Predicting Hearing Loss in Testicular Cancer Patients after Cisplatin-Based Chemotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of the Data
2.2. Treatment and Clinical Information
2.3. Assessment of Hearing Loss
2.4. DNA Preparation and Quality Control
2.5. Genetic Data Feature Selection
2.6. Statistical Analysis
2.6.1. Missing Data
2.6.2. Logistic Regression with Cross-Validated GWAS
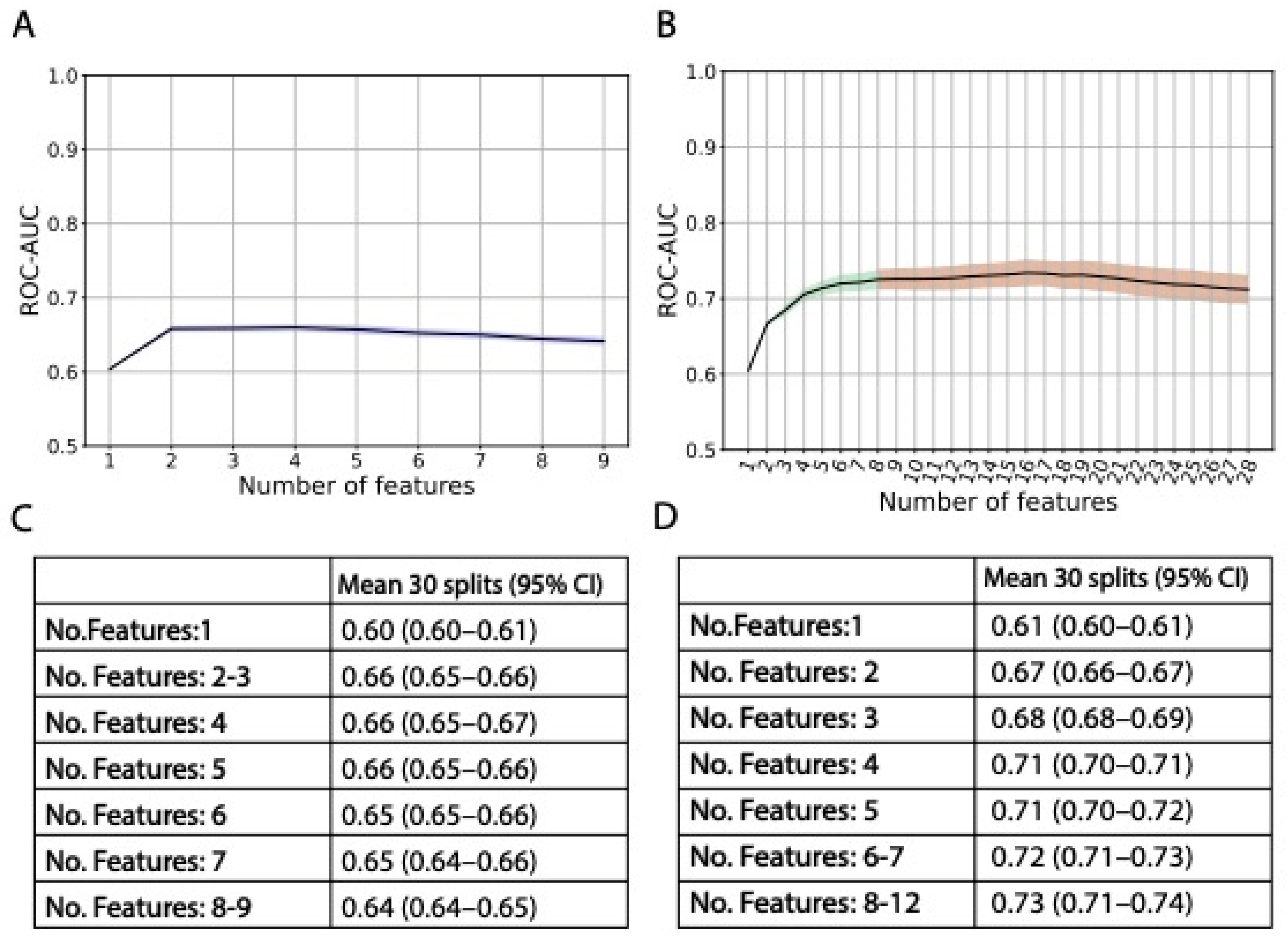
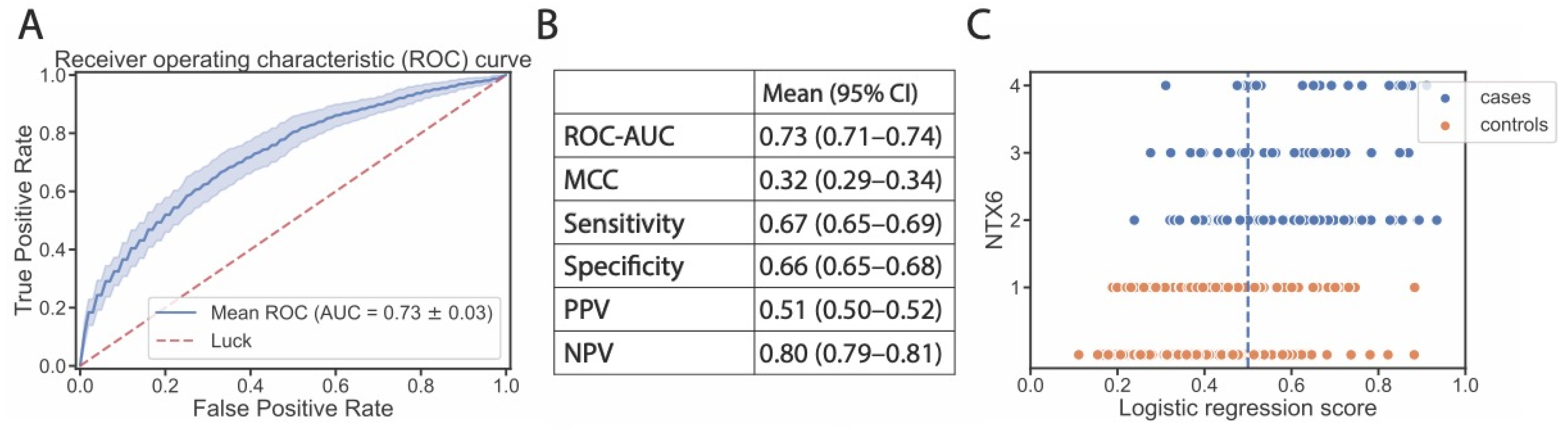
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chia, V.M.; Quraishi, S.M.; Devesa, S.S.; Purdue, M.P.; Cook, M.B.; McGlynn, K.A. International trends in the incidence of testicular cancer, 1973–2002. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1151–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanmugalingam, T.; Soultati, A.; Chowdhury, S.; Rudman, S.; Van Hemelrijck, M. Global incidence and outcome of testicular cancer. Clin. Epidemiol. 2013, 5, 417–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kier, M.G.; Lauritsen, J.; Mortensen, M.S.; Bandak, M.; Andersen, K.K.; Hansen, M.K.; Agerbaek, M.; Holm, N.V.; Dalton, S.O.; Johansen, C.; et al. Prognostic Factors and Treatment Results after Bleomycin, Etoposide, and Cisplatin in Germ Cell Cancer: A Population-based Study. Eur. Urol. 2017, 71, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Einhorn, L.H.; Donohue, J. Cis-diamminedichloroplatinum, vinblastine, and bleomycin combination chemotherapy in disseminated testicular cancer. Ann. Intern. Med. 1977, 87, 293–298. [Google Scholar] [CrossRef]
- Chovanec, M.; Lauritsen, J.; Bandak, M.; Oing, C.; Kier, G.G.; Kreiberg, M.; Rosenvilde, J.; Wagner, T.; Bokemeyer, C.; Daugaard, G. Late adverse effects and quality of life in survivors of testicular germ cell tumour. Nat. Rev. Urol. 2021, 18, 227–245. [Google Scholar] [CrossRef]
- Lauritsen, J.; Mortensen, M.S.; Kier, M.G.G.; Christensen, I.J.; Agerbaek, M.; Gupta, R.; Daugaard, G. Renal impairment and late toxicity in germ-cell cancer survivors. Ann. Oncol. 2015, 26, 173–178. [Google Scholar] [CrossRef]
- Frisina, R.D.; Wheeler, H.E.; Fossa, S.D.; Kerns, S.L.; Fung, C.; Sesso, H.D.; Monahan, P.O.; Feldman, D.R.; Hamilton, R.; Vaughn, D.J.; et al. Comprehensive audiometric analysis of hearing impairment and tinnitus after cisplatin-based chemotherapy in survivors of adult-onset cancer. J. Clin. Oncol. 2016, 34, 2712–2720. [Google Scholar] [CrossRef]
- Brydøy, M.; Oldenburg, J.; Klepp, O.; Bremnes, R.M.; Wentzel-Larsen, T.; Hauge, E.R.; Dahl, O.; Foss, S.D. Observational study of prevalence of long-term Raynaud-like phenomena and neurological side effects in testicular cancer survivors. J. Natl. Cancer Inst. 2009, 101, 1682–1695. [Google Scholar] [CrossRef]
- Lauritsen, J.; Bandak, M.; Kreiberg, M.; Skøtt, J.W.; Wagner, T.; Rosenvilde, J.J.; Dysager, L.; Agerbæk, M.; Daugaard, G. Long-term neurotoxicity and quality of life in testicular cancer survivors—A nationwide cohort study. J. Cancer Surviv. 2021, 15, 509–517. [Google Scholar] [CrossRef]
- Brock, P.R.; Knight, K.R.; Freyer, D.R.; Campbell, K.C.M.; Steyger, P.S.; Blakley, B.W.; Rassekh, S.R.; Chang, K.W.; Fligor, B.J.; Rajput, K.; et al. Platinum-induced ototoxicity in children: A consensus review on mechanisms, predisposition, and protection, including a new International Society of Pediatric Oncology Boston ototoxicity scale. J. Clin. Oncol. 2012, 30, 2408–2417. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, H.E.; Gamazon, E.R.; Frisina, R.D.; Perez-Cervantes, C.; El Charif, O.; Mapes, B.; Fossa, S.D.; Feldman, D.R.; Hamilton, R.J.; Vaughn, D.J.; et al. Variants in WFS1 and Other Mendelian Deafness Genes Are Associated with Cisplatin-Associated Ototoxicity. Clin. Cancer Res. 2017, 23, 3325–3333. [Google Scholar] [CrossRef] [Green Version]
- Rybak, L.P.; Whitworth, C.A.; Mukherjea, D.; Ramkumar, V. Mechanisms of cisplatin-induced ototoxicity and prevention. Hear. Res. 2007, 226, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Kreiberg, M.; Bandak, M.; Lauritsen, J.; Skøtt, J.W.; Johansen, N.B.; Agerbaek, M.; Holm, N.V.; Johansen, C.; Daugaard, G. Cohort profile: The Danish Testicular Cancer late treatment effects cohort (DaTeCa-LATE). Front. Oncol. 2018, 8, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daugaard, G.; Kier, M.G.G.; Bandak, M.; Mortensen, M.S.; Larsson, H.; Søgaard, M.; Toft, B.G.; Engvad, B.; Agerbæk, M.; Holm, N.V.; et al. The Danish testicular cancer database. Clin. Epidemiol. 2016, 8, 703–707. [Google Scholar] [CrossRef] [Green Version]
- Garcia, S.L.; Lauritsen, J.; Zhang, Z.; Bandak, M.; Dalgaard, M.D.; Nielsen, R.L.; Daugaard, G.; Gupta, R. Prediction of Nephrotoxicity Associated With Cisplatin-Based Chemotherapy in Testicular Cancer Patients. JNCI Cancer Spectr. 2020, 4, pkaa032. [Google Scholar] [CrossRef]
- International Germ Cell Cancer Collaborative Group. Germ Cell Consensus Classification: A prognostic factor-based staging system for metastatic germ cell cancers. J. Clin. Oncol. 1997, 15, 594–603. [Google Scholar] [CrossRef]
- Huang, H.Q.; Brady, M.F.; Cella, D.; Fleming, G.; Mackey, D. Validation and reduction of FACT/GOG-Ntx subscale for platinum/paclitaxel- induced neurologic symptoms: A gynecologic oncology group study. Int. J. Gynecol. Cancer 2007, 17, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Tserga, E.; Nandwani, T.; Edvall, N.K.; Bulla, J.; Patel, P.; Canlon, B.; Cederroth, C.R.; Baguley, D.M. The genetic vulnerability to cisplatin ototoxicity: A systematic review. Sci. Rep. 2019, 9, 3455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, S.F. A Method of Estimation of Missing Values in Multivariate Data Suitable for Use with an Electronic Computer. J. R. Stat. Soc. Ser. B 1960, 22, 302–306. [Google Scholar] [CrossRef]
- van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In NIPS’17, Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017. [Google Scholar]
- Haugnes, H.S.; Stenklev, N.C.; Brydøy, M.; Dahl, O.; Wilsgaard, T.; Laukli, E.; Fosså, S.D. Hearing loss before and after cisplatin-based chemotherapy in testicular cancer survivors: A longitudinal study. Acta Oncol. (Madr.) 2018, 57, 1075–1083. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Escamez, J.A.; Bibas, T.; Cima, R.F.F.; Van de Heyning, P.; Knipper, M.; Mazurek, B.; Szczepek, A.J.; Cederroth, C.R. Genetics of Tinnitus: An Emerging Area for Molecular Diagnosis and Drug Development. Front. Neurosci. 2016, 10, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vona, B.; Nanda, I.; Shehata-Dieler, W.; Haaf, T. Genetics of Tinnitus: Still in its Infancy. Front. Neurosci. 2017, 11, 236. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Pourvali, K.; Abbasi, M.; Mottaghi, A. Role of Superoxide Dismutase 2 Gene Ala16Val Polymorphism and Total Antioxidant Capacity in Diabetes and its Complications. Avicenna J. Med. Biotechnol. 2016, 8, 48–56. [Google Scholar] [PubMed]
- Sutton, A.; Khoury, H.; Prip-Buus, C.; Cepanec, C.; Pessayre, D.; Degoul, F. The Ala16Val genetic dimorphism modulates the import of human manganese superoxide dismutase into rat liver mitochondria. Pharmacogenetics 2003, 13, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Sutton, A.; Imbert, A.; Igoudjil, A.; Descatoire, V.; Cazanave, S.; Pessayre, D.; Degoul, F. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharmacogenet. Genom. 2005, 15, 311–319. [Google Scholar] [CrossRef]
- Brown, A.L.; Lupo, P.J.; Okcu, M.F.; Lau, C.C.; Rednam, S.; Scheurer, M.E. SOD2 genetic variant associated with treatment-related ototoxicity in cisplatin-treated pediatric medulloblastoma. Cancer Med. 2015, 4, 1679–1686. [Google Scholar] [CrossRef]
- Bastaki, M.; Huen, K.; Manzanillo, P.; Chande, N.; Chen, C.; Balmes, J.R.; Tager, I.B.; Holland, N. Genotype-activity relationship for Mn-superoxide dismutase, glutathione peroxidase 1 and catalase in humans. Pharmacogenet. Genom. 2006, 16, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Uno, Y.; Murayama, N.; Kunori, M.; Yamazaki, H. Characterization of Microsomal Glutathione S -Transferases MGST1, MGST2, and MGST3 in Cynomolgus Macaque. Drug Metab. Dispos. 2013, 41, 1621–1625. [Google Scholar] [CrossRef] [Green Version]
- Romano, A.; Capozza, M.A.; Mastrangelo, S.; Maurizi, P.; Triarico, S.; Rolesi, R.; Attinà, G.; Fetoni, A.R.; Ruggiero, A. Assessment and Management of Platinum-Related Ototoxicity in Children Treated for Cancer. Cancers 2020, 12, 1266. [Google Scholar] [CrossRef]
- Paciello, F.; Fetoni, A.R.; Mezzogori, D.; Rolesi, R.; Di Pino, A.; Paludetti, G.; Grassi, C.; Troiani, D. The dual role of curcumin and ferulic acid in counteracting chemoresistance and cisplatin-induced ototoxicity. Sci. Rep. 2020, 10, 1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Allende, G.; Chávez-Reyes, J.; Guerrero-Alba, R.; Vázquez-León, P.; Marichal-Cancino, B.A. Advances in Neurobiology and Pharmacology of GPR12. Front. Pharmacol. 2020, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Pasello, M.; Giudice, A.M.; Scotlandi, K. The ABC subfamily A transporters: Multifaceted players with incipient potentialities in cancer. Semin. Cancer Biol. 2020, 60, 57–71. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Morii, I.; Iwabuchi, Y.; Mori, S.; Suekuni, M.; Natsume, T.; Yoshida, K.; Sugimoto, N.; Kanemaki, M.T.; Fujita, M. Inhibiting the MCM8-9 complex selectively sensitizes cancer cells to cisplatin and olaparib. Cancer Sci. 2019, 110, 1044–1053. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, P.; Schmiedge, J.; Manchaiah, V.; Swapna, S.; Dhandayutham, S.; Kothandaraman, P.P. Ototoxicity: A Challenge in Diagnosis and Treatment. J. Audiol. Otol. 2018, 22, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Higginson, I.J. Measuring quality of life: Using quality of life measures in the clinical setting. BMJ 2001, 322, 1297–1300. [Google Scholar] [CrossRef]
- Kakarmath, S.; Esteva, A.; Arnaout, R.; Harvey, H.; Kumar, S.; Muse, E.; Dong, F.; Wedlund, L.; Kvedar, J. Best practices for authors of healthcare-related artificial intelligence manuscripts. NPJ Digit. Med. 2020, 3, 134. [Google Scholar] [CrossRef]
| Description | Number of Genes | ||
|---|---|---|---|
| Cisplatin metabolism | Resistance Pathway (KEGG Pathways) | Overview of genes and interactions resulting in platinum-based drugs resistance. | 46 |
| Detoxification Pathway (BioCyc Pathway) | Cisplatin is degraded via the glutathione-mediated detoxification pathway. | 9 | |
| Glutathione Transferases Cytochrome P450 Enzymes ABC Transporters (Uniprot) | The three protein groups may be associated with cisplatin-introduced neurotoxicity, since they affect the uptake and disposition. Genes associated with the groups were identified with Uniprot. | 26 61 49 | |
| Cisplatin (Uniprot) | Systematic search identifying cisplatin-related genes conducted with Uniprot. | 22 | |
| Cisplatin (DrugBank) | The DrugBank database contains information on pharmaceutical drugs including cisplatin. | 31 | |
| Ototoxicity | Sensorineural Hearing Loss | Sensorineural hearing loss-related genes conducted with Uniprot. | 155 |
| Ototoxicity | Ototoxicity-related genes conducted with Uniprot. | 2 | |
| Affected, Number (%) | Non-Affected, Number (%) | p Values a | ||
|---|---|---|---|---|
| Number of patients | 146 (34.4) | 278 (65.6) | - | |
| Age at diagnosis, median (IQR) | 34 (27–41) | 29 (26–36) | 0.002 | |
| BMI, median (IQR) Unknown: 8 Affected; 10 Non-affected | 21 (19–27) | 22 (19–26) | 0.38 | |
| BEP regimen | Normal dose | 113 (78.5) | 260 (95.6) | 6 × 10−27 |
| Double dose | 31 (21.5) | 12 (4.4) | ||
| Unknown | 2 | 6 | ||
| GFR before treatment, median (IQR), mL/min/1.73 m2 Unknown: 2 Non-affected | 122 (111–135) | 121 (110–133) | 0.68 | |
| Cisplatin, median (IQR), mg/m2 | 400 (385–403) | 400 (300–400) | p < 0.001 | |
| Treatment cycles | 3 | 30 (20.5) | 86 (30.9) | 1 × 10−5 |
| 4 | 85 (58.2) | 180 (64.7) | ||
| 5 or more | 9 (6.2) | 10 (3.6) | ||
| High-dose | 22 (15.1) | 2 (0.7) | ||
| Histology | Seminoma | 34 (23.3) | 54 (19.4) | 0.42 |
| Non-Seminoma | 112 (76.7) | 224 (80.6) | ||
| Prognostic group | Good | 103 (70.5) | 239 (86) | p < 0.001 |
| Intermediate | 32 (21.9) | 30 (10.8) | ||
| Poor | 11 (7.5) | 9 (3.2) | ||
| Alcohol consumption in number of units per week | 5 (1–10) | 5 (2–10) | 0.30 | |
| smoking | Never | 61 (41.8) | 128 (46.4) | 0.40 |
| Former | 55 (37.7) | 88 (31.9) | ||
| Current | 30 (20.5) | 60 (21.7) | ||
| Unknown | - | 2 | ||
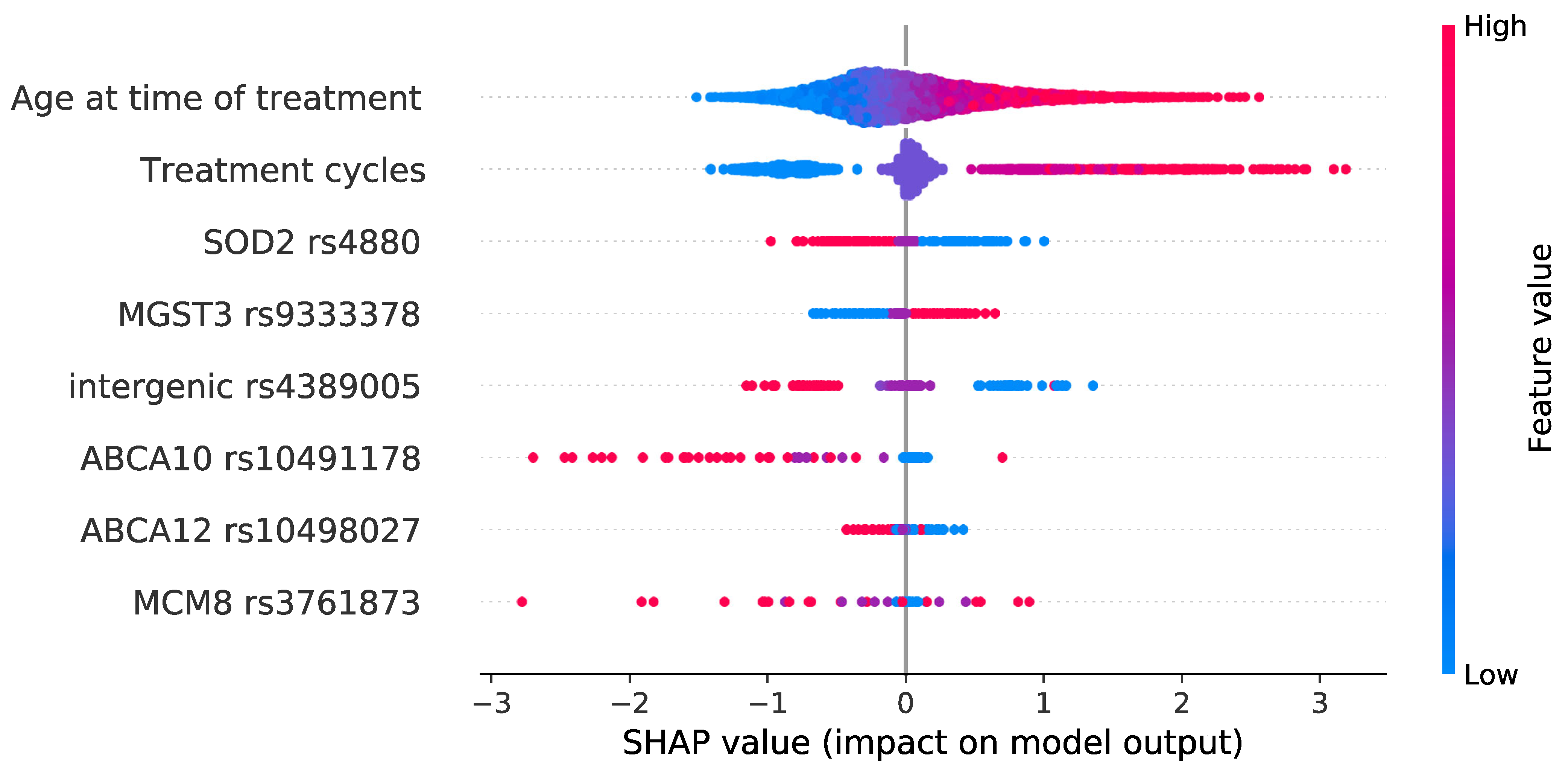
| Chr. | SNP | Genetic Position a | Gene | Reference Allele | Alternative Allele | Risk Allele | MAF (CEU) | Effect | OR (95% CI) b | p Values c |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs9333378 | 165601466 | MGST3 | G | A | A | G: 0.39 | Splice acceptor | 1.37 (1–1.86) | 0.0441 |
| 2 | rs10498027 | 215820013 | ABCA12 | G | A | G | A: 0.40 | Stop gained | 1.11 (0.81–1.51) | 0.5158 |
| 6 | rs4880 | 160113872 | SOD2 | A | G | A | G: 0.47 | Missense | 1.55 (1.13–2.13) | 0.007183 |
| 13 | rs4389005 | 27399338 | GPRR12 (nearest gene) | A | G | A | A: 0.45 | Intergenic | 2.09 (1.56–2.89) | 7 × 10−6 |
| 17 | rs10491178 | 67149973 | ABCA10 | G | A | G | A: 0.07 | Stop gained | 1.84 (0.80–4.22) | 0.1525 |
| 20 | rs3761873 | 5939214 | MCM8 | A | C | A | C: 0.06 | Stop gained | 1.35 (0.66–2.75) | 0.4148 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, S.L.; Lauritsen, J.; Christiansen, B.K.; Hansen, I.F.; Bandak, M.; Dalgaard, M.D.; Daugaard, G.; Gupta, R. Predicting Hearing Loss in Testicular Cancer Patients after Cisplatin-Based Chemotherapy. Cancers 2023, 15, 3923. https://doi.org/10.3390/cancers15153923
Garcia SL, Lauritsen J, Christiansen BK, Hansen IF, Bandak M, Dalgaard MD, Daugaard G, Gupta R. Predicting Hearing Loss in Testicular Cancer Patients after Cisplatin-Based Chemotherapy. Cancers. 2023; 15(15):3923. https://doi.org/10.3390/cancers15153923
Chicago/Turabian StyleGarcia, Sara L., Jakob Lauritsen, Bernadette K. Christiansen, Ida F. Hansen, Mikkel Bandak, Marlene D. Dalgaard, Gedske Daugaard, and Ramneek Gupta. 2023. "Predicting Hearing Loss in Testicular Cancer Patients after Cisplatin-Based Chemotherapy" Cancers 15, no. 15: 3923. https://doi.org/10.3390/cancers15153923
APA StyleGarcia, S. L., Lauritsen, J., Christiansen, B. K., Hansen, I. F., Bandak, M., Dalgaard, M. D., Daugaard, G., & Gupta, R. (2023). Predicting Hearing Loss in Testicular Cancer Patients after Cisplatin-Based Chemotherapy. Cancers, 15(15), 3923. https://doi.org/10.3390/cancers15153923






