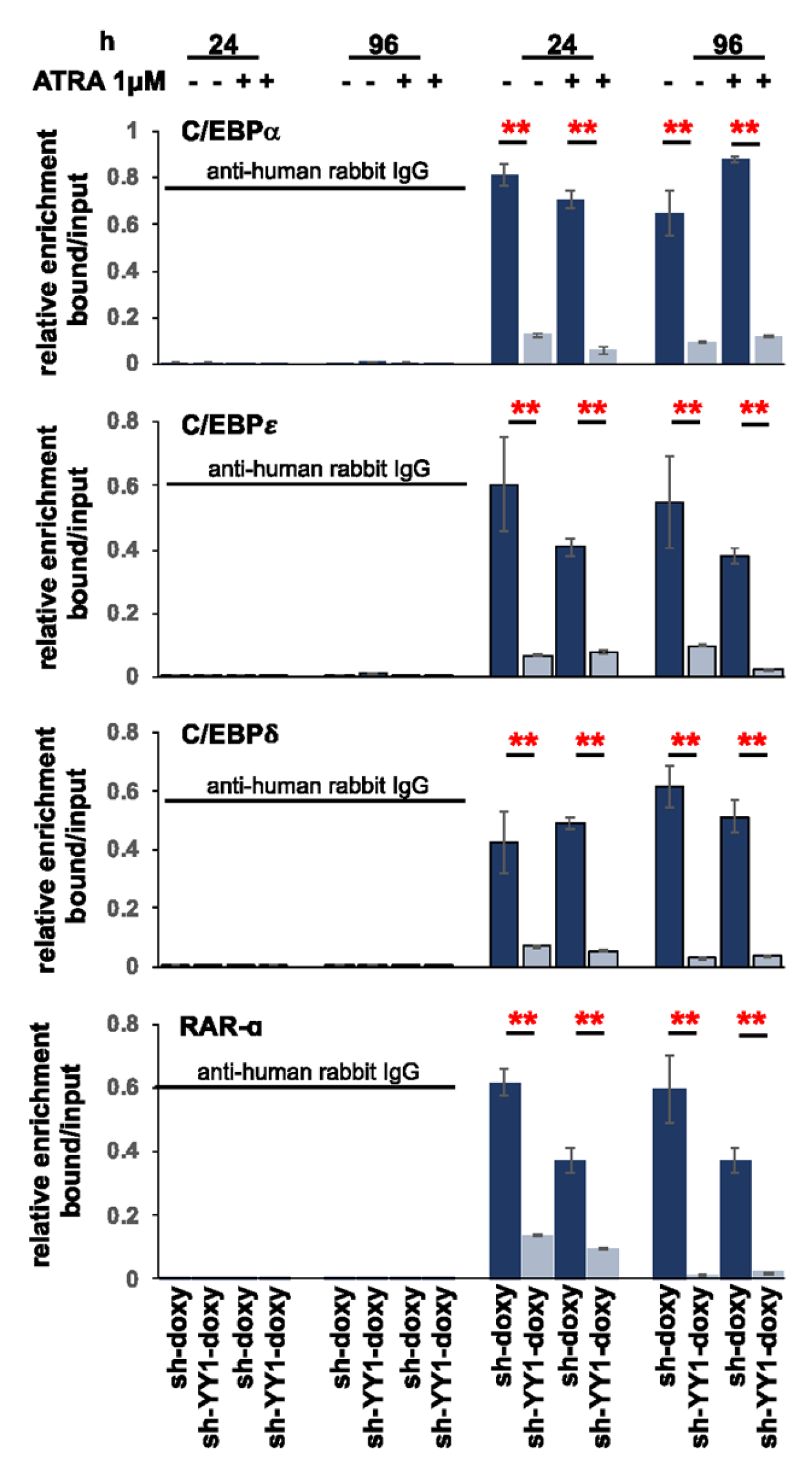
3.4. YY1 binds C/EBPα, C/EBPε, C/EBPδ, and RARα Gene Promoters
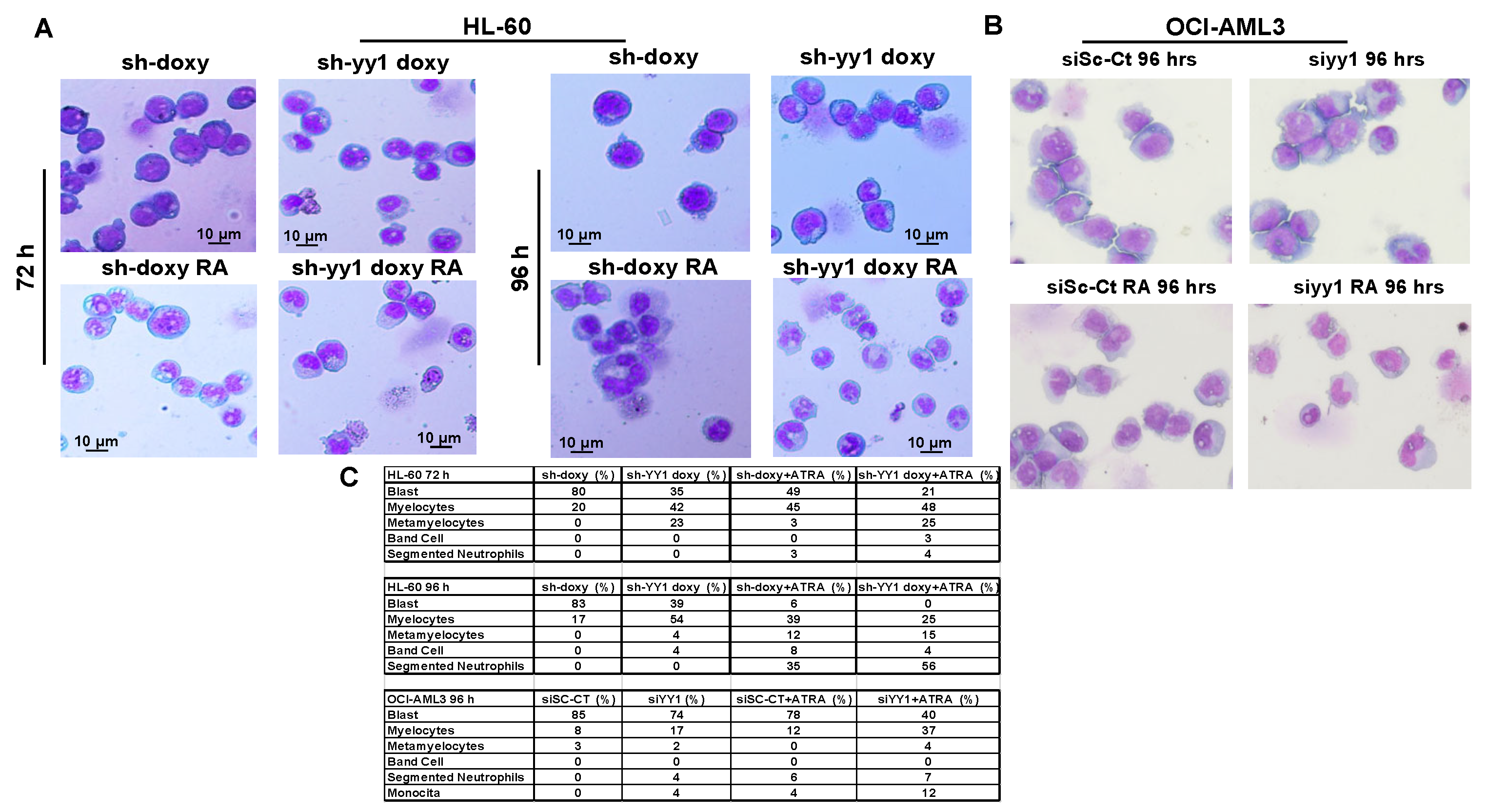
3.5. YY1 Knockdown Enhances Expression of Myeloid Differentiation Markers and Cell Morphological Differentiation Features
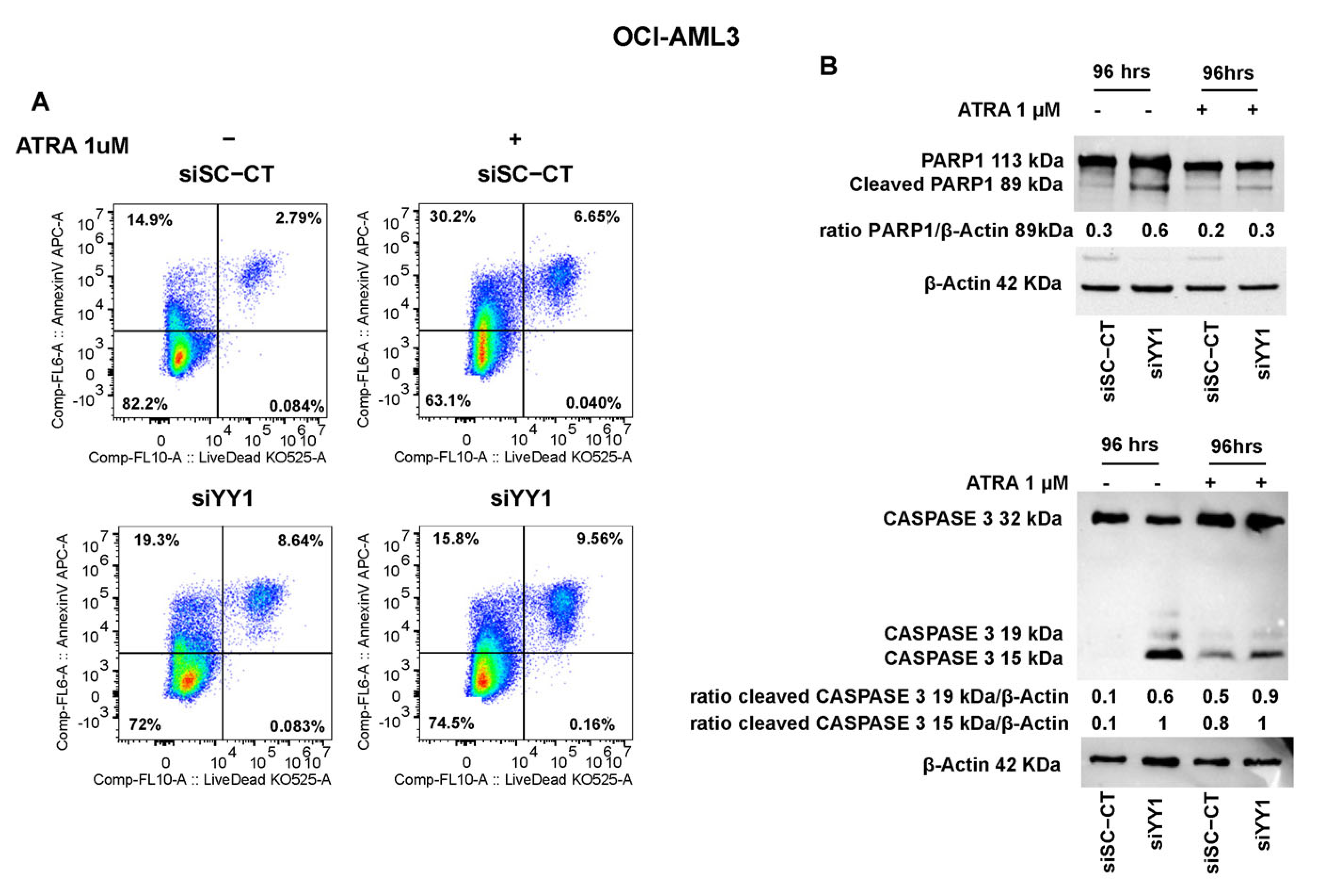
3.6. YY1 Knockdown Reduces Cell Proliferation and Promotes Apoptosis of AML Cell Lines Untreated and Treated with ATRA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, J.J.; Park, T.S.; Wan, T.S.K. Recurrent Cytogenetic Abnormalities in Acute Myeloid Leukemia. Methods Mol. Biol. 2017, 1541, 223–245. [Google Scholar] [PubMed]
- Yin, P.Y.; Wang, R.W.; Jing, R.; Li, X.; Ma, J.H.; Li, K.M.; Wang, H. Research progress on molecular biomarkers of acute myeloid leukemia. Front. Oncol. 2023, 7, 1078556. [Google Scholar] [CrossRef] [PubMed]
- Kogan, A.A.; Lapidus, R.G.; Baer, M.R.; Rassool, F.V. Exploiting epigenetically mediated changes: Acute myeloid leukemia, leukemia stem cells and the bone marrow microenvironment. Adv. Cancer Res. 2019, 141, 213–253. [Google Scholar] [PubMed]
- Yang, L.; Rau, R.; Goodell, M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 2015, 15, 152–165. [Google Scholar] [CrossRef]
- Pappalardi, M.B.; Keenan, K.; Cockerill, M.; Kellner, W.A.; Stowell, A.; Sherk, C.; Wong, K.; Pathuri, S.; Briand, J.; Steidel, M.; et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2021, 2, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Trempenau, M.L.; Schuster, M.B.; Pundhir, S.; Pereira, M.A.; Kalvisa, A.; Tapia, M.; Su, J.; Ge, Y.; de Boer, B.; Balhuizen, A.; et al. Leukemia The histone demethylase KDM5C functions as a tumor suppressor in AML by repression of bivalently marked immature genes. Leukemia 2023, 37, 593–605. [Google Scholar] [CrossRef]
- Scalea, S.; Maresca, C.; Catalanotto, C.; Marino, R.; Cogoni, C.; Reale, A.; Zampieri, M.; Zardo, G. Modifications of H3K4 methylation levels are associated with DNA hypermethylation in acute myeloid leukemia. FEBS J. 2020, 287, 1155–1175. [Google Scholar] [CrossRef]
- Khalaj, M.; Woolthuis, C.M.; Hu, W.; Durham, B.H.; Chu, S.H.; Qamar, S.; Armstrong, S.A.; Park, C.Y. miR-99 regulates normal and malignant hematopoietic stem cell self- renewal. J. Exp. Med. 2017, 214, 2453–2470. [Google Scholar] [CrossRef]
- Hu, W.; Dooley, J.; Chung, S.S.; Chandramohan, D.; Cimmino, L.; Mukherjee, S.; Mason, C.E.; de Strooper, B.; Liston, A.; Park, C.Y. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood 2015, 125, 2206–2216. [Google Scholar] [CrossRef]
- Zardo, G.; Ciolfi, A.; Vian, L.; Starnes, L.M.; Billi, M.; Racanicchi, S.; Maresca, C.; Fazi, F.; Travaglini, L.; Noguera, N.; et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 2012, 119, 4034–4046. [Google Scholar] [CrossRef]
- Mishra, S.; Liu, J.; Chai, L.; Tenen, D.G. Diverse functions of long noncoding RNAs in acute myeloid leukemia: Emerging roles in pathophysiology, prognosis, and treatment resistance. Curr. Opin. Hematol. 2022, 29, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Zardo, G.; Ciolfi, A.; Vian, L.; Billi, M.; Racanicchi, S.; Grignani, F.; Nervi, C. Transcriptional targeting by microRNA-polycomb complexes: A novel route in cell fate determination. Cell Cycle 2012, 11, 3543–3549. [Google Scholar] [CrossRef] [PubMed]
- Leone, G.; D’Alò, F.; Zardo, G.; Voso, M.T.; Nervi, C. Epigenetic treatment of myelodysplastic syndromes and acute myeloid leukemias. Curr. Med. Chem. 2008, 15, 1274–1287. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.T.; So, C.W. Epigenetic therapies by targeting aberrant histone methylome in AML: Molecular mechanisms, current preclinical and clinical development. Oncogene 2017, 36, 1753–1759. [Google Scholar] [CrossRef]
- Short, N.J.; Kantarjian, H. Hypomethylating agents for the treatment of myelodysplastic syndromes and acute myeloid leukemia: Past discoveries and future directions. Am. J. Hematol. 2022, 97, 1616–1626. [Google Scholar] [CrossRef]
- Di Croce, L.; Raker, V.A.; Corsaro, M.; Fazi, F.; Fanelli, M.; Faretta, M.; Fuks, F.; Lo Coco, F.; Kouzarides, T.; Nervi, C.; et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 2002, 295, 1079–1082. [Google Scholar] [CrossRef]
- Grignani, F.; De Matteis, S.; Nervi, C.; Tomassoni, L.; Gelmetti, V.; Cioce, M.; Fanelli, M.; Ruthardt, M.; Ferrara, F.F.; Zamir, I.; et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 1998, 391, 815–818. [Google Scholar] [CrossRef]
- Martens, J.H.; Brinkman, A.B.; Simmer, F.; Francoijs, K.J.; Nebbioso, A.; Ferrara, F.; Altucci, L.; Stunnenberg, H.G. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 2010, 17, 173–185. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Taranova, O.; He, J.; Zhang, Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 2011, 117, 6912–6922. [Google Scholar] [CrossRef]
- Bernt, K.M.; Zhu, N.; Sinha, A.U.; Vempati, S.; Faber, J.; Krivtsov, A.V.; Feng, Z.; Punt, N.; Daigle, A.; Bullinger, L.; et al. MLL-rearranged Leukemia is Dependent on Aberrant H3K79 Methylation by DOT1L. Cancer Cells 2011, 20, 66–78. [Google Scholar] [CrossRef]
- Fazi, F.; Zardo, G.; Gelmetti, V.; Travaglini, L.; Ciolfi, A.; Di Croce, L.; Rosa, A.; Bozzoni, I.; Grignani, F.; Lo-Coco, F.; et al. Heterochromatic gene repression of the retinoic acid pathway in acute myeloid leukemia. Blood 2007, 109, 4432–4440. [Google Scholar] [CrossRef] [PubMed]
- Rejeski, K.; Duque-Afonso, J.; Lübbert, M. AML1/ETO and its function as a regulator of gene transcription via epigenetic mechanisms. Oncogene 2021, 40, 5665–5676. [Google Scholar] [CrossRef] [PubMed]
- Mayle, A.; Yang, L.; Rodriguez, B.; Zhou, T.; Chang, E.; Curry, C.V.; Challen, G.A.; Li, W.; Wheeler, D.; Rebel, V.I.; et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 2015, 125, 629–663. [Google Scholar] [CrossRef] [PubMed]
- Roller, A.; Grossmann, V.; Bacher, U.; Poetzinger, F.; Weissmann, S.; Nadarajah, N.; Boeck, L.; Kern, W.; Haferlach, C.; Schnittger, S.; et al. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia 2013, 27, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Rodriguez, B.; Mayle, A.; Park, H.J.; Lin, X.; Luo, M.; Jeong, M.; Curry, C.V.; Kim, S.B.; Ruau, D.; et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD associated leukemias. Cancer Cell 2016, 29, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Kitabayashi, I. Oncogenic roles of enhancer of zeste homolog 1/2 in hematological malignancies. Cancer Sci. 2018, 109, 2342–2348. [Google Scholar] [CrossRef] [PubMed]
- Delhommeau, F.; Dupont, S.; Della Valle, V.; James, C.; Trannoy, S.; Massé, A.; Kosmider, O.; Le Couedic, J.P.; Robert, F.; Alberdi, A.; et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef]
- Langemeijer, S.M.; Kuiper, R.P.; Berends, M.; Knops, R.; Aslanyan, M.G.; Massop, M.; Stevens-Linders, E.; van Hoogen, P.; van Kessel, A.G.; Raymakers, R.A.; et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009, 41, 838–842. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Abdel-Wahab, O.; Lu, C.; Ward, P.S.; Patel, J.; Shih, A.; Li, Y.; Bhagwat, N.; Vasanthakumar, A.; Fernandez, H.F.; et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010, 18, 553–567. [Google Scholar] [CrossRef]
- Chou, W.C.; Huang, H.H.; Hou, H.A.; Chen, C.Y.; Tang, J.L.; Yao, M.; Tsay, W.; Ko, B.S.; Wu, S.J.; Huang, S.Y.; et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood 2010, 116, 4086–4094. [Google Scholar] [CrossRef] [PubMed]
- Asada, S.; Kitamura, T. Aberrant histone modifications induced by mutant ASXL1 in myeloid neoplasms. Int. J. Hematol. 2019, 110, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Raible, R.F.; Mollaaghababa, J.R.; Guyon, J.R.; Wu, C.T.; Bender, W.; Kingston, R.E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, H.K.; Erdjument-Bromage, P.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef]
- Healy, E.; Mucha, M.; Glancy, E.; Fitzpatrick, D.J.; Conway, E.; Neikes, H.K.; Monger, C.; Van Mierlo, G.; Baltissen, M.P.; Koseki, Y.; et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cell 2019, 76, 437–452. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Zeisig, B.B.; So, C.W.E. Therapeutic Opportunities of Targeting Canonical and Noncanonical PcG/TrxG Functions in Acute Myeloid Leukemia. Ann. Rev. Genom. Hum. Genet. 2021, 22, 103–125. [Google Scholar] [CrossRef]
- Rizo, A.; Olthof, S.; Han, L.; Vellenga, E.; de Haan, G.; Schuringa, J.J. Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood 2009, 114, 1498–1505. [Google Scholar] [CrossRef]
- Park, I.K.; Qian, D.; Kiel, M.; Becker, M.W.; Pihalja, M.; Weissman, I.L.; Morrison, S.J.; Clarke, M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003, 423, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Mihara, K.; Chowdhury, M.; Nakaju, N.; Hidani, S.; Ihara, A.; Hyodo, H.; Yasunaga, S.; Takihara, Y.; Kimura, A. Bmi-1 is useful as a novel molecular marker for predicting progression of myelodysplastic syndrome and patient prognosis. Blood 2006, 107, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Oguro, H.; Yuan, J.; Tanaka, S.; Miyagi, S.; Mochizuki-Kashio, M.; Ichikawa, H.; Yamazaki, S.; Koseki, H.; Nakauchi, H.; Iwama, A.; et al. Lethal myelofibrosis induced by Bmi1-deficient hematopoietic cells unveils a tumor suppressor function of the polycomb group genes. J. Exp. Med. 2012, 209, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.J.L.; Scheijen, B.; Voncken, J.-W.; Kieboom, K.; Berns, A.; van Lohuizen, M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999, 13, 2678–2690. [Google Scholar] [CrossRef]
- Beà, S.; Tort, F.; Pinyol, M.; Puig, X.; Hernández, L.; Hernández, S.; Fernandez, P.L.; van Lohuizen, M.; Colomer, D.; Campo, E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001, 61, 2409–2412. [Google Scholar]
- Klauke, K.; Radulović, V.; Broekhuis, M.; Weersing, E.; Zwart, E.; Olthof, S.; Ritsema, M.; Bruggeman, S.; Wu, X.; Helin, K.; et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 2013, 15, 353–362. [Google Scholar] [CrossRef]
- Scott, C.L.; Gil, J.; Hernando, E.; Teruya-Feldstein, J.; Narita, M.; Martínez, D.; Visakorpi, T.; Mu, D.; Cordon-Cardo, C.; Peters, G.; et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 5389–5394. [Google Scholar] [CrossRef]
- Grossmann, V.; Tiacci, E.; Holmes, A.B.; Kohlmann, A.; Martelli, M.P.; Kern, W.; Spanhol-Rosseto, A.; Klein, H.U.; Dugas, M.; Schindela, S.; et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 2011, 118, 6153–6163. [Google Scholar] [CrossRef]
- Li, M.; Collins, R.; Jiao, Y.; Ouillette, P.; Bixby, D.; Erba, H.; Vogelstein, B.; Kinzler, K.W.; Papadopoulos, N.; Malek, S.N.; et al. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood 2011, 118, 5914–5917. [Google Scholar] [CrossRef]
- Damm, F.; Chesnais, V.; Nagata, Y.; Yoshida, K.; Scourzic, L.; Okuno, Y.; Itzykson, R.; Sanada, M.; Shiraishi, Y.; Gelsi-Boyer, V.; et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood 2013, 122, 3169–3177. [Google Scholar] [CrossRef]
- Pettirossi, V.; Venanzi, A.; Spanhol-Rosseto, A.; Schiavoni, G.; Santi, A.; Tasselli, L.; Naccari, M.; Pensato, V.; Pucciarini, A.; Martelli, M.P.; et al. The gene mutation landscape of acute myeloid leukemia cell lines and its exemplar use to study the BCOR tumor suppressor. Leukemia 2023, 37, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Ziemin-van der Poel, S.; McCabe, N.R.; Gill, H.J.; Espinosa, R., 3rd; Patel, Y.; Harden, A.; Rubinelli, P.; Smith, S.D.; LeBeau, M.M.; Rowley, J.D.; et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. USA 1991, 88, 10735–10739. [Google Scholar] [CrossRef] [PubMed]
- Faber, J.; Krivtsov, A.V.; Stubbs, M.C.; Wright, R.; Davis, T.N.; van den Heuvel-Eibrink, M.; Zwaan, C.M.; Kung, A.L.; Armstrong, S.A. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 2009, 113, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, P.; Su, R.; Hu, C.; Li, Y.; Elkahloun, A.G.; Zuo, Z.; Gurbuxani, S.; Arnovitz, S.; Weng, H.; et al. PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute Myeloid Leukemias Characterized by a Core Transcriptome of the MLL-Rearranged Disease. Cancer Res. 2016, 76, 619–629. [Google Scholar] [CrossRef]
- Stasik, S.; Middeke, J.M.; Kramer, M.; Röllig, C.; Krämer, A.; Scholl, S.; Hochhaus, A.; Crysandt, M.; Brümmendorf, T.H.; Naumann, R.; et al. EZH2 mutations and impact on clinical outcome: An analysis in 1604 patients with newly diagnosed acute myeloid leukemia. Haematologica 2020, 105, e228–e231. [Google Scholar] [CrossRef]
- Score, J.; Hidalgo-Curtis, C.; Jones, A.V.; Winkelmann, N.; Skinner, A.; Ward, D.; Zoi, K.; Ernst, T.; Stegelmann, F.; Döhner, K.; et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood 2012, 119, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-L.; Deng, T.-R.; Shang, Z.; Xiao, Y. JARID2 inhibits leukemia cell proliferation by regulating CCND1 expression. Int. J. Hematol. 2015, 102, 76–85. [Google Scholar] [CrossRef]
- Puda, A.; Milosevic, J.D.; Berg, T.; Klampfl, T.; Harutyunyan, A.S.; Gisslinger, B.; Rumi, E.; Pietra, D.; Malcovati, L.; Elena, C.; et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am. J. Hematol. 2012, 87, 245–250. [Google Scholar] [CrossRef]
- Ueda, T.; Nakata, Y.; Nagamachi, A.; Yamasaki, N.; Kanai, A.; Sera, Y.; Sasaki, M.; Matsui, H.; Honda, Z.; Oda, H.; et al. Propagation of trimethylated H3K27 regulated by polycomb protein EED is required for embryogenesis, hematopoietic maintenance, and tumor suppression. Proc. Natl. Acad. Sci. USA 2016, 113, 10370–10375. [Google Scholar] [CrossRef]
- Klymenko, T.; Papp, B.; Fischle, W.; Köcher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Müller, J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef]
- Brown, J.L.; Mucci, D.; Whiteley, M.; Dirksen, M.L.; Kassis, J.A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1998, 1, 1057–1064. [Google Scholar] [CrossRef]
- Atchison, L.; Ghias, A.; Wilkinson, F.; Bonini, N.; Atchison, M.L. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003, 22, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brown, J.L.; Cao, R.; Zhang, Y.; Kassis, J.A.; Jones, R.S. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 2004, 14, 637–646. [Google Scholar] [CrossRef]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 2010, 140, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, F.H.; Park, K.; Atchison, M.L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA 2006, 103, 19296–19301. [Google Scholar] [CrossRef]
- Caretti, G.; Di Padova, M.; Micales, B.; Lyons, G.E.; Sartorelli, V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004, 18, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Li, G.; Wang, W.; Sun, Y.; Zhang, Y.; Zhong, C.; Stovall, D.B.; Li, D.; Shi, J.; Sui, G.; et al. Disruption of YY1-EZH2 Interaction Using Synthetic Peptides Inhibits Breast Cancer Development. Cancers 2021, 13, 2402. [Google Scholar] [CrossRef]
- Kim, S.Y.; Paylor, S.W.; Magnuson, T.; Schumacher, A. Juxtaposed Polycomb complexes co-regulate vertebral identity. Development 2006, 133, 4957–4968. [Google Scholar] [CrossRef]
- Yao, Y.L.; Yang, W.M.; Seto, E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell Biol. 2001, 21, 5979–5991. [Google Scholar] [CrossRef]
- Seto, E.; Lewis, B.; Shenk, T. Interaction between transcription factors Sp1 and YY1. Nature 1993, 365, 462–464. [Google Scholar] [CrossRef]
- Shrivastava, A.; Saleque, S.; Kalpana, G.V.; Artandi, S.; Goff, S.P.; Calame, K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 1993, 262, 1889–1892. [Google Scholar] [CrossRef] [PubMed]
- Morales-Rosado, J.A.; Kaiwar, C.; Smith, B.E.; Klee, E.W.; Dhamija, R. A case of YY1-associated syndromic learning disability or Gabriele-de vries syndrome with myasthenia gravis. Am. J. Med. Genet. A 2018, 176, 2846–2849. [Google Scholar] [CrossRef]
- Gabriele, M.; Vulto-van Silfhout, A.T.; Germain, P.L.; Vitriolo, A.; Kumar, R.; Douglas, E.; Haan, E.; Kosaki, K.; Takenouchi, T.; Rauch, A.; et al. YY1 Haploinsufficiency Causes an Intellectual Disability Syndrome Featuring Transcriptional and Chromatin Dysfunction. Am. J. Hum. Genet. 2017, 100, 907–925. [Google Scholar] [CrossRef]
- Aubry, S.; Shin, W.; Crary, J.F.; Lefort, R.; Qureshi, Y.H.; Lefebvre, C.; Califano, A.; Shelanski, M.L. Assembly and interrogation of Alzheimer’s disease genetic networks reveal novel regulators of progression. PLoS ONE 2015, 10, e0120352. [Google Scholar] [CrossRef]
- Pal, R.; Tiwari, P.C.; Nath, R.; Pant, K.K. Role of neuroinflammation and latent transcription factors in pathogenesis of Parkinson’s disease. Neurol. Res. 2016, 38, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Meliala, I.T.S.; Hosea, R.; Kasim, V.; Wu, S. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics 2020, 10, 4183–4200. [Google Scholar] [CrossRef] [PubMed]
- Varum, S.; Baggiolini, A.; Zurkirchen, L.; Atak, Z.K.; Cantù, C.; Marzorati, E.; Bossart, R.; Wouters, J.; Häusel, J.; Tuncer, E.; et al. Yin Yang 1 Orchestrates a Metabolic Program Required for Both Neural Crest Development and Melanoma Formation. Cell Stem Cell 2019, 24, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, L.; Wu, Z.; Xuan, C.; Zhang, J.; You, Y.; Chen, X. miR218 inhibits the proliferation of human glioma cells through downregulation of Yin Yang 1. Mol. Med. Rep. 2018, 17, 1926–1932. [Google Scholar]
- Li, H.; Li, T.; Huang, D.; Zhang, P. Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506- 3p/CTNNB1 axis to activate Wnt/beta-catenin signaling pathway. Cancer Cell Int. 2020, 20, 29. [Google Scholar] [CrossRef]
- Galloway, N.R.; Ball, K.F.; Stiff, T.; Wall, N.R. Yin Yang 1 (YY1): Regulation of Survivin and Its Role in Invasion and Metastasis. Crit. Rev. Oncog. 2017, 22, 23–36. [Google Scholar] [CrossRef]
- Kaufhold, S.; Garban, H.; Bonavida, B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J. Exp. Clin. Cancer Res. 2016, 35, 84. [Google Scholar] [CrossRef] [PubMed]
- Petkova, V.; Romanowski, M.J.; Sulijoadikusumo, I.; Rohne, D.; Kang, P.; Shenk, T.; Usheva, A. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J. Biol. Chem. 2001, 276, 7932–7936. [Google Scholar] [CrossRef] [PubMed]
- Sui, G.; Affarel, B.; Shi, Y.; Brignone, C.; Wall, N.R.; Yin, P.; Donohoe, M.; Luke, M.P.; Calvo, D.; Grossman, S.R.; et al. Yin Yang 1 is a negative regulator of p53. Cell 2004, 117, 859–872. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, X.; Xu, L.; Pan, H.; Zhu, S.; Liang, Q.; Huang, B.; Lu, J. The transcription factor ZBP-89 suppresses p16 expression through a histone modification mechanism to affect cell senescence. FEBS J. 2009, 276, 4197–4206. [Google Scholar] [CrossRef] [PubMed]
- Riggs, K.J.; Saleque, S.; Wong, K.K.; Merrell, K.T.; Lee, J.S.; Shi, Y.; Calame, K. Yin-yang 1 activates the c-myc promoter. Mol. Cell Biol. 1993, 13, 7487–7495. [Google Scholar] [PubMed]
- Pan, X.; Jones, M.; Jiang, J.; Zaprazna, K.; Yu, D.; Pear, W.; Maillard, I.; Atchison, M.L. Increased expression of PcG protein YY1 negatively regulates B cell development while allowing accumulation of myeloid cells and LT-HSC cells. PLoS ONE 2012, 7, e30656. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hong, C.C.; Kong, G.; Assumpção, A.L.F.V.; Ong, I.M.; Bresnick, E.H.; Zhang, J.; Pan, X. Polycomb group protein YY1 is an essential regulator of hematopoietic stem cell quiescence. Cell Rep. 2018, 22, 1545–1559. [Google Scholar] [CrossRef]
- Grubach, L.; Juhl-Christensen, C.; Rethmeier Olesen, L.H.; Aggerholm, A.; Hokland, P.; Ostergaard, M. Gene expression profiling of Polycomb, Hox and Meis genes in patients with acute myeloid leukaemia. Eur. J. Haematol. 2008, 81, 112–122. [Google Scholar] [CrossRef]
- Erkeland, S.J.; Valkhof, M.; Heijmans-Antonissen, C.; Delwel, R.; Valk, P.J.; Hermans, M.H.; Touw, I.P. The gene encoding the transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood 2003, 101, 1111–1117. [Google Scholar] [CrossRef]
- Antonio-Andres, G.; Rangel-Santiago, J.; Tirado-Rodriguez, B.; Martinez-Ruiz, G.U.; Klunder-Klunder, M.; Vega, M.I.; Lopez-Martinez, B.; Jiménez-Hernández, E.; Torres Nava, J.; Medina-Sanson, A.; et al. Role of Yin Yang-1 (YY1) in the transcription regulation of the multi-drug resistance (MDR1) gene. Leuk. Lymphoma. 2018, 59, 2628–2638. [Google Scholar] [CrossRef]
- Roboz, G.J.; Mandrekar, S.J.; Desai, P.; Laumann, K.; Walker, A.R.; Wang, E.S.; Kolitz, J.E.; Powell, B.L.; Attar, E.C.; Stock, W.; et al. Randomized trial of 10 days of decitabine ± bortezomib in untreated older patients with AML: CALGB 11002 (Alliance). Blood Adv. 2018, 2, 3608–3617. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Kim, E. Efficacy of epigenetic agents for older patients with acute myeloid leukemia and myelodysplastic syndrome in randomized controlled trials: A systematic review and network meta-analysis. Clin. Exp. Med. 2023, 1–10. [Google Scholar] [CrossRef]
- Hideshima, T.; Chauhan, D.; Richardson, P.; Mitsiades, C.; Mitsiades, N.; Hayashi, T.; Munshi, N.; Dang, L.; Castro, A.; Palombella, V.; et al. NF-kappa B as a therapeutic target in multiple myeloma. J. Biol. Chem. 2002, 10, 16639–16647. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.L.; Neering, S.J.; Upchurch, D.; Grimes, B.; Howard, D.S.; Rizzieri, D.A.; Luger, S.M.; Jordan, C.T. Nuclear Factor-KappaB Is Constitutively Activated in Primitive Human Acute Myelogenous Leukemia Cells. Blood 2001, 98, 2301–2307. [Google Scholar] [CrossRef]
- Baumgartner, B.; Weber, M.; Quirling, M.; Fischer, C.; Page, S.; Adam, M.; Von Schilling, C.; Waterhouse, C.; Schmid, C.; Neumeier, D.; et al. Increased IkappaB Kinase Activity Is Associated with Activated NF-KappaB in Acute Myeloid Blasts. Leukemia 2002, 16, 2062–2071. [Google Scholar] [CrossRef]
- Wang, H.; Hertlein, E.; Bakkar, N.; Sun, H.; Acharyya, S.; Wang, J.; Carathers, M.; Davuluri, R.; Guttridge, D.C. NF-κB regulation of YY1 inhibits skeletal myo-genesis through transcriptional silencing of myofibrillar genes. Mol. Cell Biol. 2007, 27, 4374–4387. [Google Scholar] [CrossRef]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-κB–YY1–miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef]
- Pulikkan, J.A.; Tenen, D.G.; Behre, G. C/EBPα deregulation as a paradigm for leukemogenesis. Leukemia 2017, 31, 2279–2285. [Google Scholar] [CrossRef]
- Larsen, M.T.; Häger, M.; Glenthøj, A.; Asmar, F.; Clemmensen, S.N.; Mora-Jensen, H.; Borregaard, N.; Cowland, J.B. miRNA-130a regulates C/EBP-ε expression during granulopoiesis. Blood 2014, 123, 1079–1089. [Google Scholar] [CrossRef]
- Agrawal, S.; Hofmann, W.K.; Tidow, N.; Ehrich, M.; van den Boom, D.; Koschmieder, S.; Berdel, W.E.; Serve, H.; Müller-Tidow, C. The C/EBPdelta tumor suppressor is silenced by hypermethylation in acute myeloid leukemia. Blood 2007, 109, 3895–3905. [Google Scholar] [CrossRef]
- Tolomeo, M.; Grimaudo, S. The “Janus” Role of C/EBPs Family Members in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 4308. [Google Scholar] [CrossRef]
- Tasseff, R.; Jensen, H.A.; Congleton, J. An Effective Model of the Retinoic Acid Induced HL-60 Differentiation Program. Sci. Rep. 2017, 7, 14327. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Huang, M.; Thomas, D.; Li, M.X.; Feng, W.; Chan, S.M.; Majeti, R.; Mitchell, B.S. Role of cysteine 288 in nucleophosmin cytoplasmic mutations: Sensitization to toxicity induced by arsenic trioxide and bortezomib. Leukemia 2013, 27, 1970–1980. [Google Scholar] [CrossRef]
- Martelli, M.P.; Gionfriddo, I.; Mezzasoma, F.; Milano, F.; Pierangeli, S.; Mulas, F.; Pacini, R.; Tabarrini, A.; Pettirossi, V.; Rossi, R.; et al. Arsenic trioxide and all-trans retinoic acid target NPM1 mutant oncoprotein levels and induce apoptosis in NPM1-mutated AML cells. Blood 2015, 125, 3455–3465. [Google Scholar] [CrossRef] [PubMed]
- Schlenk, R.F.; Dohner, K.; Kneba, M.; Götze, K.; Hartmann, F.; Del Valle, F.; Kirchen, H.; Koller, E.; Fischer, J.T.; Bullinger, L.; et al. Gene mutations and response to treatment with all-trans retinoic acid in elderly patients with acute myeloid leukemia. Results from the AMLSG Trial AML HD98B. Haematologica 2009, 94, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Lanotte, M.; Martin-Thouvenin, V.; Najman, S.; Balerini, P.; Valensi, F.; Berger, R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 1991, 77, 1080–1086. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B.; Kaufhold, S. Prognostic significance of YY1 protein expression and mRNA levels by bioinformatics analysis in human cancers: A therapeutic target. Pharmacol. Ther. 2015, 150, 149–168. [Google Scholar]
- Yin, Q.; Shen, X.; Xu, H.; Feng, W.; Shi, X.; Ju, S. YY1-induced LncRNA-TUG1 elevates YOD1 to promote cell proliferation and inhibit bortezomib sensitivity in multiple myeloma. Leuk. Lymphoma. 2023, 64, 1–14. [Google Scholar] [CrossRef]
- Tameni, A.; Sauta, E.; Mularoni, V.; Torricelli, F.; Manzotti, G.; Inghirami, G.; Bellazzi, R.; Fragliasso, V.; Ciarrocchi, A. The DNA-helicase HELLS drives ALK-ALCL proliferation by the transcriptional control of a cytokinesis-related program. Cell Death Dis. 2021, 12, 130. [Google Scholar] [CrossRef]
- Morales-Martinez, M.; Vega, G.G.; Neri, N.; Nambo, M.J.; Alvarado, I.; Cuadra, I.; Duran-Padilla, M.A.; Huerta-Yepez, S.; Vega, M.I. MicroRNA-7 Regulates Migration and Chemoresistance in Non-Hodgkin Lymphoma Cells Through Regulation of KLF4 and YY1. Front. Oncol. 2020, 10, 588893. [Google Scholar] [CrossRef] [PubMed]
- Morales-Martinez, M.; Valencia-Hipolito, A.; Vega, G.G.; Neri, N.; Nambo, M.J.; Alvarado, I.; Cuadra, I.; Duran-Padilla, M.A.; Martinez-Maza, O.; Huerta-Yepez, S.; et al. Regulation of Krüppel-Like Factor 4 (KLF4) expression through the transcription factor Yin-Yang 1 (YY1) in non-Hodgkin B-cell lymphoma. Oncotarget 2019, 10, 2173–2188. [Google Scholar] [CrossRef] [PubMed]
- Antonio-Andres, G.; Martinez-Ruiz, G.U.; Morales-Martinez, M.; Jiménez-Hernandez, E.; Martinez-Torres, E.; Lopez-Perez, T.V.; Estrada-Abreo, L.A.; Patino-Lopez, G.; Juarez-Mendez, S.; Davila-Borja, V.M.; et al. Transcriptional Regulation of Yin-Yang 1 Expression through the Hypoxia Inducible Factor-1 in Pediatric Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2022, 23, 1728. [Google Scholar] [CrossRef] [PubMed]
- Marchwicka, A.; Marcinkowska, E. Regulation of Expression of CEBP Genes by Variably Expressed Vitamin D Receptor and Retinoic Acid Receptor α in Human Acute Myeloid Leukemia Cell Lines. Int. J. Mol. Sci. 2018, 19, 1918. [Google Scholar] [CrossRef]
- Konopleva, M.; Pollyea, D.A.; Potluri, J.; Chyla, B.; Hogdal, L.; Busman, T.; McKeegan, E.; Salem, A.H.; Zhu, M.; Ricker, J.L.; et al. Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov. 2016, 6, 1106–1117. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noguera, N.I.; Travaglini, S.; Scalea, S.; Catalanotto, C.; Reale, A.; Zampieri, M.; Zaza, A.; Ricciardi, M.R.; Angelini, D.F.; Tafuri, A.; et al. YY1 Knockdown Relieves the Differentiation Block and Restores Apoptosis in AML Cells. Cancers 2023, 15, 4010. https://doi.org/10.3390/cancers15154010
Noguera NI, Travaglini S, Scalea S, Catalanotto C, Reale A, Zampieri M, Zaza A, Ricciardi MR, Angelini DF, Tafuri A, et al. YY1 Knockdown Relieves the Differentiation Block and Restores Apoptosis in AML Cells. Cancers. 2023; 15(15):4010. https://doi.org/10.3390/cancers15154010
Chicago/Turabian StyleNoguera, Nelida Ines, Serena Travaglini, Stefania Scalea, Caterina Catalanotto, Anna Reale, Michele Zampieri, Alessandra Zaza, Maria Rosaria Ricciardi, Daniela Francesca Angelini, Agostino Tafuri, and et al. 2023. "YY1 Knockdown Relieves the Differentiation Block and Restores Apoptosis in AML Cells" Cancers 15, no. 15: 4010. https://doi.org/10.3390/cancers15154010
APA StyleNoguera, N. I., Travaglini, S., Scalea, S., Catalanotto, C., Reale, A., Zampieri, M., Zaza, A., Ricciardi, M. R., Angelini, D. F., Tafuri, A., Ottone, T., Voso, M. T., & Zardo, G. (2023). YY1 Knockdown Relieves the Differentiation Block and Restores Apoptosis in AML Cells. Cancers, 15(15), 4010. https://doi.org/10.3390/cancers15154010









