Integrated Analysis Identifies DPP7 as a Prognostic Biomarker in Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Genetic Alteration Analysis
2.2. The Genetic Alteration of DPP7 and the Clinical Roles of DPP7 in CRC
2.3. Biological Functional Analysis and Gene Set Enrichment Analysis (GSEA)
2.4. Correlation between DPP7 and Drug Response
2.5. RNA Extraction and Quantitative Real-Time RT-PCR
2.6. Tissue Microarray and IHC
3. Results
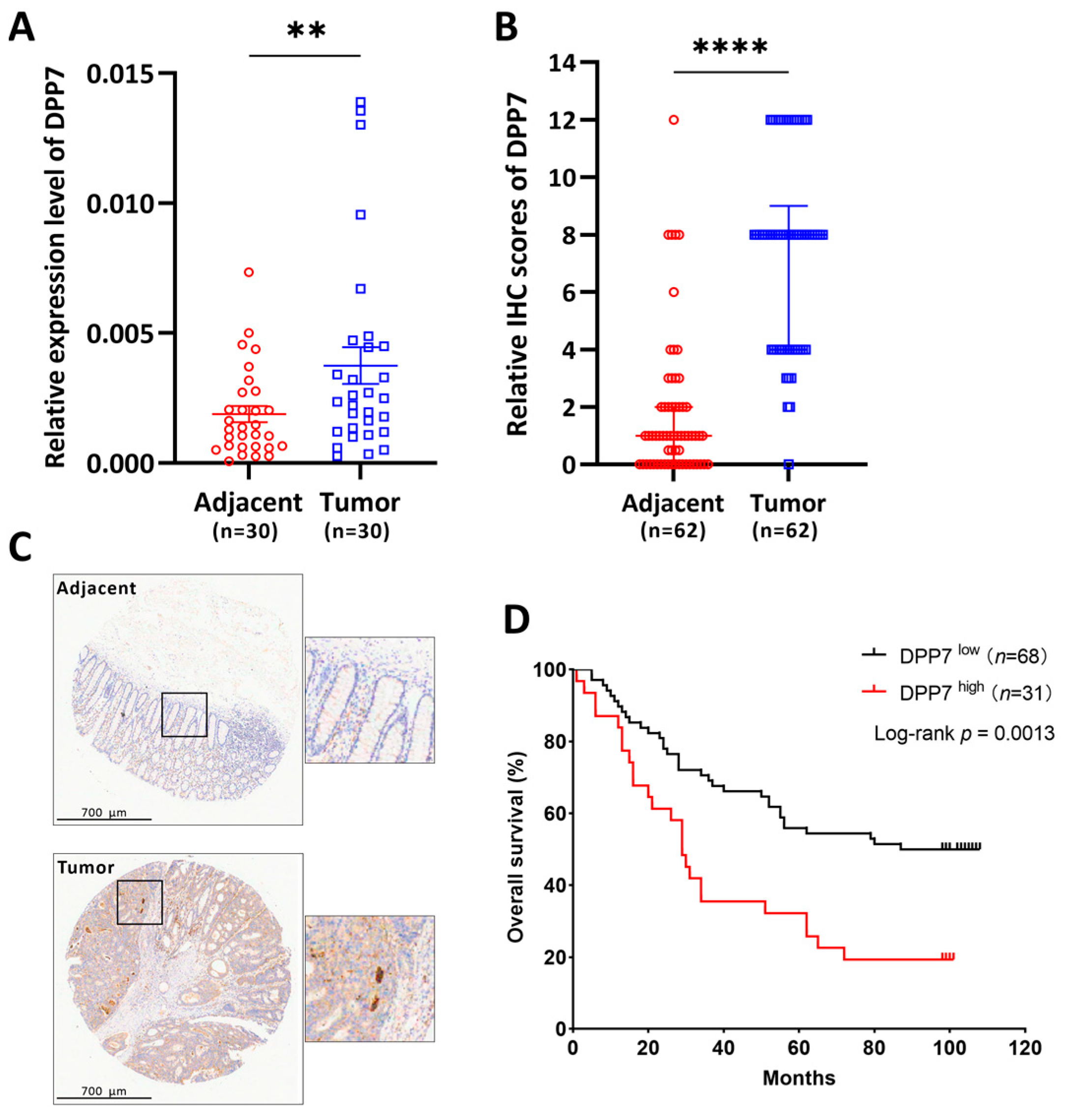
3.1. The Expression of DPP7 in CRC Was Elevated
3.2. Diagnostic and Prognostic Value of DPP7 Expression in CRC
3.3. Construction and Evaluation of a Predictive Nomogram with DPP7
3.4. The PPI Network and DPP7-Related Pathways and Biological Functions
3.5. Correlation between DPP7 and Drug Response
3.6. DPP7 Is Highly Expressed in Patients with Colorectal Cancer and Associated with a Poor Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiser, M.R.; Hsu, M.; Bauer, P.S.; Chapman, W.C., Jr.; Gonzalez, I.A.; Chatterjee, D.; Lingam, D.; Mutch, M.G.; Keshinro, A.; Shia, J.; et al. Clinical Calculator Based on Molecular and Clinicopathologic Characteristics Predicts Recurrence Following Resection of Stage I-III Colon Cancer. J. Clin. Oncol. 2021, 39, 911–919. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.J.; Campbell, M.E.; Goldberg, R.M.; Grothey, A.; Seitz, J.F.; Benedetti, J.K.; Andre, T.; Haller, D.G.; Sargent, D.J. Survival following recurrence in stage II and III colon cancer: Findings from the ACCENT data set. J. Clin. Oncol. 2008, 26, 2336–2341. [Google Scholar] [CrossRef]
- Lone, A.M.; Nolte, W.M.; Tinoco, A.D.; Saghatelian, A. Peptidomics of the prolyl peptidases. AAPS J. 2010, 12, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danilova, O.; Li, B.; Szardenings, A.K.; Huber, B.T.; Rosenblum, J.S. Synthesis and activity of a potent, specific azabicyclo[3.3.0]-octane-based DPP II inhibitor. Bioorganic Med. Chem. Lett. 2007, 17, 507–510. [Google Scholar] [CrossRef] [Green Version]
- Chiravuri, M.; Schmitz, T.; Yardley, K.; Underwood, R.; Dayal, Y.; Huber, B.T. A novel apoptotic pathway in quiescent lymphocytes identified by inhibition of a post-proline cleaving aminodipeptidase: A candidate target protease, quiescent cell proline dipeptidase. J. Immunol. 1999, 163, 3092–3099. [Google Scholar] [CrossRef]
- Miettinen, J.J.; Kumari, R.; Traustadottir, G.A.; Huppunen, M.E.; Sergeev, P.; Majumder, M.M.; Schepsky, A.; Gudjonsson, T.; Lievonen, J.; Bazou, D.; et al. Aminopeptidase Expression in Multiple Myeloma Associates with Disease Progression and Sensitivity to Melflufen. Cancers 2021, 13, 1527. [Google Scholar] [CrossRef]
- Danilov, A.V.; Danilova, O.V.; Brown, J.R.; Rabinowitz, A.; Klein, A.K.; Huber, B.T. Dipeptidyl peptidase 2 apoptosis assay determines the B-cell activation stage and predicts prognosis in chronic lymphocytic leukemia. Exp. Hematol. 2010, 38, 1167–1177. [Google Scholar] [CrossRef] [Green Version]
- Danilov, A.V.; Klein, A.K.; Lee, H.J.; Baez, D.V.; Huber, B.T. Differential control of G0 programme in chronic lymphocytic leukaemia: A novel prognostic factor. Br. J. Haematol. 2005, 128, 472–481. [Google Scholar] [CrossRef]
- Choy, T.K.; Wang, C.Y.; Phan, N.N.; Khoa Ta, H.D.; Anuraga, G.; Liu, Y.H.; Wu, Y.F.; Lee, K.H.; Chuang, J.Y.; Kao, T.J. Identification of Dipeptidyl Peptidase (DPP) Family Genes in Clinical Breast Cancer Patients via an Integrated Bioinformatics Approach. Diagnostics 2021, 11, 1204. [Google Scholar] [CrossRef]
- Hu, X.; Jiang, J.; Ni, C.; Xu, Q.; Ye, S.; Wu, J.; Ge, F.; Han, Y.; Mo, Y.; Huang, D.; et al. HBV Integration-mediated Cell Apoptosis in HepG2.2.15. J. Cancer 2019, 10, 4142–4150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Gomez, N.C.; Adam, R.C.; Nikolova, M.; Yang, H.; Verma, A.; Lu, C.P.; Polak, L.; Yuan, S.; Elemento, O.; et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell 2017, 169, 636–650.e14. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Cannistra, A.; Fried, I.; Lim, T.; Schaffner, T.; Crovella, M.; Hescott, B.; Leiserson, M.D.M. Functional protein representations from biological networks enable diverse cross-species inference. Nucleic Acids Res. 2019, 47, e51. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wang, J.; Liang, W.J.; Min, G.T.; Wang, H.P.; Chen, W.; Yao, N. LTBP2 promotes the migration and invasion of gastric cancer cells and predicts poor outcome of patients with gastric cancer. Int. J. Oncol. 2018, 52, 1886–1898. [Google Scholar] [CrossRef] [Green Version]
- Ahluwalia, P.; Mondal, A.K.; Bloomer, C.; Fulzele, S.; Jones, K.; Ananth, S.; Gahlay, G.K.; Heneidi, S.; Rojiani, A.M.; Kota, V.; et al. Identification and Clinical Validation of a Novel 4 Gene-Signature with Prognostic Utility in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 3818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, W.; Zeng, Z.; Pan, R.; Wu, H.; Zhang, X.; Chen, H.; Nie, Y.; Yu, Z.; Lei, S. Hypoxia-Related Gene FUT11 Promotes Pancreatic Cancer Progression by Maintaining the Stability of PDK1. Front. Oncol. 2021, 11, 675991. [Google Scholar] [CrossRef] [PubMed]
- Tuna, M.; Smid, M.; Zhu, D.; Martens, J.W.; Amos, C.I. Association between acquired uniparental disomy and homozygous mutations and HER2/ER/PR status in breast cancer. PLoS ONE 2010, 5, e15094. [Google Scholar] [CrossRef] [PubMed]
- Kumari, K.; Das, B.; Adhya, A.K.; Rath, A.K.; Mishra, S.K. Genome-wide expression analysis reveals six contravened targets of EZH2 associated with breast cancer patient survival. Sci. Rep. 2019, 9, 1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mather, R.L.; Loveson, K.F.; Fillmore, H.L. Human Sialic acid O-acetyl esterase (SIAE)—Mediated changes in sensitivity to etoposide in a medulloblastoma cell line. Sci. Rep. 2019, 9, 8609. [Google Scholar] [CrossRef] [Green Version]
- Kumari, R.; Majumder, M.M.; Lievonen, J.; Silvennoinen, R.; Anttila, P.; Nupponen, N.N.; Lehmann, F.; Heckman, C.A. Prognostic significance of esterase gene expression in multiple myeloma. Br. J. Cancer 2021, 124, 1428–1436. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Tsan, Y.T.; Chan, W.C.; Sheu, W.H.; Chen, P.C. Use of an alpha-Glucosidase Inhibitor and the Risk of Colorectal Cancer in Patients with Diabetes: A Nationwide, Population-Based Cohort Study. Diabetes Care 2015, 38, 2068–2074. [Google Scholar] [CrossRef] [Green Version]
- Deschuyter, M.; Pennarubia, F.; Pinault, E.; Legardinier, S.; Maftah, A. Functional Characterization of POFUT1 Variants Associated with Colorectal Cancer. Cancers 2020, 12, 1430. [Google Scholar] [CrossRef]
- Korkmaz, B.; Lesner, A.; Letast, S.; Mahdi, Y.K.; Jourdan, M.L.; Dallet-Choisy, S.; Marchand-Adam, S.; Kellenberger, C.; Viaud-Massuard, M.C.; Jenne, D.E.; et al. Neutrophil proteinase 3 and dipeptidyl peptidase I (cathepsin C) as pharmacological targets in granulomatosis with polyangiitis (Wegener granulomatosis). Semin. Immunopathol. 2013, 35, 411–421. [Google Scholar] [CrossRef]
- Chalmers, J.D.; Haworth, C.S.; Metersky, M.L.; Loebinger, M.R.; Blasi, F.; Sibila, O.; O’Donnell, A.E.; Sullivan, E.J.; Mange, K.C.; Fernandez, C.; et al. Phase 2 Trial of the DPP-1 Inhibitor Brensocatib in Bronchiectasis. N. Engl. J. Med. 2020, 383, 2127–2137. [Google Scholar] [CrossRef]
- Chalmers, J.D.; Usansky, H.; Rubino, C.M.; Teper, A.; Fernandez, C.; Zou, J.; Mange, K.C. Pharmacokinetic/Pharmacodynamic Evaluation of the Dipeptidyl Peptidase 1 Inhibitor Brensocatib for Non-cystic Fibrosis Bronchiectasis. Clin. Pharmacokinet. 2022, 61, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, D.; Zhang, J.; Korkmaz, B.; Chalmers, J.D.; Basso, J.; Lasala, D.; Fernandez, C.; Teper, A.; Mange, K.C.; Perkins, W.R.; et al. Dipeptidyl peptidase-1 inhibition with brensocatib reduces the activity of all major neutrophil serine proteases in patients with bronchiectasis: Results from the WILLOW trial. Respir. Res. 2023, 24, 133. [Google Scholar] [CrossRef] [PubMed]
- Lecot, P.; Sarabi, M.; Pereira Abrantes, M.; Mussard, J.; Koenderman, L.; Caux, C.; Bendriss-Vermare, N.; Michallet, M.C. Neutrophil Heterogeneity in Cancer: From Biology to Therapies. Front. Immunol. 2019, 10, 2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.; Fu, S.; Mastio, J.; Dominguez, G.A.; Purohit, A.; Kossenkov, A.; Lin, C.; Alicea-Torres, K.; Sehgal, M.; Nefedova, Y.; et al. Unique pattern of neutrophil migration and function during tumor progression. Nat. Immunol. 2018, 19, 1236–1247. [Google Scholar] [CrossRef]







| Characteristic | Low Expression of DPP7 | High Expression of DPP7 | p |
|---|---|---|---|
| n | 322 | 322 | |
| Age, meidan (IQR) | 68 (57, 75) | 68 (58.25, 77) | 0.210 |
| Gender, n (%) | 0.752 | ||
| Female | 148 (46.0%) | 153 (47.5%) | |
| Male | 174 (54.0%) | 169 (52.5%) | |
| Race, n (%) | 0.628 | ||
| Asian | 6 (2.8%) | 6 (3.3%) | |
| Black or African American | 41 (19.2%) | 28 (15.6%) | |
| White | 167 (78.0%) | 146 (81.1%) | |
| Age, n (%) | 0.474 | ||
| ≤65 | 143 (44.4%) | 133 (41.3%) | |
| >65 | 179 (55.6%) | 189 (58.7%) | |
| T stage, n (%) | 0.377 | ||
| T1 | 11 (3.4%) | 9 (2.8%) | |
| T2 | 58 (18.2%) | 53 (16.5%) | |
| T3 | 220 (69.0%) | 216 (67.1%) | |
| T4 | 30 (9.4%) | 44 (13.7%) | |
| N stage, n (%) | 0.038 | ||
| N0 | 195 (61.3%) | 173 (53.7%) | |
| N1 | 76 (23.9%) | 77 (23.9%) | |
| N2 | 47 (14.8%) | 72 (22.4%) | |
| M stage, n (%) | 0.138 | ||
| M0 | 236 (86.8%) | 239 (81.8%) | |
| M1 | 36 (13.2%) | 53 (18.2%) | |
| Pathologic stage, n (%) | 0.076 | ||
| Stage I | 57 (18.4%) | 54 (17.3%) | |
| Stage II | 131 (42.3%) | 107 (34.2%) | |
| Stage III | 86 (27.7%) | 98 (31.3%) | |
| Stage IV | 36 (11.6%) | 54 (17.3%) | |
| CEA level, n (%) | 0.830 | ||
| ≤5 | 133 (63.6%) | 128 (62.1%) | |
| >5 | 76 (36.4%) | 78 (37.9%) | |
| Residual tumor, n (%) | 0.082 | ||
| R0 | 228 (92.7%) | 240 (90.9%) | |
| R1 | 5 (2.0%) | 1 (0.4%) | |
| R2 | 13 (5.3%) | 23 (8.7%) | |
| Perineural invasion, n (%) | 0.977 | ||
| No | 91 (74.0%) | 84 (75.0%) | |
| Yes | 32 (26.0%) | 28 (25.0%) | |
| Lymphatic invasion, n (%) | <0.001 | ||
| No | 198 (67.3%) | 152 (52.8%) | |
| Yes | 96 (32.7%) | 136 (47.2%) | |
| Neoplasm type, n (%) | 0.787 | ||
| Colon adenocarcinoma | 237 (73.6%) | 241 (74.8%) | |
| Rectum adenocarcinoma | 85 (26.4%) | 81 (25.2%) |
| Characteristics | Total (n) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| T stage (T3 and T4 vs. T1 and T2) | 640 | 2.468 (1.327–4.589) | 0.004 | 3.669 (0.433–31.068) | 0.233 |
| N stage (N1 and N2 vs. N0) | 639 | 2.627 (1.831–3.769) | <0.001 | 1.113 (0.439–2.821) | 0.821 |
| M stage (M1 vs. M0) | 563 | 3.989 (2.684–5.929) | <0.001 | 1.720 (0.661–4.481) | 0.267 |
| Gender (Male vs. Female) | 643 | 1.054 (0.744–1.491) | 0.769 | ||
| Age (>65 vs. ≤65) | 643 | 1.939 (1.320–2.849) | <0.001 | 2.927 (1.369–6.258) | 0.006 |
| CEA level (>5 vs. ≤5) | 414 | 2.620 (1.611–4.261) | <0.001 | 1.536 (0.763–3.092) | 0.229 |
| Perineural invasion (Yes vs. No) | 235 | 1.692 (0.907–3.156) | 0.099 | ||
| Lymphatic invasion (Yes vs. No) | 581 | 2.144 (1.476–3.114) | <0.001 | 2.845 (1.289–6.281) | 0.010 |
| DPP7 (High vs. Low) | 643 | 2.094 (1.462–2.998) | <0.001 | 2.704 (1.327–5.512) | 0.006 |
| Residual tumor (R0 vs. R1 and R2) | 509 | 0.217 (0.132–0.357) | <0.001 | 0.274 (0.112–0.671) | 0.005 |
| Neoplasm type (rectum adenocarcinoma vs. colon adenocarcinoma) | 643 | 0.799 (0.519–1.230) | 0.308 | ||
| Race (White vs. Black or African American and Asian) | 394 | 0.933 (0.541–1.607) | 0.802 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Wang, H.; Wang, H.; Xu, C.; Zhao, R.; Yao, J.; Zhai, C.; Han, W.; Pan, H.; Sheng, J. Integrated Analysis Identifies DPP7 as a Prognostic Biomarker in Colorectal Cancer. Cancers 2023, 15, 3954. https://doi.org/10.3390/cancers15153954
Zhang W, Wang H, Wang H, Xu C, Zhao R, Yao J, Zhai C, Han W, Pan H, Sheng J. Integrated Analysis Identifies DPP7 as a Prognostic Biomarker in Colorectal Cancer. Cancers. 2023; 15(15):3954. https://doi.org/10.3390/cancers15153954
Chicago/Turabian StyleZhang, Wei, Haidong Wang, Huadi Wang, Chuchu Xu, Rongjie Zhao, Junlin Yao, Chongya Zhai, Weidong Han, Hongming Pan, and Jin Sheng. 2023. "Integrated Analysis Identifies DPP7 as a Prognostic Biomarker in Colorectal Cancer" Cancers 15, no. 15: 3954. https://doi.org/10.3390/cancers15153954
APA StyleZhang, W., Wang, H., Wang, H., Xu, C., Zhao, R., Yao, J., Zhai, C., Han, W., Pan, H., & Sheng, J. (2023). Integrated Analysis Identifies DPP7 as a Prognostic Biomarker in Colorectal Cancer. Cancers, 15(15), 3954. https://doi.org/10.3390/cancers15153954






