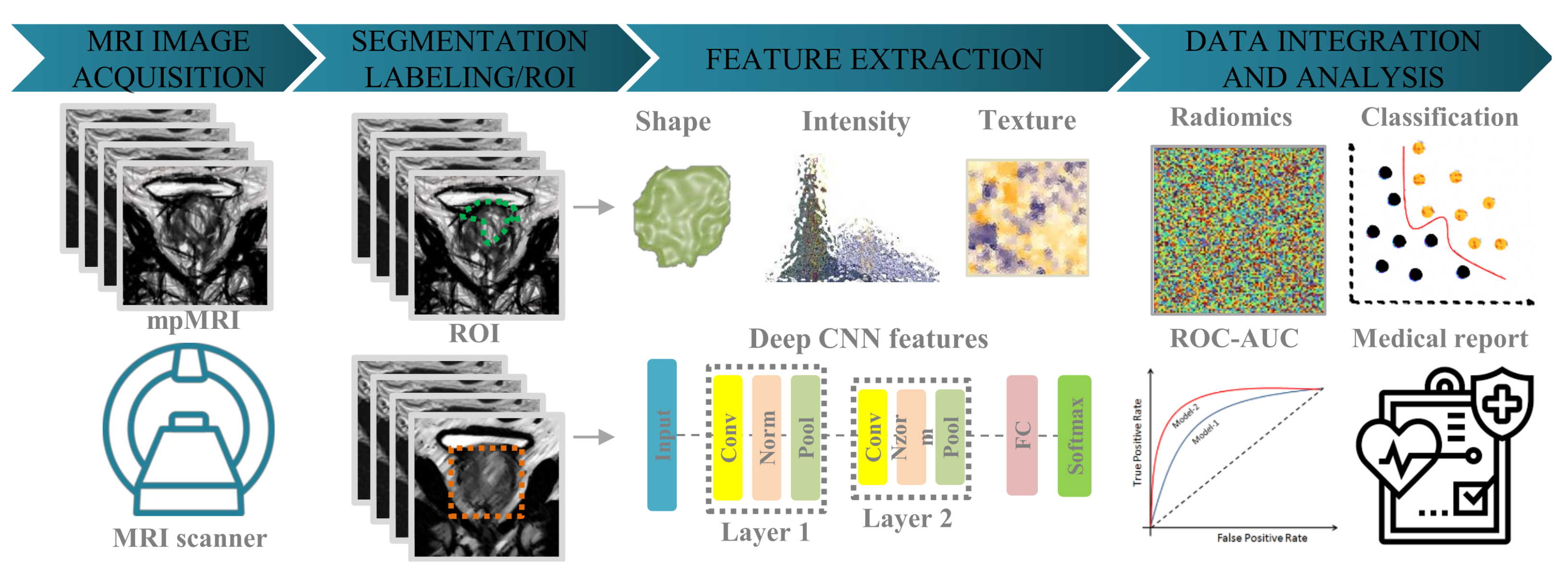
1. Introduction
Prostate cancer (PCa) is a malignant tumor of the male genitourinary system, characterized by epithelial cells. It ranks as the most prevalent malignant tumor in men, the second most common cancer globally, and the fifth leading cause of cancer-related deaths in men. The disease is the primary cancer in 112 nations and is responsible for the majority of cancer deaths in 48 countries [
1]. According to the latest statistics, in 2020, there were approximately 1.4 million newly diagnosed PCa cases and 375,000 deaths worldwide [
2]. Since PCa develops slowly in its early stages, older men, who are at high risk, may not realize that they are affected. Therefore, timely detection is important to reduce mortality rates. Furthermore, early detection and prompt treatment can significantly reduce PCa-related deaths.
PCa can be examined using several methods, including (1) digital rectal examination (DRE), which is the most straightforward and effective method, and critical for the diagnosis; (2) prostate-specific antigen (PSA) test, including total PSA and free PSA; (3) non-invasive ultrasound examination of the prostate, which can detect early internal nodular changes; (4) Computed Tomography (CT) examination of prostate lesions; (5) magnetic resonance examination of prostate lesions; and (6) prostate biopsy, usually performed with transrectal ultrasound guidance. A biopsy involves taking tissue samples from at least 12 sites, which are then examined for pathological changes [
3].
The existing detection methods for PCa have limitations and room for improvement. DRE suffers from variability between reviewers, low reproducibility, sensitivity, and specificity, as well as high false positive rates [
4]. PSA is a nonspecific blood marker and lacks sensitivity, leading to false negatives and many unnecessary biopsies [
5]. Transrectal ultrasound-guided biopsy is prone to random sampling errors and may cause bleeding or infection, making PCa detection more challenging [
3]. Furthermore, combining multiple methods may be more effective than using them individually. In this context, there is a need for more precise, accurate, and non-invasive detection methods to improve PCa diagnosis.
PCa patients are typically classified according to their test results (i.e., PSA, DRE, TRUS, and biopsy), and treatment plans are determined accordingly. In addition, the Gleason score (GS) is a widely used classification method. Unlike other cancer grading systems, the GS does not use the worst morphological grade but instead sums up the primary and secondary morphological grades to determine the overall grade. This method provides better prognostic information for PCa patients and is therefore more appropriate for PCa diagnosis [
6]. For example, Gleason grade 1 (rare) shows large glands with consistent rules and dense back-to-back arrangement. Gleason grade 2 presents relatively irregular large glands forming small nodules that are not fused. Gleason grade 3 exhibits small acinic glands with infiltrative growth or small cribriform glands. Gleason grade 4 presents fused glands, large cribriform glands, or renal clear cell carcinoma-like morphology. Gleason grade 5 has no adenoid structure, single cancer cell infiltration, or acne-like appearance with cancer cell necrosis [
6].
As GS has certain limitations, the International Society of Urological Pathology (ISUP) has proposed a new grading system based on five grade groups (GG) to address these limitations. The morphological definition of the five GGs is as follows: GG 1 (GS
): cancer composed of a single, discrete, and well-structured gland; GG 2 (GS 3 + 4 = 7): mainly composed of discrete and well-structured glands, with a small part composed of poorly shaped, fused, renal globular, cribriform glands; GG 3 (GS 4 + 3 = 7): mainly composed of poorly shaped, fused, renal small globular, cribriform glands, and a small part composed of suitable discrete glands; GG 4 (GS 4 + 4, GS 3 + 5, GS 5 + 3): composed of poorly shaped, fused, renal globular, cribriform glands or discontinuous glands and glands lacking a small part, or glands lacking a small part of discontinuous and well-formed glands; GG 5 (GS 9, GS 10): no glandular cavity formation or glandular cavity necrosis [
7]. Proper classification and grading of patients can help clinicians formulate personalized treatment plans and evaluate the prognosis of patients. While prostate biopsy remains the gold standard for detecting PCa, mpMRI is emerging as a useful method in early screening, especially as early clinical symptoms may not be apparent. mpMRI allows a detailed anatomical evaluation of the prostate, provides a clear description of the regional anatomy and acceptable resolution of soft tissue, and has many MRI scan options that are superior to other imaging methods [
8]. Specifically, mpMRI is a non-invasive imaging technique that has several applications in PCa detection, localization, staging, risk classification, and biopsy guidance [
9]. However, radiologists interpret mpMRI images to diagnose illnesses, including PCa. However, like any medical imaging, the interpretation of mpMRI images is subjective and can be influenced by the radiologist’s experience and expertise. This subjectivity can potentially lead to errors in interpretation. Therefore, obtaining high-quality images and ensuring proper patient preparation is important for obtaining accurate diagnoses. With an increasing emphasis on identifying and treating high-risk tumors and reducing overtreatment of low-risk tumors, mpMRI plays a critical role in PCa diagnosis [
8]. Additionally, mpMRI can be used for quantitative imaging (radiomics) to predict clinical outcomes of PCa [
9].
Radiomics is a quantitative method used to analyze data obtained from medical images, including mpMRI, to evaluate cancer (e.g., PCa) and other diseases. Radiomics aims to extract a large number of quantitative features from medical images and use these features to establish models that can classify and predict various aspects of cancer, such as diagnosis, prognosis, and response to treatment. Traditional radiology extracts features from a single modality, such as Computed Tomography. However, with the development of artificial intelligence (AI) technology, radiomics is becoming more applicable in the medical field. It can be used to predict the prognosis of multiple cancers, the response to various treatment methods, distinguish benign treatment confounding factors and progression, identify abnormal tumor response, and predict mutations and molecular characteristics. Radiomics is moving towards a multi-parameter approach, enabling tumors to be characterized more quantitatively and objectively to overcome the variability between observers. This may result in helpful predictive biomarkers that cannot be recognized by visual analysis [
10]. However, one of the obstacles in translating radiomics from research to clinical practice is the interpretability of the data [
11]. Furthermore, the challenges of texture image variability must also be addressed [
12,
13]. Despite these challenges, radiomics studies have been widely distributed and published, as shown in
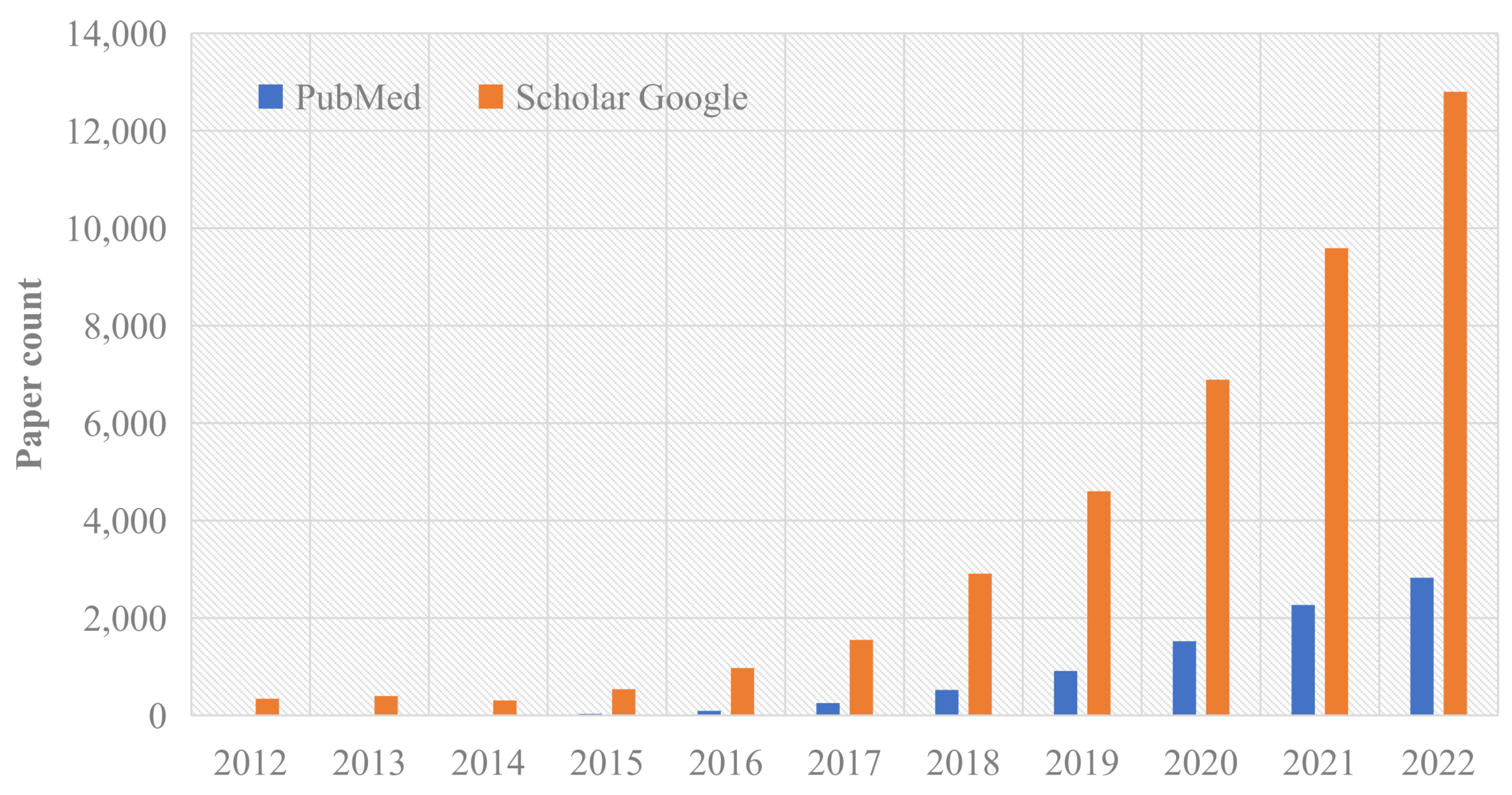
Figure 1.
In summary, the contributions of this survey can be listed as follows:
We provide a brief overview of radiomics models used for PCa. A detailed analysis of the key motivations for radiomics applications using current feature extraction, feature selection, and machine learning techniques is also included.
We commonly analyze the clinical value of mpMRI used in PCa, such as guidance for treatment, showing the pathological areas of tumors, and stating the current challenges with mpMRI.
We present the development of radiogenomics and multi-omics with PCa applications.
We discuss the recent challenges related to the current PCa radiomics, radiogenomics, and multi-omics with future directions in these topics.
The remainder of this paper is structured as follows.
Section 2 briefly describes the impact of mpMRI for PCa.
Section 3 presents the standard radiomic model.
Section 4 discusses the stability of radiomics.
Section 5 introduces the predictive models for classifying PCa with MRI scans.
Section 6 and
Section 7 highlight the research value of radiogenomics and multi-omics for analyzing PCa, respectively.
Section 8 discusses the future perspective and limitations.
Section 9 summarizes the work and contribution of this paper.
5. Radiomics Related to Prostate Cancer
Many machine learning algorithms have been used for classifying prostate lesions using MRI images. For example, the PCa classifications may be related to malignant versus (vs.) benign, csPCa vs. clinically insignificant prostate cancer (ciPCa), multi-class of invasiveness (aggressive, indolent, and indeterminate), GS groups, etc. In [
102], they combined texture features derived from T2W images and ADC maps using a support vector machine to classify between low and high aggressive cases of PCa, which showed a higher AUC value with 0.96 compared to the use-only ADC map with 0.55. In [
102], a fully automatic computer-aided diagnosis system has been developed, which can correctly identify patients with invasive PCa, and it can eliminate the need for manual segmentation and analyze data sets from multiple centers. In [
103], they presented an algorithm model that combines radiomics and pathology to differentiate between indolent and aggressive cancers on MRI-CorrSigNIA, which achieved an accuracy of 80%. Another study aimed to predict GS and established a radiomics model using T2WI, ADC, and diffusion kurtosis imaging (DKI) sequences. The radiomic model using imaging features with lesion size and PI-RADS score predicted PCa with
[
70]. Using MRI images, the radiomics model could distinguish between csPCa and ciPCa [
71]. In addition, the radiomics model using DCE-MRI sequences with logistic regression in predicting the aggressiveness of PCa showed a feasible diagnostic performance [
72]. Compared with T2WI and DWI sequences, prostate DCE-MRI could better display the tumor boundary, which is beneficial to the segmentation of the ROI. However, the study only focused on the radiomics features of the DCE-MRI sequence, and future studies need to be combined with other sequences to improve the diagnostic performance of the radiomics model [
72]. Chaddad et al. proposed a new radiomic signature based on the joint intensity matrix (JIM) to predict the Gleason score (GS) of prostate cancer (PCa) patients. The predictive model achieves an AUC value of 78.40% for
, 82.35% for
, and 64.76% for
[
55]. In another study, texture features used with a random forest model achieved an average of AUC of 83.40%, 72.71%, and 77.35% to predict
and
, respectively [
54]. The performance metric in predicting the GS is significantly improved when the imaging features are extracted from CNN layers, known as deep radiomic features [
56]. With the related approaches to PCa, more investigation is still needed to consider all MRI sequences with AI models in monitoring patients with PCa. To reduce the gap between the academic research of AI in PCa and the improvement of the interpretability of AI models in clinical diagnosis support. It is suggested to solve the limited labeled data, complete the further development and validation of multi-reader research and prospective evaluation, and formulate and improve the standard evaluation criteria [
104].
6. Radiogenomics in Prostate Cancer
The improvement of gene expression levels has strongly promoted the rapid development of genomics. By combining imaging and genomics data, radiogenomics provides a more accurate method for diagnosing and avoiding overtreatment of low-risk tumors [
56]. Specifically, radiogenomics may use imaging features to predict (or combine) the status of genes and guide the diagnosis, treatment, and prognostic process of PCa [
105]. For example, the combination of mpMRI and gene expression data can detect the radioactive signature of PCa. Because of the susceptibility of gene mutations in PCa, many genes are included in gene testing guidelines to assess the risk of PCa and provide guidance for targeted personalized therapy. Common genes used as biomarkers include the breast cancer (BRCA) gene, E-twenty six(ETS)-related gene (ERG), hypoxia gene, ATM gene, etc. Identification of BRCA mutations can be used for PCa screening strategies, in which BRCA 1 and BRCA 2 are key genes associated with PCa susceptibility and are related to hereditary breast cancer and ovarian cancer syndrome [
106]. ERG is the result of a fusion of the androgen receptor-regulated transmembrane protease serine 2 (TMPRSS2) with proto-oncogenes. Hypoxia is an essential feature of the tumor microenvironment, which affects the treatment and prognosis of PCa. Hypoxic gene signatures are usually based on gene expression responses in cell lines exposed to hypoxia. In [
107], the risk marker constructed by two hypoxia and immune-related genes, ISG15 and ZFP36, showed significant PCa prediction ability and was helpful to the prognosis of PCa.
Table 5 presents recent radiogenomic studies of PCa and their findings. In [
108], PTEN and ERG were found to be correlated with PCa visibility on MRI. In clinical trials, prophylactic PCa resection is the primary prevention choice for BRCA 2 carriers [
109]. Detecting BRCA gene mutations in PCa patients helps guide treatment and further genetic detection [
110]. HP
-MRI can distinguish inactive from aggressive PCa based on unique metabolic features [
111]. The visibility of mpMRI increased when the tumor evolution resulted in numerous protein groups different from normal PCa [
112]. Ragnum-signature has been further developed as a biopsy-derived hypoxia biomarker for PCa [
113]. The combination of sSelectMDx and PI-RADS is more sensitive in detecting PCa and may avoid unnecessary biopsy [
114]. Early gene mutation detection, including BRCA 1/2, can improve the survival rate of patients [
115]. Furthermore, the RNA sequencing of benign biopsies revealed the upregulation of NKX3-1 and HOXB13 in the absence of T cells, which may help identify a higher risk of PCa [
116]. Hyperlipidemia is associated with invasive features of PCa without TMPRSS2-ERG fusion or PTEN deletion/mutation [
117]. Eleven miRNAs were identified as sensitive biomarkers for early detection of clinically significant PCa [
118]. Additionally, recent research has found that ANGPTL4, VEGFA, and P4HA1 (hypoxia-related genes) are related to PCa texture features [
119]. In [
120], Fischer et al. identified four biomarkers belonging to genes and miRNAs that play important roles in PCa, which have the ability to differentiate between T2c and T3b stages. Benafif et al. [
121] demonstrated the feasibility of using germline SNPs in targeted PCa population screening in the UK community through the BARCODE1 study. So far, these studies demonstrate the importance of radiogenomic research in understanding PCa and identifying potential biomarkers for early detection, risk assessment, and treatment guidance.
Furthermore, genomic measurements are typically assessed on a small tumor. They reflect only one aspect of tumor heterogeneity. With the ability to determine tumor heterogeneity, radiogenomics offers a personalized approach to risk stratification in patients with PCa [
122]. It can also guide clinical treatment strategies based on individual clinical risk factors. For example, one of the personalized methods of PCa risk calculation is to include clinical data of patients, consisting of PSA levels, and PCa Antigen 3 (PCa3) and TMPRSS2-ERG (T2:ERG) expression [
105]. Due to the limited medical datasets, the short-term solution is to use transfer learning or data augmentation, and the long-term solution is to use multi-institutional data by facilitating the development of online databases [
123]. Personalized treatment requires sequencing a patient’s genome, transcriptome, or proteome [
124]. Using genome sequencing to classify cancers and identify tumor patients with actionable goals may help clinicians make more accurate treatment decisions. Targeted sequencing is currently used to detect genetic changes. The development of next-generation sequencing (NGS) technology is a major advance in a different aspect. It will help in recording unique genetic alterations, enabling the generation of large datasets of genomic, transcriptomics, and/or epigenetic features of tumor cells. As known, DNA or RNA sequencing can help detect changes in gene expression features and gene mutations in cancer. RNA sequencing can help identify and produce new long non-coding RNA and gene fusion in PCa. DNA sequencing becomes more sensitive and scalable with the help of NGS. Genome-wide association studies (GWAS) generate large amounts of genomic data and link these data to related cancers like PCa. Thus, integrating data from genomes and radiomics helps to understand their correlation. In [
125], a web-based platform ImaGene analyzes the correlation between oncology and imaging data sets by inputting them and building an AI model. Although radiogenomics improves model performance by combining genomic and imaging data, data heterogeneity mainly coming from data source inconsistencies between radioactivity and genomes may be considered a challenge.
7. Multi-Omics for PCa
Omics is the comprehensive and quantitative analysis of molecular classes in biological samples. It includes genomics, epigenomics, transcriptomics, proteomics, and metabolomics analyses. Omics is the holistic study of a medical problem from a biological point of view to better achieve a predetermined clinical effect through a single model or a specific feature. It can be used to understand and define changes in biomolecules as complex diseases develop and change. Scientists can search for associations between organisms by analyzing these complex biological macromolecules and constructing accurate disease biomarkers. Multi-omics is to combine these different types of omics data to determine the universal disease–pheno–envirotype relationship or association. Gene expression signatures are the gold standard to guide clinical decision-making, but some questions remain about their clinical utility and interpretability. In 2003, the human genome project was completed, and the information contained in the DNA sequence was deciphered [
126]. Thus, omics data associated with the genome, transcriptome, proteome, epigenome, and metabolome rapidly increased. Furthermore, as the technology matures and costs decrease, the likelihood of using omics data to guide clinical practice increases.
We note that epigenomics studies genome modifications, which affect gene expression without altering the DNA sequence. Epigenetic regulatory mechanisms controlling gene expression in PCa mainly include DNA methylation and histone post-translational modifications. DNA methylation is predominantly seen at GPG dinucleotides and leads to gene silencing [
127]. Histone post-translational modifications can enhance or attenuate gene expression [
128]. These studies facilitate the discovery of new biomarkers or new targeted drugs. In contrast, transcriptomics aims to study the situation of gene expression at the RNA level. Gene signatures of the PCa cell lines LNCaP and VCaP with pre-existing or treatment-induced resistance have been established using single-cell sequencing [
129]. For example, a single-cell transcriptomic study identified a population of luminal cells with progenitor functions as a possible contributor to prostate carcinogenesis [
130].
In addition, proteomics essentially refers to a protein at a large-scale level, including the expression level of the protein, post-translational modifications, and protein–protein interactions. It provides knowledge about disease occurrence that is gained at the protein level. Proteomics also can discover new molecular biomarkers, which have high clinical potential, especially for routine monitoring because their expression can reflect disease activity in real-time [
131].
Metabolomics is a way to quantify metabolites in an organism and find a relationship with physiopathological changes. Analytical techniques are mainly based on nuclear magnetic resonance spectroscopy and mass spectrometry. For example, metabolomics has led to a renewed focus on urine as a valuable biomarker because PCa cells or their substances can be found in prostate fluid. This leads to detecting PCa in urine samples [
132]. Moreover, metabolomics studies can lead to a better understanding of disease pathogenesis and therefore better interventions [
133]. For example, 26 metabolites were significantly altered in PCa tissues, indicating dysregulation of 13 metabolic pathways associated with PCa development. The most affected metabolic pathways were amino acid metabolism, nicotinate, nicotinamide metabolism, purine metabolism, and glycerophospholipid metabolism [
134]. In contrast, the multi-omics study can better describe cancer progress [
135], help us to have a more comprehensive view of factors leading to pathological changes [
136], develop new biomarkers, and improve clinical management of patients [
137,
138]. Despite advances in multi-omics analysis, radiomic with multi-omic topics is still limited. More investigation in this direction will detect more biomarkers of PCa.
Table 6 lists the recent literature on multi-omics in PCa, including the specific type of omics, research objectives, and experimental results. As reported, we observe that the results of multi-omics studies are superior to the single omics, multi-omics are very extensive, and the specific methods are also quite different. For example, multimodal molecular analysis based on cell network biology provides robust prognostic biomarkers to detect and identify high-level diseases [
139]. While in other studies, multi-omics analysis, integrating genomics, methylomics, and transcriptomics are used to assess the risk correlation between DNA methylation and PCa [
140]. Therefore, we suggest explaining the incidence and prognosis of PCA from multi-omics dimensions.
8. Future Perspective and Limitations
Radiomics with PCa is gaining increasing attention as a research direction. While MRI is the primary modality used in current radiomics studies of PCa due to its broad clinical application, there remain numerous challenges in future research and application.
Firstly, the majority of current radiomics studies on PCa are single-center, retrospective studies with small sample sizes, which can limit the accuracy of the research results. Therefore, there is a need for multi-center, prospective studies with larger sample sizes to further validate the research findings.
Secondly, DCE-MRI sequences are commonly included in clinical prostate MRI scans, but most current radiomics studies of PCa do not incorporate these sequences. It is suggested to include DCE-MRI sequences to improve the efficiency of image data utilization.
Thirdly, while manual segmentation is currently the primary method used for delineating the region of interest, automatic segmentation algorithms for the prostate could be improved. This is significant in the clinical practice of oncology, as automatic segmentation can enhance the accuracy of biopsy positioning and allow for more precise and repeatable evaluation of metastatic lesions [
148].
Finally, since most prostate lesions have low malignancy, prostatectomy is not typically performed, and the diagnosis of suspicious lesions relies heavily on pathological examination. However, inaccurate pathological results from a needle biopsy can negatively impact the diagnostic performance of radiomics models, which rely on pathological findings.
Early detection of most cases of PCa is highly challenging. Currently, the primary means of diagnosing suspected PCa is through pathological examination. However, this process relies heavily on needle biopsy, which carries a risk of missed or incorrect diagnoses, leading to inaccurate results. These errors in pathology directly impact the diagnostic accuracy of radiomics. Therefore, enhancing the precision of pathological examination can help improve the performance of radiomics models.
As is widely acknowledged, the approach to treating tumors depends on a range of factors, including the tumor’s pathological type, disease stage, patient condition, cytogenetic changes, and other considerations. The efficacy of treatment can vary from patient to patient, and before administering genomic targeted therapy, a gene test is typically required. Additional assessments, such as radiomics, may also be necessary during treatment to monitor the development of drug resistance. It is noted that gene testing remains a costly, invasive, and time-consuming procedure [
149], whereas radiomics is a relatively inexpensive and non-invasive alternative. As a result, genomic targeting may not be a viable treatment option for all patients with PCa.
Radiogenomics associates imaging data with genome maps, but the availability of these data is affected by the databases (e.g., TCGA, TCIA) and the heterogeneity of tumors. It also requires standardization of imaging and biochemical techniques for analysis to identify stable and repeatable biomarkers. Obtaining reliable results requires many queues and biological sample collection to ensure stability.
Advances in omics, such as genomics, transcriptomics, proteomics, and metabolomics, have begun to enable personalized medicine at the highly detailed molecular level. However, omics alone cannot capture the entire biological complexity of most human diseases. Integrating multiple omics features (radiomics + radiogenomics + omics) provides a more comprehensive view of biology and disease [
135]. In addition, few studies performed biomarker validation, but few used independent sample cohorts to exclude false positives caused by sample collection and processing. Moreover, the discovery cohorts were small due to the need for standardized methods for sample collection and processing, data acquisition, and bioinformatics analysis. Finally, few studies shared the same biomarker candidates [
131]. These challenges require a massive investigation and a collaborative way to share the findings between federated hospital systems.
AI techniques rely increasingly on large datasets, especially when the data are suitable. It is important to note that data sets have varying feature distributions, and differences arise across various techniques when multiple data sources are used. Therefore, the process of identifying and preprocessing appropriate data can result in more valuable research outcomes.
Table 7 lists the most common and public data sets that can be used for PCa studies, such as MRI images containing benign and malignant labels acquired by different types of scanners, consisting of manual labels, distinguishing csPCa from ciPCa, clinical variables, examination, diagnosis, and treatment including PSA and other biochemical data, microscopic scans of prostate biopsy samples with imperfect labels and large images.
In the past, open-source datasets were typically constructed to meet specific research needs, which may not align with current research requirements. To better serve a wider range of communities, it is preferable to provide clean data in multiple formats. However, current datasets face several challenges such as low data reading rates, the presence of multiple data types, and complex data processing requirements. In the near future, researchers are likely to adopt a responsible approach to data collection and annotation, as well as data set maintenance and problem formulation, in order to mitigate these challenges [
162].
The translation of radiomics models constructed from medical images into clinical applications faces challenges in terms of interpretability. Specifically, there is a lack of transparent explanation regarding the relationship between selected features and clinical outcomes. In order to ensure interpretability and assist clinicians in making clinical decisions, it is important to have a thorough understanding of the decision-making process behind the radiomics process, especially before incorporating AI fields like DL methods. Without adequate interpretability, the decision-making process and possible biases are not well accounted for, leading to the limited clinical utility of radiomics features and models. The General Data Protection Regulation (GDPR) law in the European Union requires an explanation of an algorithm’s decision-making process, and data subjects are entitled to meaningful information about the logic involved [
163]. Explainable Artificial Intelligence (XAI) can help to interpret the information behind the “black box” model, showing how the decision was made transparently, thus enhancing the credibility of the model. Different XAI techniques, such as class activation map (CAM), local interpretable model-agnostic explanation (LIME), Shapley additive explanations (SHAP), Gradient-weighted class activation mapping (Grad-CAM), Attention, and Saliency, can be used to improve algorithm performance [
90,
164]. The explanation forms generated by XAI can be categorized as feature-based, text-based, and example-based, and improve the credibility of AI from different levels. Interpretable methods have been associated with various tasks in radiomics, including image segmentation, lesion and organ detection, image registration, computer-aided diagnosis and staging, prognosis, radiotherapy planning, disease progression monitoring, classification, and image reconstruction [
165]. In one study, multi-modal volumetric concept activation was used to provide an explanation, which showed that the detection was mainly based on the location of metastatic PCa in CT anatomy, and the reliability of PET detection was high [
166]. In another study, a model fused with multiple DL methods was used to examine PCa with MRI images, and then XAI explained how the model differentiated benign or malignant PCa [
167]. We note that many other radiomics, AI, and radiogenomics works could be also discussed. However, this study collects the most common models that are used for PCa analysis.
Radiomics analysis based on mpMRI can not only improve the detection rate of PCa but also predict prognosis, its texture features can reflect the heterogeneity of lesions. After radiation therapy, radiomics during the follow-up process can be used to evaluate the efficacy of treatment and tumor recurrence. When comparing the radiomic results before and after treatment, tumor shrinkage, tissue recovery, and the presence of residual or new lesions can be evaluated, helping determine whether further treatment is needed, thus improving the survival rate. The combination of mpMRI and Prostate Health Index (PHI) in radiomics may help to better estimate the risk categories of prostate cancer at the initial diagnosis, thus achieving personalized treatment methods [
168]. We note that the prediction of cancer prognosis is based on statistical data and models, which provide a probability estimate rather than an absolute prediction. Everyone’s cancer situation is unique, including pathological features, health status, and personal factors, all of which may have an impact on prognosis. Therefore, predicting cancer prognosis should serve as a reference for auxiliary decision-making, rather than the only basis. The final treatment decision should comprehensively consider multiple factors and make personalized choices based on individual circumstances.
As PCa is increasingly being diagnosed at an early stage, with excellent survival rates, the rationale for patients’ primary treatment selection has switched to health-related quality of life (HRQOL). Use mpMRI to detect suspicious PCa before biopsy, thus reducing the number of unnecessary biopsies and avoiding the risk of overdiagnosis and overtreatment. At the same time, through the combined method of systematic and fusion targeted biopsy, the detection rate of PCa can be further improved and the risk of missing csPCa can be reduced [
169]. Studies usually conduct follow-ups or send questionnaires (e.g., the Expanded Prostate Cancer Index Composite (EPIC) questionnaire and the Short-Form 12 Item Health (SF-12)) at baseline 3, 6, 12, and 24 months after treatment to collect patient-reported QOL outcomes. The EPIC complements existing instruments by measuring a broad range of urinary, bowel, sexual and hormonal symptoms, allowing for a more comprehensive assessment of important HRQOL issues in contemporary PCa management [
170]. In [
171], they examined a prospective serial cohort of low-dose-rate (LDR) brachytherapy for PCa using MRI and explored factors associated with toxicity and QOL, as assessed by EPIC and the International Prostate Symptom Score (IPSS). In [
172], a prospective phase II clinical study was developed to evaluate outcomes in patients treated with MRI-guided whole-gland prostate high-dose-rate brachytherapy (HDR-BT) augmentation with an assessment of toxicity and HRQOL outcomes. In [
173], the HRQOL of early PCa patients who did not receive hormone therapy within 3 years after radiotherapy was examined using the 15D instrument and the FACT-P questionnaire, and the HRQOL was the same in the radiotherapy group and the age-standardized general male population. The treatment of PCa is mainly for curative purposes, but the treatment options are usually accompanied by high morbidity of urinary problems and/or erectile dysfunction, significant loss of quality of life, and high treatment costs. Curing PCa, solving possible complications during treatment, and improving quality of life are the common pursuits of doctors and PCa patients. In [
174], palliative transurethral resection of the prostate (pTURP) combined with intermittent androgen deprivation therapy (ADT) can be used in the treatment of elderly patients with localized PCa to resolve dysuria and improve QOL. The personalized treatment of PCa remains one of the challenging areas that require further investigation.