Mutational Signatures in Gastric Cancer and Their Clinical Implications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Mutational Signatures
3. Mutational Signatures in Gastric Cancer
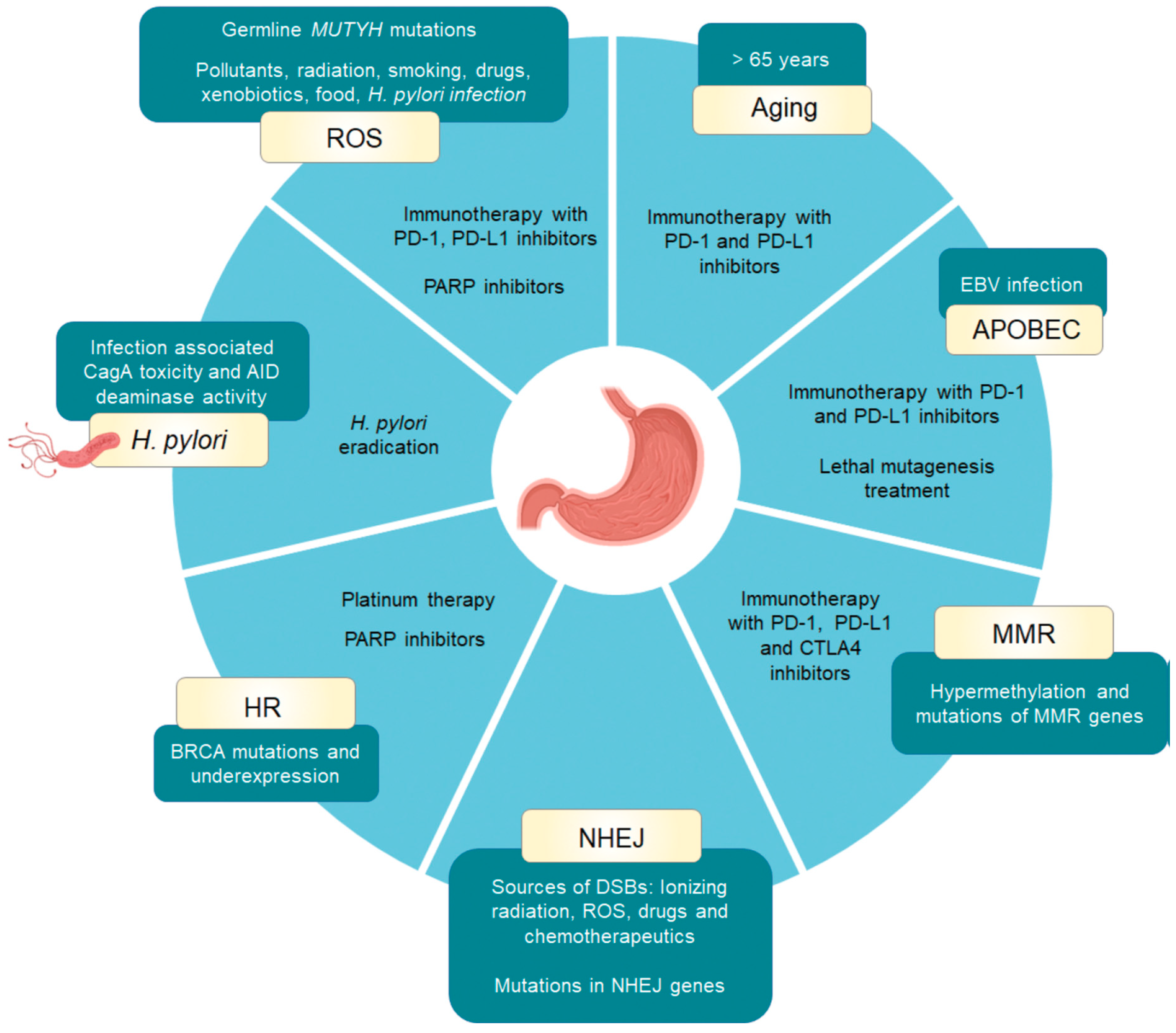
3.1. Aging
3.2. APOBEC Activity
3.3. DNA Integrity Machinery
3.3.1. Homologous Recombination DNA Repair
3.3.2. DNA Mismatch Repair Deficiency
| SBS | Base Substitution Subtype 1 | Associated ID | Transcriptional (T) and Replicational (R) Strand Asymmetry in Stomach Cancer |
|---|---|---|---|
| SBS6 | C > T (ACA, ACG, CCG, GCN) | ID1, ID2 | T:/ R:/ |
| SBS14 | C > A (ACT, CCT, GCT, TCT) | ID1, ID2 | T:/ R:/ |
| SBS15 | C > A (CCA) C > T (ACG, GCN) | ID1, ID2 | T:/ R:/ |
| SBS20 2 | C > A (CCC, CCT) C > T (ACA, GCA, GCC) | ID1, ID2 | T: no significance R: lagging, C > A |
| SBS21 | T > C (GTN, TTA, TTC, TTT) | ID1, ID2 | T: no significance R: lagging strand, T > C |
| SBS26 | T > C (ATA, ATC, CTA, CTG, CTT, GTA, GTG, GTT, TTT) | ID1, ID2 | T: untranscribed strand, T > C R: lagging strand, T > C |
| SBS44 | C > A (CCT) C > T (ACA, GCN) | ID1, ID2 | T: no significance R: lagging strand, C > A; leading strand, C > T and T > A |
MLH1 Promoter Hypermethylation and MSI Phenotype
POLD1/POLE Mutations and MSI Phenotype
Mutational Status of ARID1A and MSI Phenotype
3.3.3. Double Strand Break Repair by Nonhomologous End Joining
3.4. Reactive Oxygen Species
3.5. Helicobacter Pylori Infection
3.6. Mutational Signatures of Unknown Origin
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACRG | Asian Cancer Research Group |
| BER | Base Excision Repair |
| CEA | Carcinoembryonic Antigen |
| CN | Copy Number Variations Signatures |
| ctDNA | Circulating Tumour DNA |
| DBS | Doublet Base Substitution |
| DSBs | Double-Strand Breaks |
| EBV | Epstein–Barr Virus |
| FDA | Food and Drug Administration |
| GS | Genomically Stable |
| HBV | Hepatitis B Virus |
| HCV | Hepatitis C Virus |
| HRR | Homologous Recombination Repair |
| HTLV-1 | Human T-Lymphotropic Virus Type 1 |
| ICIs | Immune Checkpoint Inhibitors |
| ID | Insertion and Deletion Mutational Signatures |
| MMR | Mismatch Repair |
| MSI | Microsatellite Instability |
| MSS | Microsatellite Stable |
| NER | Nucleotide Excision Repair |
| NHEJ | Nonhomologous End Joining |
| NMF | Nonnegative Matrix Factorisation |
| ROS | Reactive Oxygen Species |
| SBS | Single Base Substitutions |
| TCGA | The Cancer Genome Atlas |
| TMB | Tumour Mutational Burden |
| TNM | Tumour Node Metastasis |
| WES | Whole Exome Sequencing |
| WGS | Whole-Genome Sequencing |
References
- Thrift, A.P.; Wenker, T.N.; El-Serag, H.B. Global burden of gastric cancer: Epidemiological trends, risk factors, screening and prevention. Nat. Rev. Clin. Oncol. 2023, 20, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Stowe, T.A.; McClung, C.A. How Does Chronobiology Contribute to the Development of Diseases in Later Life. Clin. Interv. Aging 2023, 18, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, K.; Inoue, D.; Sugimoto, K.; Tanaka, C.; Murofushi, K.; Okuyama, T.; Watanuki, S.; Imamura, C.K.; Sakai, D.; Sakurai, N.; et al. Significance of the comprehensive geriatric assessment in the administration of chemotherapy to older adults with cancer: Recommendations by the Japanese Geriatric Oncology Guideline Committee. J. Geriatr. Oncol. 2023, 14, 101485. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef]
- Thrift, A.P.; El-Serag, H.B. Burden of Gastric Cancer. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2020, 18, 534–542. [Google Scholar] [CrossRef]
- Morgan, E.; Arnold, M.; Camargo, M.C.; Gini, A.; Kunzmann, A.T.; Matsuda, T.; Meheus, F.; Verhoeven, R.H.A.; Vignat, J.; Laversanne, M.; et al. The current and future incidence and mortality of gastric cancer in 185 countries, 2020-40: A population-based modelling study. EClinicalMedicine 2022, 47, 101404. [Google Scholar] [CrossRef]
- Jensen, E.; Kristensen, J.K.; Bjerglund, R.T.; Johnsen, S.P.; Thomsen, J.L. The pathway and characteristics of patients with non-specific symptoms of cancer: A systematic review. BMC Cancer 2022, 22, 574. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, X.; Tang, H.; Yan, J.; Li, J.; Bao, H.; Wu, X.; Shao, Y.; Luo, C.; Wen, H.; et al. Exploring potential molecular resistance and clonal evolution in advanced HER2-positive gastric cancer under trastuzumab therapy. Oncogenesis 2023, 12, 21. [Google Scholar] [CrossRef]
- Sirody, J.; Kaji, A.H.; Hari, D.M.; Chen, K.T. Patterns of gastric cancer metastasis in the United States. Am. J. Surg. 2022, 224, 445–448. [Google Scholar] [CrossRef]
- Arnold, M.; Rutherford, M.J.; Bardot, A.; Ferlay, J.; Andersson, T.M.; Myklebust, T.; Tervonen, H.; Thursfield, V.; Ransom, D.; Shack, L.; et al. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995–2014 (ICBP SURVMARK-2): A population-based study. Lancet. Oncol. 2019, 20, 1493–1505. [Google Scholar] [CrossRef] [Green Version]
- Van Cutsem, E.; Bang, Y.J.; Feng-Yi, F.; Xu, J.M.; Lee, K.W.; Jiao, S.C.; Chong, J.L.; López-Sanchez, R.I.; Price, T.; Gladkov, O.; et al. HER2 screening data from ToGA: Targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2015, 18, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Brar, G.; Shah, M.A. The role of pembrolizumab in the treatment of PD-L1 expressing gastric and gastroesophageal junction adenocarcinoma. Ther. Adv. Gastroenterol. 2019, 12, 1756284819869767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muro, K.; Chung, H.C.; Shankaran, V.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Golan, T.; Le, D.T.; Burtness, B.; et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): A multicentre, open-label, phase 1b trial. Lancet. Oncol. 2016, 17, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.S.; Doi, T.; Jang, R.W.; Muro, K.; Satoh, T.; Machado, M.; Sun, W.; Jalal, S.I.; Shah, M.A.; Metges, J.P.; et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients with Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol. 2018, 4, e180013. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; An, M.; Klempner, S.J.; Lee, H.; Kim, K.M.; Sa, J.K.; Cho, H.J.; Hong, J.Y.; Lee, T.; Min, Y.W.; et al. Determinants of Response and Intrinsic Resistance to PD-1 Blockade in Microsatellite Instability-High Gastric Cancer. Cancer Discov. 2021, 11, 2168–2185. [Google Scholar] [CrossRef]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; Dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Choi, S.; Park, S.; Kim, H.; Kang, S.Y.; Ahn, S.; Kim, K.M. Gastric Cancer: Mechanisms, Biomarkers, and Therapeutic Approaches. Biomedicines 2022, 10, 543. [Google Scholar] [CrossRef]
- Niclauss, N.; Gütgemann, I.; Dohmen, J.; Kalff, J.C.; Lingohr, P. Novel Biomarkers of Gastric Adenocarcinoma: Current Research and Future Perspectives. Cancers 2021, 13, 5660. [Google Scholar] [CrossRef]
- Waddell, T.; Chau, I.; Cunningham, D.; Gonzalez, D.; Okines, A.F.; Okines, C.; Wotherspoon, A.; Saffery, C.; Middleton, G.; Wadsley, J.; et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): A randomised, open-label phase 3 trial. Lancet Oncol. 2013, 14, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Sahin, U.; Koslowski, M.; Dhaene, K.; Usener, D.; Brandenburg, G.; Seitz, G.; Huber, C.; Türeci, O. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 7624–7634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. p53 mutations in human cancers. Science 1991, 253, 49–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giglia-Mari, G.; Sarasin, A. TP53 mutations in human skin cancers. Hum. Mutat. 2003, 21, 217–228. [Google Scholar] [CrossRef]
- Pfeifer, G.P.; Denissenko, M.F.; Olivier, M.; Tretyakova, N.; Hecht, S.S.; Hainaut, P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 2002, 21, 7435–7451. [Google Scholar] [CrossRef] [Green Version]
- Nik-Zainal, S.; Alexandrov, L.B.; Wedge, D.C.; Van Loo, P.; Greenman, C.D.; Raine, K.; Jones, D.; Hinton, J.; Marshall, J.; Stebbings, L.A.; et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012, 149, 979–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucab, J.E.; Zou, X.; Morganella, S.; Joel, M.; Nanda, A.S.; Nagy, E.; Gomez, C.; Degasperi, A.; Harris, R.; Jackson, S.P.; et al. A Compendium of Mutational Signatures of Environmental Agents. Cell 2019, 177, 821–836.e816. [Google Scholar] [CrossRef] [Green Version]
- Thatikonda, V.; Islam, S.M.A.; Autry, R.J.; Jones, B.C.; Gröbner, S.N.; Warsow, G.; Hutter, B.; Huebschmann, D.; Fröhling, S.; Kool, M.; et al. Comprehensive analysis of mutational signatures reveals distinct patterns and molecular processes across 27 pediatric cancers. Nat. Cancer 2023, 4, 276–289. [Google Scholar] [CrossRef]
- Landi, M.T.; Synnott, N.C.; Rosenbaum, J.; Zhang, T.; Zhu, B.; Shi, J.; Zhao, W.; Kebede, M.; Sang, J.; Choi, J.; et al. Tracing Lung Cancer Risk Factors through Mutational Signatures in Never-Smokers. Am. J. Epidemiol. 2021, 190, 962–976. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.D.; Abbasi, A.; Islam, S.M.A.; Bowes, A.L.; Khandekar, A.; Haase, K.; Hames-Fathi, S.; Ajayi, D.; Verfaillie, A.; Dhami, P.; et al. Signatures of copy number alterations in human cancer. Nature 2022, 606, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Lehner, B. Clustered Mutation Signatures Reveal that Error-Prone DNA Repair Targets Mutations to Active Genes. Cell 2017, 170, 534–547 e523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasar, S.; Brown, J.R. Mutational landscape and underlying mutational processes in chronic lymphocytic leukemia. Mol. Cell Oncol. 2016, 3, e1157667. [Google Scholar] [CrossRef] [Green Version]
- Blokzijl, F.; de Ligt, J.; Jager, M.; Sasselli, V.; Roerink, S.; Sasaki, N.; Huch, M.; Boymans, S.; Kuijk, E.; Prins, P.; et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016, 538, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Jones, P.H.; Wedge, D.C.; Sale, J.E.; Campbell, P.J.; Nik-Zainal, S.; Stratton, M.R. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015, 47, 1402–1407. [Google Scholar] [CrossRef] [Green Version]
- Panda, A.; Mehnert, J.M.; Hirshfield, K.M.; Riedlinger, G.; Damare, S.; Saunders, T.; Kane, M.; Sokol, L.; Stein, M.N.; Poplin, E.; et al. Immune Activation and Benefit from Avelumab in EBV-Positive Gastric Cancer. J. Natl. Cancer Inst. 2018, 110, 316–320. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Niu, Z.; Wang, Y.; Zheng, Y.; Zhu, Y.; Wang, C.; Gao, X.; Gao, L.; Zhang, W.; Zhang, K.; et al. Senescence as a dictator of patient outcomes and therapeutic efficacies in human gastric cancer. Cell Death Discov. 2022, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.M.; Loeb, L.A. Effect of DNA-repair enzymes on mutagenesis by oxygen free radicals. Mutat. Res. 1993, 289, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Bobrovnitchaia, I.; Valieris, R.; Drummond, R.D.; Lima, J.P.; Freitas, H.C.; Bartelli, T.F.; de Amorim, M.G.; Nunes, D.N.; Dias-Neto, E.; da Silva, I.T. APOBEC-mediated DNA alterations: A possible new mechanism of carcinogenesis in EBV-positive gastric cancer. Int. J. Cancer 2020, 146, 181–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Wen, W.; Bao, J.; Kuhs, K.L.; Cai, Q.; Long, J.; Shu, X.O.; Zheng, W.; Guo, X. Integrative genomic analyses of APOBEC-mutational signature, expression and germline deletion of APOBEC3 genes, and immunogenicity in multiple cancer types. BMC Med. Genom. 2019, 12, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petljak, M.; Alexandrov, L.B.; Brammeld, J.S.; Price, S.; Wedge, D.C.; Grossmann, S.; Dawson, K.J.; Ju, Y.S.; Iorio, F.; Tubio, J.M.C.; et al. Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell 2019, 176, 1282–1294 e1220. [Google Scholar] [CrossRef] [Green Version]
- Fox, E.J.; Loeb, L.A. Lethal mutagenesis: Targeting the mutator phenotype in cancer. Semin. Cancer Biol. 2010, 20, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Kidd, J.M.; Newman, T.L.; Tuzun, E.; Kaul, R.; Eichler, E.E. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007, 3, e63. [Google Scholar] [CrossRef]
- Wang, S.; Jia, M.; He, Z.; Liu, X.S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene 2018, 37, 3924–3936. [Google Scholar] [CrossRef]
- Boichard, A.; Pham, T.V.; Yeerna, H.; Goodman, A.; Tamayo, P.; Lippman, S.; Frampton, G.M.; Tsigelny, I.F.; Kurzrock, R. APOBEC-related mutagenesis and neo-peptide hydrophobicity: Implications for response to immunotherapy. Oncoimmunology 2019, 8, 1550341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewing, A.D.; Gacita, A.; Wood, L.D.; Ma, F.; Xing, D.; Kim, M.S.; Manda, S.S.; Abril, G.; Pereira, G.; Makohon-Moore, A.; et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015, 25, 1536–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Siu, H.C.; Leung, S.Y.; Stratton, M.R. A mutational signature in gastric cancer suggests therapeutic strategies. Nat. Commun. 2015, 6, 8683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franzese, E.; Centonze, S.; Diana, A.; Carlino, F.; Guerrera, L.P.; Di Napoli, M.; De Vita, F.; Pignata, S.; Ciardiello, F.; Orditura, M. PARP inhibitors in ovarian cancer. Cancer Treat. Rev. 2019, 73, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keung, M.Y.T.; Wu, Y.; Vadgama, J.V. PARP Inhibitors as a Therapeutic Agent for Homologous Recombination Deficiency in Breast Cancers. J. Clin. Med. 2019, 8, 435. [Google Scholar] [CrossRef] [Green Version]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietanza, M.C.; Waqar, S.N.; Krug, L.M.; Dowlati, A.; Hann, C.L.; Chiappori, A.; Owonikoko, T.K.; Woo, K.M.; Cardnell, R.J.; Fujimoto, J.; et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination with Either Veliparib or Placebo in Patients with Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 2386–2394. [Google Scholar] [CrossRef]
- Bang, Y.J.; Xu, R.H.; Chin, K.; Lee, K.W.; Park, S.H.; Rha, S.Y.; Shen, L.; Qin, S.; Xu, N.; Im, S.A.; et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet. Oncol. 2017, 18, 1637–1651. [Google Scholar] [CrossRef]
- Chen, W.; Wang, J.; Li, X.; Li, J.; Zhou, L.; Qiu, T.; Zhang, M.; Liu, P. Prognostic significance of BRCA1 expression in gastric cancer. Med. Oncol. 2013, 30, 423. [Google Scholar] [CrossRef]
- Kim, J.W.; Cho, H.J.; Kim, M.; Lee, K.H.; Kim, M.A.; Han, S.W.; Oh, D.Y.; Lee, H.J.; Im, S.A.; Kim, T.Y.; et al. Differing effects of adjuvant chemotherapy according to BRCA1 nuclear expression in gastric cancer. Cancer Chemother. Pharmacol. 2013, 71, 1435–1443. [Google Scholar] [CrossRef]
- Buttura, J.R.; Provisor Santos, M.N.; Valieris, R.; Drummond, R.D.; Defelicibus, A.; Lima, J.P.; Calsavara, V.F.; Freitas, H.C.; Cordeiro de Lima, V.C.; Fernanda Bartelli, T.; et al. Mutational Signatures Driven by Epigenetic Determinants Enable the Stratification of Patients with Gastric Cancer for Therapeutic Intervention. Cancers 2021, 13, 490. [Google Scholar] [CrossRef] [PubMed]
- Togasaki, K.; Sukawa, Y.; Kanai, T.; Takaishi, H. Clinical efficacy of immune checkpoint inhibitors in the treatment of unresectable advanced or recurrent gastric cancer: An evidence-based review of therapies. Onco Targets Ther. 2018, 11, 8239–8250. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhang, F.; Zhou, N.; Gu, Y.M.; Zhang, Y.T.; He, Y.D.; Wang, L.; Yang, L.X.; Zhao, Y.; Li, Y.M. Efficacy and safety of immune checkpoint inhibitors in advanced gastric or gastroesophageal junction cancer: A systematic review and meta-analysis. Oncoimmunology 2019, 8, e1581547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Cui, H.; Zhang, L.; Zhao, K.; Jia, X.; Jin, H. Assessment of POLE and POLD1 mutations as prognosis and immunotherapy biomarkers for stomach adenocarcinoma. Transl. Cancer Res. 2022, 11, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Wei, X.; Li, S.; Liu, C.; Chen, H.; Gong, J.; Li, J.; Zhang, X.; Wang, X.; Peng, Z.; et al. A genomic mutation signature predicts the clinical outcomes of immunotherapy and characterizes immunophenotypes in gastrointestinal cancer. NPJ Precis. Oncol. 2021, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Chida, K.; Kawazoe, A.; Kawazu, M.; Suzuki, T.; Nakamura, Y.; Nakatsura, T.; Kuwata, T.; Ueno, T.; Kuboki, Y.; Kotani, D.; et al. A Low Tumor Mutational Burden and PTEN Mutations Are Predictors of a Negative Response to PD-1 Blockade in MSI-H/dMMR Gastrointestinal Tumors. Clin. Cancer Res. 2021, 27, 3714–3724. [Google Scholar] [CrossRef]
- Kim, C.G.; Hong, M.; Jeung, H.C.; Lee, G.; Chung, H.C.; Rha, S.Y.; Kim, H.S.; Lee, C.K.; Lee, J.H.; Han, Y.; et al. Hyperprogressive disease during PD-1 blockade in patients with advanced gastric cancer. Eur. J. Cancer 2022, 172, 387–399. [Google Scholar] [CrossRef]
- Wang, F.; Zhao, Q.; Wang, Y.N.; Jin, Y.; He, M.M.; Liu, Z.X.; Xu, R.H. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol. 2019, 5, 1504–1506. [Google Scholar] [CrossRef] [Green Version]
- Pietrantonio, F.; Miceli, R.; Raimondi, A.; Kim, Y.W.; Kang, W.K.; Langley, R.E.; Choi, Y.Y.; Kim, K.M.; Nankivell, M.G.; Morano, F.; et al. Individual Patient Data Meta-Analysis of the Value of Microsatellite Instability As a Biomarker in Gastric Cancer. J. Clin. Oncol. 2019, 37, 3392–3400. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, P.; Wang, J.; Lin, C.; Liu, H.; Li, H.; He, H.; Li, R.; Zhang, H.; Zhang, W. Somatic ARID1A mutation stratifies patients with gastric cancer to PD-1 blockade and adjuvant chemotherapy. Cancer Immunol. Immunother. 2023, 72, 1199–1208. [Google Scholar] [CrossRef]
- Wang, M.; Huang, Y.K.; Kong, J.C.; Sun, Y.; Tantalo, D.G.; Yeang, H.X.A.; Ying, L.; Yan, F.; Xu, D.; Halse, H.; et al. High-dimensional analyses reveal a distinct role of T-cell subsets in the immune microenvironment of gastric cancer. Clin. Transl. Immunol. 2020, 9, e1127. [Google Scholar] [CrossRef] [PubMed]
- Puliga, E.; Corso, S.; Pietrantonio, F.; Giordano, S. Microsatellite instability in Gastric Cancer: Between lights and shadows. Cancer Treat. Rev. 2021, 95, 102175. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, M.; Choudhary, B.; Raghavan, S.C. SCR7, a potent cancer therapeutic agent and a biochemical inhibitor of nonhomologous DNA end-joining. Cancer Rep. 2021, 4, e1341. [Google Scholar] [CrossRef]
- Wang, Y.; Kuramitsu, Y.; Baron, B.; Kitagawa, T.; Tokuda, K.; Akada, J.; Maehara, S.-I.; Maehara, Y.; Nakamura, K. PI3K inhibitor LY294002, as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells. Int. J. Oncol. 2017, 50, 606–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niazi, M.T.; Mok, G.; Heravi, M.; Lee, L.; Vuong, T.; Aloyz, R.; Panasci, L.; Muanza, T. Effects of dna-dependent protein kinase inhibition by NU7026 on dna repair and cell survival in irradiated gastric cancer cell line N87. Curr. Oncol. 2014, 21, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Xie, C.; Yang, Z.; Chen, J.; Lu, N.H. Abnormal DNA-PKcs and Ku 70/80 expression may promote malignant pathological processes in gastric carcinoma. World J. Gastroenterol. 2013, 19, 6894–6901. [Google Scholar] [CrossRef]
- Jin, S.G.; Meng, Y.; Johnson, J.; Szabó, P.E.; Pfeifer, G.P. Concordance of hydrogen peroxide-induced 8-oxo-guanine patterns with two cancer mutation signatures of upper GI tract tumors. Sci. Adv. 2022, 8, eabn3815. [Google Scholar] [CrossRef]
- Volkov, N.M.; Yanus, G.A.; Ivantsov, A.O.; Moiseenko, F.V.; Matorina, O.G.; Bizin, I.V.; Moiseyenko, V.M.; Imyanitov, E.N. Efficacy of immune checkpoint blockade in MUTYH-associated hereditary colorectal cancer. Investig. New Drugs 2020, 38, 894–898. [Google Scholar] [CrossRef]
- Mouw, K.W.; Goldberg, M.S.; Konstantinopoulos, P.A.; D’Andrea, A.D. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 2017, 7, 675–693. [Google Scholar] [CrossRef] [Green Version]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Kanter-Smoler, G.; Bjork, J.; Fritzell, K.; Engwall, Y.; Hallberg, B.; Karlsson, G.; Gronberg, H.; Karlsson, P.; Wallgren, A.; Wahlstrom, J.; et al. Novel findings in Swedish patients with MYH-associated polyposis: Mutation detection and clinical characterization. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2006, 4, 499–506. [Google Scholar] [CrossRef]
- Vogt, S.; Jones, N.; Christian, D.; Engel, C.; Nielsen, M.; Kaufmann, A.; Steinke, V.; Vasen, H.F.; Propping, P.; Sampson, J.R.; et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology 2009, 137, 1976–1985.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Fan, Y.; Ding, J.; Xu, A.; Zhou, X.; Hu, X.; Zhu, M.; Zhang, X.; Li, S.; et al. Germline mutations and polymorphic variants in MMR, E-cadherin and MYH genes associated with familial gastric cancer in Jiangsu of China. Int. J. Cancer 2006, 119, 2592–2596. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Ooki, T.; Murata-Kamiya, N.; Komura, D.; Tahmina, K.; Wu, W.; Takahashi-Kanemitsu, A.; Knight, C.T.; Kunita, A.; Suzuki, N.; et al. Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe 2021, 29, 941–958.e910. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Marusawa, H.; Endo, Y.; Chiba, T. Inflammation-mediated genomic instability: Roles of activation-induced cytidine deaminase in carcinogenesis. Cancer Sci. 2012, 103, 1201–1206. [Google Scholar] [CrossRef]
- Pfeifer, G.P. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 2006, 301, 259–281. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.N.; Mort, M.; Stenson, P.D.; Ball, E.V.; Chuzhanova, N.A. Methylation-mediated deamination of 5-methylcytosine appears to give rise to mutations causing human inherited disease in CpNpG trinucleotides, as well as in CpG dinucleotides. Hum. Genom. 2010, 4, 406–410. [Google Scholar] [CrossRef] [Green Version]
- Sassa, A.; Kanemaru, Y.; Kamoshita, N.; Honma, M.; Yasui, M. Mutagenic consequences of cytosine alterations site-specifically embedded in the human genome. Genes. Environ. Off. J. Jpn. Environ. Mutagen. Soc. 2016, 38, 17. [Google Scholar] [CrossRef] [Green Version]
- Brittan, M.; Wright, N.A. Stem cell in gastrointestinal structure and neoplastic development. Gut 2004, 53, 899–910. [Google Scholar] [CrossRef] [Green Version]
- Demaria, M.; O’Leary, M.N.; Chang, J.; Shao, L.; Liu, S.; Alimirah, F.; Koenig, K.; Le, C.; Mitin, N.; Deal, A.M.; et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discov. 2017, 7, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Vieira, V.C.; Soares, M.A. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed. Res. Int. 2013, 2013, 683095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conticello, S.G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008, 9, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.A.; Sterling, J.; Thompson, C.; Harris, S.; Mav, D.; Shah, R.; Klimczak, L.J.; Kryukov, G.V.; Malc, E.; Mieczkowski, P.A.; et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 2012, 46, 424–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helleday, T.; Eshtad, S.; Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014, 15, 585–598. [Google Scholar] [CrossRef]
- Roberts, S.A.; Gordenin, D.A. Hypermutation in human cancer genomes: Footprints and mechanisms. Nat. Rev. Cancer 2014, 14, 786–800. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wei, W.; Jin, H.C.; Ying, R.C.; Zhu, A.K.; Zhang, F.J. The roles of APOBEC3B in gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 5089–5096. [Google Scholar]
- Middlebrooks, C.D.; Banday, A.R.; Matsuda, K.; Udquim, K.I.; Onabajo, O.O.; Paquin, A.; Figueroa, J.D.; Zhu, B.; Koutros, S.; Kubo, M.; et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet. 2016, 48, 1330–1338. [Google Scholar] [CrossRef]
- Goodier, J.L.; Kazazian, H.H., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Kouno, T.; Silvas, T.V.; Hilbert, B.J.; Shandilya, S.M.D.; Bohn, M.F.; Kelch, B.A.; Royer, W.E.; Somasundaran, M.; Kurt Yilmaz, N.; Matsuo, H.; et al. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 2017, 8, 15024. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.; Demir, O.; Carpenter, M.A.; Banerjee, S.; Harki, D.A.; Amaro, R.E.; Harris, R.S.; Aihara, H. Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B. FASEB Bioadv 2020, 2, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.; Carpenter, M.A.; Banerjee, S.; Shaban, N.M.; Kurahashi, K.; Salamango, D.J.; McCann, J.L.; Starrett, G.J.; Duffy, J.V.; Demir, O.; et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017, 24, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Mullan, P.B.; Quinn, J.E.; Harkin, D.P. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 2006, 25, 5854–5863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.P. Sensing and repairing DNA double-strand breaks. Carcinogenesis 2002, 23, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Casadei, S.; Coats, K.H.; Swisher, E.; Stray, S.M.; Higgins, J.; Roach, K.C.; Mandell, J.; Lee, M.K.; Ciernikova, S.; et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 2006, 295, 1379–1388. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zheng, K.; Huang, Y.; Xiong, H.; Su, J.; Chen, R.; Zou, Y. PARP inhibitors in gastric cancer: Beacon of hope. J. Exp. Clin. Cancer Res. 2021, 40, 211. [Google Scholar] [CrossRef]
- Gotwals, P.; Cameron, S.; Cipolletta, D.; Cremasco, V.; Crystal, A.; Hewes, B.; Mueller, B.; Quaratino, S.; Sabatos-Peyton, C.; Petruzzelli, L.; et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 2017, 17, 286–301. [Google Scholar] [CrossRef]
- Hee Sang, H.; Deokhoon, K.; Jene, C. Distinct mutational profile and immune microenvironment in microsatellite-unstable and POLE-mutated tumors. J. ImmunoTherapy Cancer 2021, 9, e002797. [Google Scholar] [CrossRef]
- Li, H.; van der Merwe, P.A.; Sivakumar, S. Biomarkers of response to PD-1 pathway blockade. Br. J. Cancer 2022, 126, 1663–1675. [Google Scholar] [CrossRef]
- Cheng, Y.; Bu, D.; Zhang, Q.; Sun, R.; Lyle, S.; Zhao, G.; Dong, L.; Li, H.; Zhao, Y.; Yu, J.; et al. Genomic and transcriptomic profiling indicates the prognosis significance of mutational signature for TMB-high subtype in Chinese patients with gastric cancer. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- Elizabeth, D.T.; Marianna, Z.; Adrian, M.; Toby, C.; Nathan, C.; Eihab, A.; Stephen, Y.; Mark, D.; Nita, A.; Janis, M.T.; et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 2017, 66, 794. [Google Scholar] [CrossRef]
- Nicolas, E.; Golemis, E.A.; Arora, S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene 2016, 590, 128–141. [Google Scholar] [CrossRef]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef] [Green Version]
- Secrier, M.; Li, X.; de Silva, N.; Eldridge, M.D.; Contino, G.; Bornschein, J.; MacRae, S.; Grehan, N.; O’Donovan, M.; Miremadi, A.; et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat. Genet. 2016, 48, 1131–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janjigian, Y.Y.; Sanchez-Vega, F.; Jonsson, P.; Chatila, W.K.; Hechtman, J.F.; Ku, G.Y.; Riches, J.C.; Tuvy, Y.; Kundra, R.; Bouvier, N.; et al. Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer. Cancer Discov. 2018, 8, 49–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, J.M.; Panda, A.; Zhong, H.; Hirshfield, K.; Damare, S.; Lane, K.; Sokol, L.; Stein, M.N.; Rodriguez-Rodriquez, L.; Kaufman, H.L.; et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J. Clin. Investig. 2016, 126, 2334–2340. [Google Scholar] [CrossRef] [Green Version]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; Morse, M.A.; Van Cutsem, E.; McDermott, R.; Hill, A.; et al. Durable Clinical Benefit with Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [Green Version]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef]
- Thompson, S.L.; Bakhoum, S.F.; Compton, D.A. Mechanisms of chromosomal instability. Curr. Biol. 2010, 20, R285–R295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sishc, B.J.; Davis, A.J. The Role of the Core Non-Homologous End Joining Factors in Carcinogenesis and Cancer. Cancers 2017, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Jain, U.; Saxena, K.; Chauhan, N. Helicobacter pylori induced reactive oxygen Species: A new and developing platform for detection. Helicobacter 2021, 26, e12796. [Google Scholar] [CrossRef] [PubMed]
- Swenberg, J.A.; Lu, K.; Moeller, B.C.; Gao, L.; Upton, P.B.; Nakamura, J.; Starr, T.B. Endogenous versus exogenous DNA adducts: Their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. Off. J. Soc. Toxicol. 2011, 120 (Suppl. S1), S130–S145. [Google Scholar] [CrossRef]
- Mazzei, F.; Viel, A.; Bignami, M. Role of MUTYH in human cancer. Mutat. Res. 2013, 743–744, 33–43. [Google Scholar] [CrossRef]
- Al-Tassan, N.; Chmiel, N.H.; Maynard, J.; Fleming, N.; Livingston, A.L.; Williams, G.T.; Hodges, A.K.; Davies, D.R.; David, S.S.; Sampson, J.R.; et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 2002, 30, 227–232. [Google Scholar] [CrossRef]
- Viel, A.; Bruselles, A.; Meccia, E.; Fornasarig, M.; Quaia, M.; Canzonieri, V.; Policicchio, E.; Urso, E.D.; Agostini, M.; Genuardi, M.; et al. A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer. EBioMedicine 2017, 20, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, M.; van Steenbergen, L.N.; Jones, N.; Vogt, S.; Vasen, H.F.; Morreau, H.; Aretz, S.; Sampson, J.R.; Dekkers, O.M.; Janssen-Heijnen, M.L.; et al. Survival of MUTYH-associated polyposis patients with colorectal cancer and matched control colorectal cancer patients. J. Natl. Cancer Inst. 2010, 102, 1724–1730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahm, S.H.; Park, J.H.; Ko, S.I.; Lee, Y.R.; Chung, I.S.; Chung, J.H.; Kang, L.W.; Han, Y.S. Knock-down of human MutY homolog (hMYH) decreases phosphorylation of checkpoint kinase 1 (Chk1) induced by hydroxyurea and UV treatment. BMB Rep. 2011, 44, 352–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Miao, Z.H.; Zhu, H.; Cai, Y.J.; Lu, W.; Ding, J. Chk1 and Chk2 are differentially involved in homologous recombination repair and cell cycle arrest in response to DNA double-strand breaks induced by camptothecins. Mol. Cancer Ther. 2008, 7, 1440–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, C.; Pinheiro, H.; Figueiredo, J.; Seruca, R.; Carneiro, F. Familial gastric cancer: Genetic susceptibility, pathology, and implications for management. Lancet. Oncol. 2015, 16, e60–e70. [Google Scholar] [CrossRef] [PubMed]
- Peek, R.M., Jr.; Crabtree, J.E. Helicobacter infection and gastric neoplasia. J. Pathol. 2006, 208, 233–248. [Google Scholar] [CrossRef]
- Hatakeyama, M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 2004, 4, 688–694. [Google Scholar] [CrossRef]
- Murata-Kamiya, N.; Hatakeyama, M. Helicobacter pylori-induced DNA double-stranded break in the development of gastric cancer. Cancer Sci. 2022, 113, 1909–1918. [Google Scholar] [CrossRef]
- Hatakeyama, M. Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 2014, 15, 306–316. [Google Scholar] [CrossRef] [Green Version]
- Byrum, A.K.; Vindigni, A.; Mosammaparast, N. Defining and Modulating ‘BRCAness’. Trends Cell Biol. 2019, 29, 740–751. [Google Scholar] [CrossRef]
- Hu, Y.; Guo, M. Synthetic lethality strategies: Beyond BRCA1/2 mutations in pancreatic cancer. Cancer Sci. 2020, 111, 3111–3121. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Marusawa, H.; Matsumoto, Y.; Inuzuka, T.; Ikeda, A.; Fujii, Y.; Minamiguchi, S.; Miyamoto, S.; Kou, T.; Sakai, Y.; et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 2014, 147, 407–417.e403. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014, 26, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Chu, H.; Gu, D.; Xu, M.; Xu, Z.; Gong, Y.; Gong, W.; Tang, Y.; Zhou, J.; Tong, N.; Zhang, Z.; et al. A genetic variant in ERCC2 is associated with gastric cancer prognosis in a Chinese population. Mutagenesis 2013, 28, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Xue, H.; Lu, Y.; Lin, B.; Chen, J.; Tang, F.; Huang, G. The effect of XPD/ERCC2 polymorphisms on gastric cancer risk among different ethnicities: A systematic review and meta-analysis. PLoS ONE 2012, 7, e43431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.J.; Huang, M.Y.; Cheng, T.L.; Kuo, S.H.; Ke, C.C.; Chen, Y.T.; Hsieh, Y.C.; Wang, J.Y.; Cheng, C.M.; Chuang, C.H. ERCC1 Overexpression Increases Radioresistance in Colorectal Cancer Cells. Cancers 2022, 14, 4798. [Google Scholar] [CrossRef] [PubMed]
| Underlying Mechanism | Signature Type: Base Substitution Subtype 1 [References] | Molecular Consequences | Treatment Implications [References] | Additional Biomarkers [References] |
|---|---|---|---|---|
| Aging | SBS1: C > T (NCG) [29,31,39,40] | Deamination of 5-methylcytosine | immune checkpoint inhibition (PD-L1 and PD-1 inhibitors) [16,41,42] | senescence score, EBV+, TMB [43] |
| APOBEC overactivity | SBS2: C > T (TCN) SBS13: C > A (TCA), C > G (TCA, TCC, TCT) [29,31,44,45,46,47] | High mutational load in transcriptionally active genes | APOBEC3B inhibitors, immune checkpoint inhibition (PD-L1 and PD-1 inhibitors), lethal mutagenesis treatment [48,49,50,51] | EBV+, TMB, L1-sequencing, APOBEC3B expression levels [31,45,50,52] |
| Homologous recombination repair (BRCA1/2 mutations) | SBS3: C > A, C > G, C > T, T > A, T > C, T > G (NNN) 3 ID6: microhomology—deletion length: 5+ (microhomology length: 1, 2, 3, 4, 5+) [27,29,53] | Higher mutational burden due to alternative error-prone DSBs repair by NHEJ | Pt-based therapy, PARP inhibitors [54,55,56,57] | BRCA1/2 expression levels, ATM loss [58,59,60] |
| Mismatch repair | SBS20 (POLD1 mutations): C > A (CCC, CCT), C > T (ACA, GCA, GCC) SBS6, SBS14 (POLE mutations), SBS15, SBS21, SBS26, SBS44 2 DBS7: AC > NN (CA), CT > NN (TC), GC > NN (AT), TA > NN (AT), TT > NN (AA, AG, CA, GA) DBS10: CG > NN (TA), TT > NN (GG) ID1: 1 bp insertion (T, homopolymer length: 5+) ID2: 1 bp deletion (C, homopolymer length: 5, 6+) [31,61] | Microsatellite instability, middle to high tumour mutational burden | Immune checkpoint inhibition (PD-L1, PD-1 and CTLA4 inhibitors) [62,63,64,65,66,67] | MMR mutations, MSI status, PD-L1 status, EBV+, T-cell inflamed score, TMB status, POLD1 mutations, ARID1A mutations [22,68,69,70,71,72] |
| Nonhomologous end joining repair | ID6: microhomology—deletion length: 5+ (microhomology length: 1, 2, 3, 4, 5+) ID8: > 1 bp deletion at repeats—deletion length: 5+ (number of repeat units: 1); microhomology—deletion length: 5+ (microhomology length: 1, 2, 3) [31] | Chromosomal instability | NHEJ inhibitors [73,74,75] | CIN, KUs, DNA-PKcs, DNA ligase IV, XRCC4 expression levels [22,76] |
| Reactive oxygen species DNA damage | SBS18: C > A (ACA, CCA, GCA, GCT, TCA, TCC, TCT) [31,77] | DNA damage | Immune checkpoint inhibition therapy (PD-L1 and PD-1 inhibitors) PARP inhibitors [78,79,80] | MUTYH mutations and expression levels, CHEK1 mutations [77,81,82,83] |
| H. pylori infection | SBS3: C > A, C > G, C > T, T > A, T > C, T > G (NNN) 3 ID6: microhomology—deletion length: 5+ (microhomology length: 1, 2, 3, 4, 5+) [31,53,84] | Inflammation, DNA damage | H. pylori eradication | AID expression levels [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pužar Dominkuš, P.; Hudler, P. Mutational Signatures in Gastric Cancer and Their Clinical Implications. Cancers 2023, 15, 3788. https://doi.org/10.3390/cancers15153788
Pužar Dominkuš P, Hudler P. Mutational Signatures in Gastric Cancer and Their Clinical Implications. Cancers. 2023; 15(15):3788. https://doi.org/10.3390/cancers15153788
Chicago/Turabian StylePužar Dominkuš, Pia, and Petra Hudler. 2023. "Mutational Signatures in Gastric Cancer and Their Clinical Implications" Cancers 15, no. 15: 3788. https://doi.org/10.3390/cancers15153788
APA StylePužar Dominkuš, P., & Hudler, P. (2023). Mutational Signatures in Gastric Cancer and Their Clinical Implications. Cancers, 15(15), 3788. https://doi.org/10.3390/cancers15153788







