miR-4284 Functions as a Tumor Suppressor in Renal Cell Carcinoma Cells by Targeting Glutamate Decarboxylase 1
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Animals
2.2. miRNA Mimics and Plasmid Vector Transfection
2.2.1. Real-Time Reverse Transcription-PCR Analysis of mRNA
2.2.2. Real-Time Reverse Transcription-PCR Analysis of miRNA
2.3. Cell Viability Assay
2.4. Colony-Forming Assay
2.5. Apoptotic Cell Analysis
2.6. Wound-Healing Assay
2.7. Transwell Assay
2.8. Sphere-Forming Assay
2.9. Selection of Target Genes
2.10. Western Blotting
2.11. Reporter Assay
2.12. In Vivo Xenograft Experiments
2.13. Immunohistochemistry Analysis
2.14. Overall Survival Analysis
2.15. Statistical Analyses
3. Results
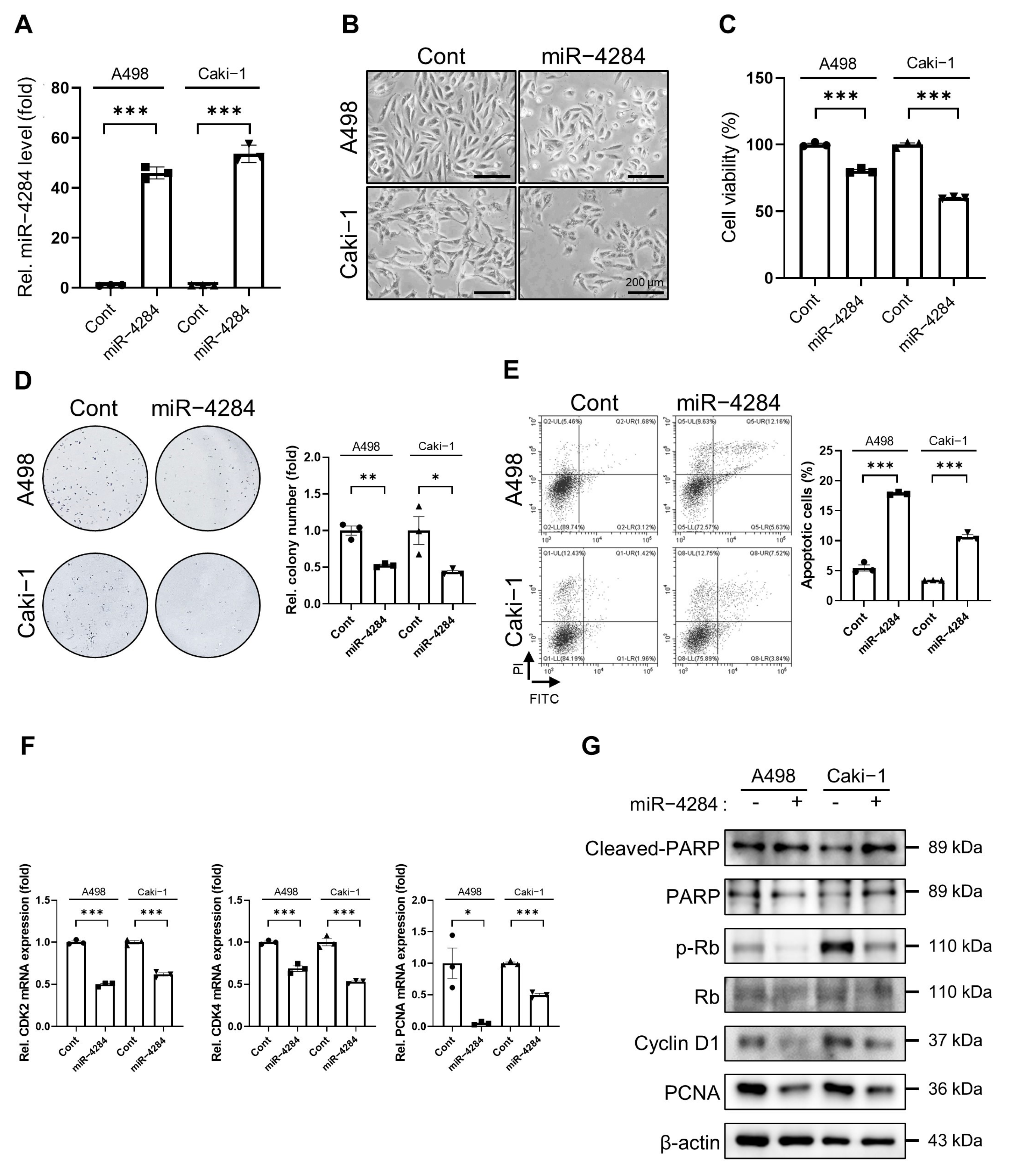
3.1. Overexpression of miR-4284 Inhibits Growth and Promotes Apoptosis in Renal Cancer Cells
3.2. Overexpressed miR-4284 Inhibits the Tumorigenic Characterization of Renal Cancer Cells
3.3. Glutamate Decarboxylase 1 (GAD1) Is a Direct Target of miR-4284 in Renal Cancer Cells
3.4. Enhanced miR-4284 Level Suppresses Tumor Growth In Vivo
3.5. miR-4284 or GAD1 Expression Level Correlates with the Overall Survival of Patients with Renal Cell Carcinoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Porta, C.; Schmidinger, M.; Rioux-Leclercq, N.; Bex, A.; Khoo, V.; Grunwald, V.; Gillessen, S.; Horwich, A.; ESMO Guidelines Committee. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann. Oncol. 2019, 30, 706–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorko, M.; Pacik, D.; Wasserbauer, R.; Juracek, J.; Varga, G.; Ghazal, M.; Nussir, M.I. MicroRNAs in the pathogenesis of renal cell carcinoma and their diagnostic and prognostic utility as cancer biomarkers. Int. J. Biol. Mark. 2016, 31, e26–e37. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Neal, C.S.; Michael, M.Z.; Rawlings, L.H.; Van der Hoek, M.B.; Gleadle, J.M. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 2010, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Brannon, A.R.; Reddy, A.R.; Alexe, G.; Seiler, M.W.; Arreola, A.; Oza, J.H.; Yao, M.; Juan, D.; Liou, L.S.; et al. Identifying mRNA targets of microRNA dysregulated in cancer: With application to clear cell Renal Cell Carcinoma. BMC Syst. Biol. 2010, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, H.; Enokida, H.; Itesako, T.; Tatarano, S.; Kinoshita, T.; Fuse, M.; Kojima, S.; Nakagawa, M.; Seki, N. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J. Hum. Genet. 2013, 58, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, X.; Wang, R.; Xiao, P.; Xu, Z.; Chen, L.; Hang, W.; Ruan, A.; Yang, H.; Zhang, X. microRNA-200c modulates the epithelial-to-mesenchymal transition in human renal cell carcinoma metastasis. Oncol. Rep. 2013, 30, 643–650. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Mollenkopf, H.J.; Grimm, C.; Wagner, I.; Albrecht, M.; Waller, T.; Pilarsky, C.; Johannsen, M.; Stephan, C.; Lehrach, H.; et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J. Cell. Mol. Med. 2009, 13, 3918–3928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juan, D.; Alexe, G.; Antes, T.; Liu, H.; Madabhushi, A.; Delisi, C.; Ganesan, S.; Bhanot, G.; Liou, L.S. Identification of a microRNA panel for clear-cell kidney cancer. Urology 2010, 75, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Chow, T.F.; Youssef, Y.M.; Lianidou, E.; Romaschin, A.D.; Honey, R.J.; Stewart, R.; Pace, K.T.; Yousef, G.M. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clin. Biochem. 2010, 43, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, W.; Luan, Q.; Liu, Y. miR-4284 Promotes Cell Proliferation, Migration, and Invasion in Non-Small Cell Lung Cancer Cells and is Associated with Postoperative Prognosis. Cancer Manag. Res. 2021, 13, 5865–5872. [Google Scholar] [CrossRef] [PubMed]
- Xiaofei, M.; Zengyao, L.; Ye, Z.; Tong, W. MicroRNA-4284 inhibits colon cancer epithelial-mesenchymal transition by down-regulating Perilipin 5. STEMedicine 2021, 2, e85. [Google Scholar]
- Sun, D.; Shang, D.; Miao, P.; Jiang, Z.; Chen, Y.; Gao, J. MiR-4284 inhibits sensitivity to paclitaxel in human ovarian carcinoma SKOV3ip1 and HeyA8 cells by targeting DMC1. Anticancer Drugs 2022, 33, 701–709. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, W.; Ding, Z.; Xu, T.; Zhang, X.; Xu, K. Comprehensive exploration of the expression and prognostic value of AQPs in clear cell renal cell carcinoma. Medicine 2022, 101, e29344. [Google Scholar] [CrossRef]
- Bahadoram, S.; Davoodi, M.; Hassanzadeh, S.; Bahadoram, M.; Barahman, M.; Mafakher, L. Renal cell carcinoma: An overview of the epidemiology, diagnosis, and treatment. G. Ital. Nefrol. 2022, 3, 39. [Google Scholar]
- Hogner, A.; Krause, H.; Jandrig, B.; Kasim, M.; Fuller, T.F.; Schostak, M.; Erbersdobler, A.; Patzak, A.; Kilic, E. PBRM1 and VHL expression correlate in human clear cell renal cell carcinoma with differential association with patient’s overall survival. Urol. Oncol. 2018, 36, 94.e1–94.e14. [Google Scholar] [CrossRef]
- Munari, E.; Marchionni, L.; Chitre, A.; Hayashi, M.; Martignoni, G.; Brunelli, M.; Gobbo, S.; Argani, P.; Allaf, M.; Hoque, M.O.; et al. Clear cell papillary renal cell carcinoma: Micro-RNA expression profiling and comparison with clear cell renal cell carcinoma and papillary renal cell carcinoma. Hum. Pathol. 2014, 45, 1130–1138. [Google Scholar] [CrossRef] [Green Version]
- Barth, D.A.; Slaby, O.; Klec, C.; Juracek, J.; Drula, R.; Calin, G.A.; Pichler, M. Current Concepts of Non-Coding RNAs in the Pathogenesis of Non-Clear Cell Renal Cell Carcinoma. Cancers 2019, 11, 1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaravinos, A.; Lambrou, G.I.; Mourmouras, N.; Katafygiotis, P.; Papagregoriou, G.; Giannikou, K.; Delakas, D.; Deltas, C. New miRNA profiles accurately distinguish renal cell carcinomas and upper tract urothelial carcinomas from the normal kidney. PLoS ONE 2014, 9, e91646. [Google Scholar] [CrossRef]
- Tamaddon, G.; Geramizadeh, B.; Karimi, M.H.; Mowla, S.J.; Abroun, S. miR-4284 and miR-4484 as Putative Biomarkers for Diffuse Large B-Cell Lymphoma. Iran J. Med. Sci. 2016, 41, 334–339. [Google Scholar] [PubMed]
- Zhang, L.; Zhou, F.; Garcia de Vinuesa, A.; de Kruijf, E.M.; Mesker, W.E.; Hui, L.; Drabsch, Y.; Li, Y.; Bauer, A.; Rousseau, A.; et al. TRAF4 promotes TGF-beta receptor signaling and drives breast cancer metastasis. Mol. Cell. 2013, 51, 559–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, H.; Elksnis, A.; Lundkvist, P.; Ubhayasekera, K.; Bergquist, J.; Birnir, B.; Carlsson, P.O.; Espes, D. Endogenous Levels of Gamma Amino-Butyric Acid Are Correlated to Glutamic-Acid Decarboxylase Antibody Levels in Type 1 Diabetes. Biomedicines 2021, 10, 91. [Google Scholar] [CrossRef]
- Yan, H.; Tang, G.; Wang, H.; Hao, L.; He, T.; Sun, X.; Ting, A.H.; Deng, A.; Sun, S. DNA methylation reactivates GAD1 expression in cancer by preventing CTCF-mediated polycomb repressive complex 2 recruitment. Oncogene 2016, 35, 3995–4008. [Google Scholar] [CrossRef]
- Soejima, S.; Kondo, K.; Tsuboi, M.; Muguruma, K.; Tegshee, B.; Kawakami, Y.; Kajiura, K.; Kawakita, N.; Toba, H.; Yoshida, M.; et al. GAD1 expression and its methylation as indicators of malignant behavior in thymic epithelial tumors. Oncol. Lett. 2021, 21, 483. [Google Scholar] [CrossRef]
- Schnepp, P.M.; Lee, D.D.; Guldner, I.H.; O’Tighearnaigh, T.K.; Howe, E.N.; Palakurthi, B.; Eckert, K.E.; Toni, T.A.; Ashfeld, B.L.; Zhang, S. GAD1 Upregulation Programs Aggressive Features of Cancer Cell Metabolism in the Brain Metastatic Microenvironment. Cancer Res. 2017, 77, 2844–2856. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.Y.; Chao, T.B.; Sheu, M.J.; Tian, Y.F.; Chen, T.J.; Lee, S.W.; He, H.L.; Chang, I.W.; Hsing, C.H.; Lin, C.Y.; et al. Glutamate Decarboxylase 1 Overexpression as a Poor Prognostic Factor in Patients with Nasopharyngeal Carcinoma. J. Cancer 2016, 7, 1716–1723. [Google Scholar] [CrossRef] [Green Version]
- Maemura, K.; Yamauchi, H.; Hayasaki, H.; Kanbara, K.; Tamayama, T.; Hirata, I.; Watanabe, M. Gamma-amino-butyric acid immunoreactivity in intramucosal colonic tumors. J. Gastroenterol. Hepatol. 2003, 18, 1089–1094. [Google Scholar] [CrossRef]
- Samborska, B.; Griss, T.; Ma, E.H.; Jones, N.; Williams, K.S.; Sergushichev, A.; Johnson, R.M.; Esaulova, E.; Loginicheva, E.; Flynn, B.; et al. A non-canonical role for glutamate decarboxylase 1 in cancer cell amino acid homeostasis, independent of the GABA shunt. bioRxiv 2021. [Google Scholar] [CrossRef]




| Gene | Sequence (5′–3′) | |
|---|---|---|
| CDK2 | F | CCAGGAGTTACTTCTATGCCTGA |
| R | TTCATCCAGGGGAGGTACAAC | |
| CDK4 | F | ATGGCTACCTCTCGATATGAGC |
| R | CATTGGGGACTCTCACACTCT | |
| PCNA | F | ACACTAAGGGCCGAAGATAACG |
| R | ACAGCATCTCCAATATGGCTGA | |
| HDAC1 | F | CTACTACGACGGGGATGTTGG |
| R | GAGTCATGCGGATTCGGTGAG | |
| GAD1 | F | GCTTCCGGCTAAGAACGGT |
| R | TTGCGGACATAGTTGAGGAGT | |
| RSBN1L | F | GCGGAGAGTGAACGGAGAAG |
| R | GAGGGGCAAAGCTCCAAGAC | |
| SIGLEC11 | F | CTACTGCTGCTTATGGCTACTG |
| R | CGAAAGAAGTACCATGCCTCATC | |
| GAPDH | F | GGAGCGAGATCCCTCCAAAAT |
| R | GGCTGTTGTCATACTTCTCATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, S.; Kim, K.; Yeo, H.; Lee, G.; Kim, I.; Oh, J.; An, H.-J.; Lee, S. miR-4284 Functions as a Tumor Suppressor in Renal Cell Carcinoma Cells by Targeting Glutamate Decarboxylase 1. Cancers 2023, 15, 3888. https://doi.org/10.3390/cancers15153888
Choi S, Kim K, Yeo H, Lee G, Kim I, Oh J, An H-J, Lee S. miR-4284 Functions as a Tumor Suppressor in Renal Cell Carcinoma Cells by Targeting Glutamate Decarboxylase 1. Cancers. 2023; 15(15):3888. https://doi.org/10.3390/cancers15153888
Chicago/Turabian StyleChoi, Sujin, Kyeongmi Kim, Hyunjeong Yeo, Gyurim Lee, Isaac Kim, Jisu Oh, Hyun-Ju An, and Soonchul Lee. 2023. "miR-4284 Functions as a Tumor Suppressor in Renal Cell Carcinoma Cells by Targeting Glutamate Decarboxylase 1" Cancers 15, no. 15: 3888. https://doi.org/10.3390/cancers15153888
APA StyleChoi, S., Kim, K., Yeo, H., Lee, G., Kim, I., Oh, J., An, H.-J., & Lee, S. (2023). miR-4284 Functions as a Tumor Suppressor in Renal Cell Carcinoma Cells by Targeting Glutamate Decarboxylase 1. Cancers, 15(15), 3888. https://doi.org/10.3390/cancers15153888






