Unveiling the Therapeutic Potential of Squalene Synthase: Deciphering Its Biochemical Mechanism, Disease Implications, and Intriguing Ties to Ferroptosis
Abstract
Simple Summary
Abstract
1. Introduction
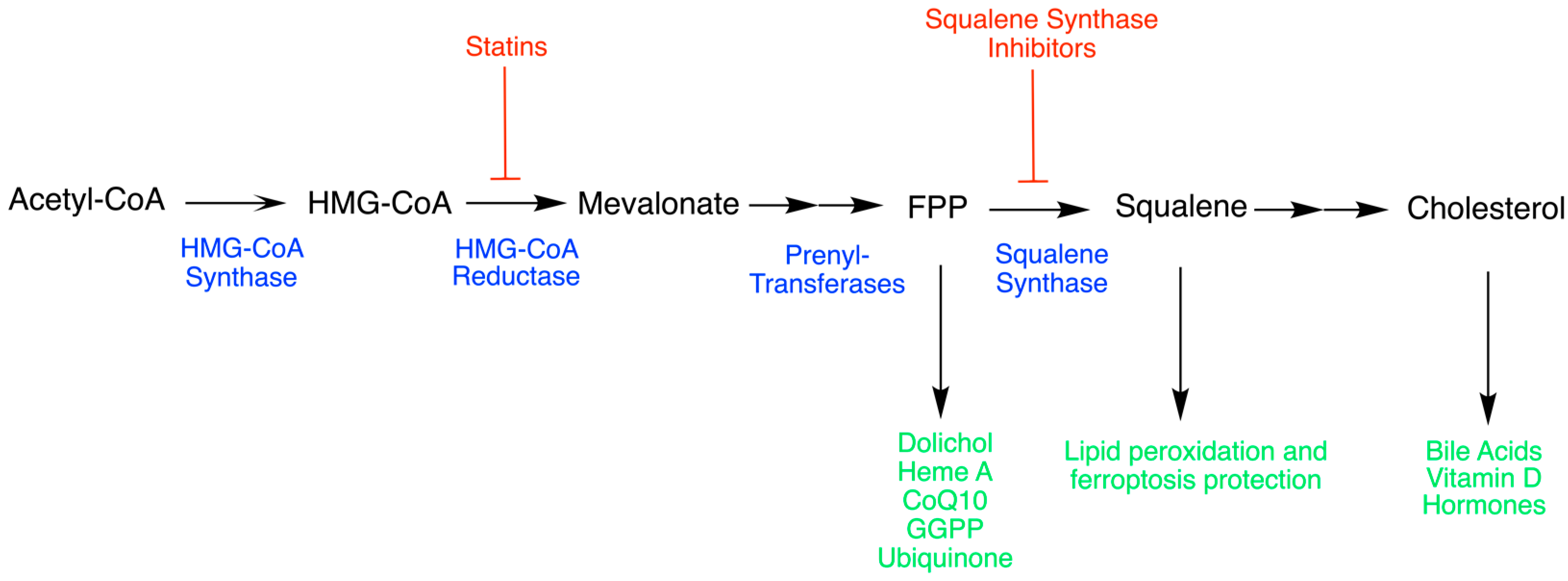
2. Squalene Synthase (SQS)
2.1. The Origins of SQS
2.2. Isoprenoid Synthase Superfamily
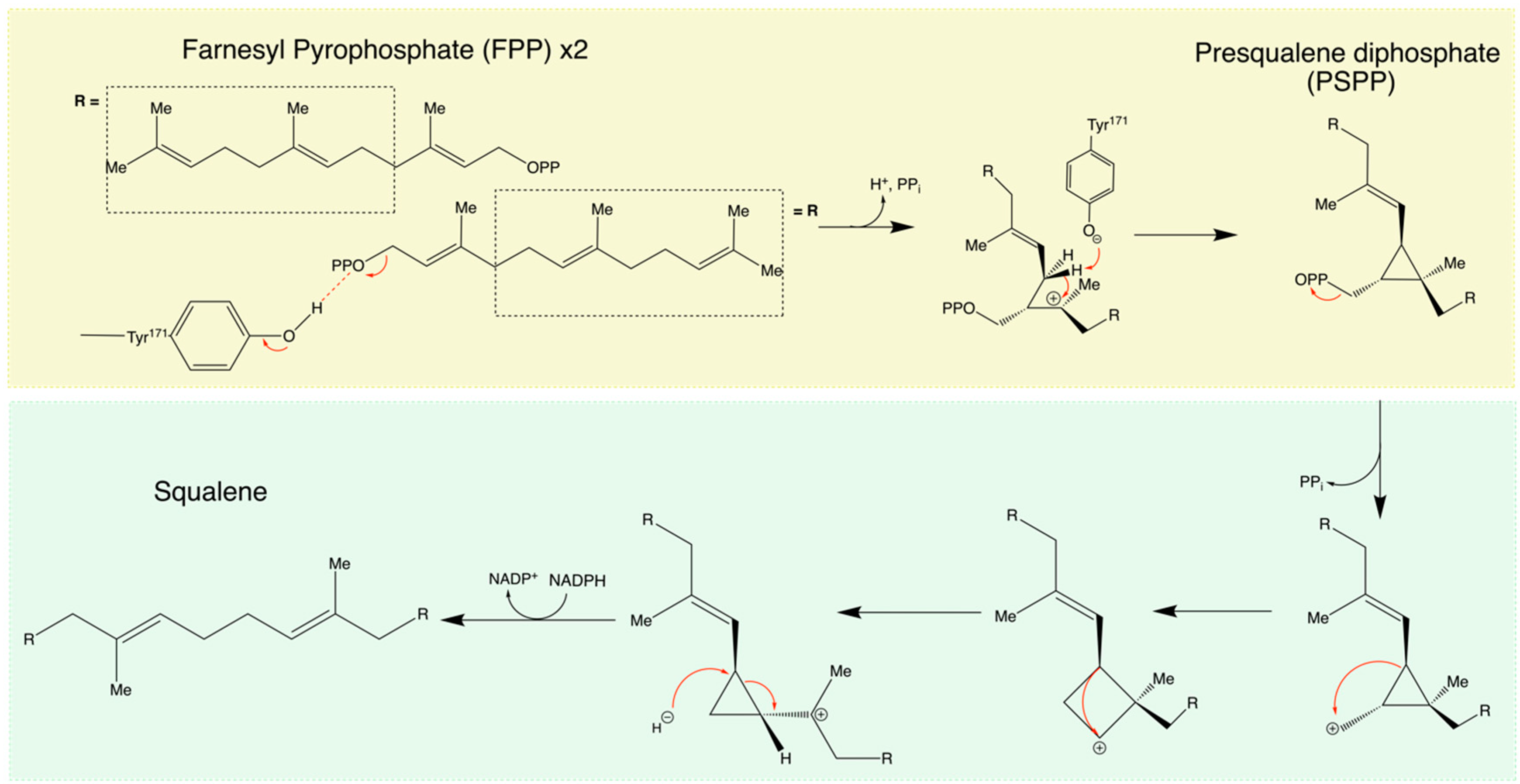
2.3. General Mechanism of SQS
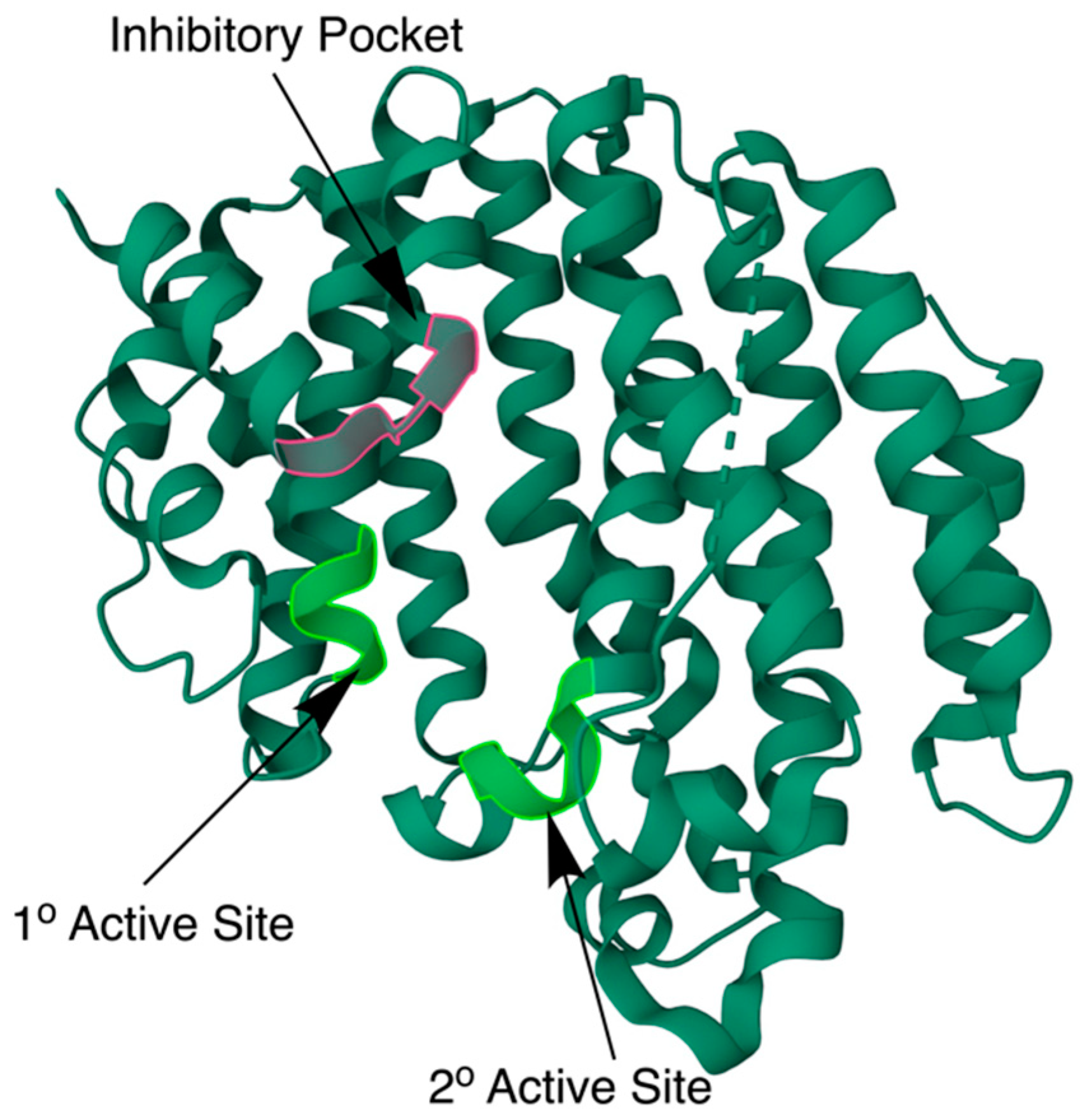
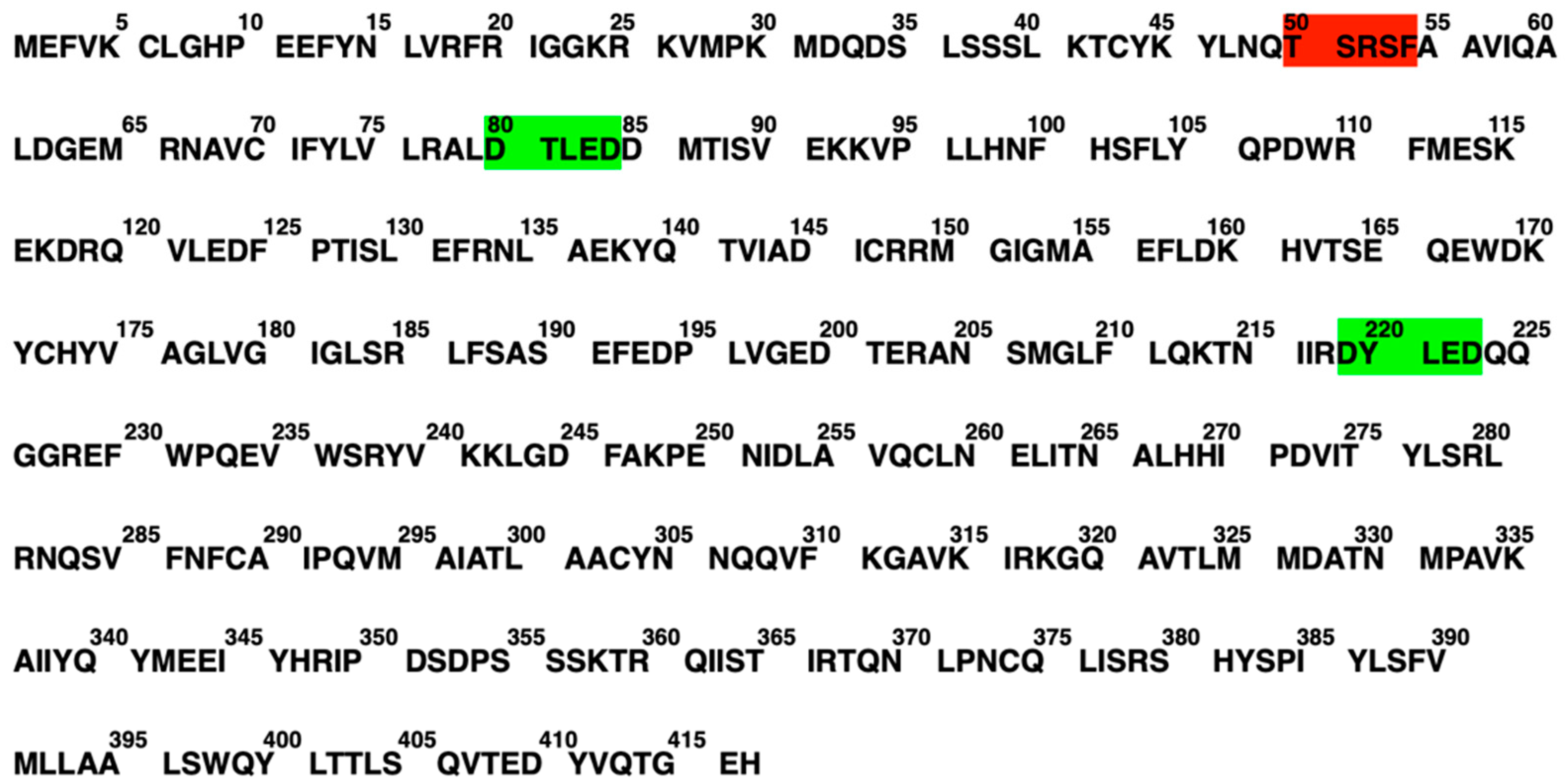
2.4. The Biochemical Structure of Human SQS
2.5. Mutations
2.5.1. Natural Mutations of SQS
2.5.2. Artificial Mutations of SQS
2.6. SQS Regulation
2.7. SQS Relevance in Health and Diseases
2.7.1. Anticancer Therapeutic Strategies
Non-Small Cell Lung Cancer
Colorectal Cancer
Prostate Cancer
2.7.2. Interference Therapeutic Strategies
Chagas Disease
Hepatitis C
High Cholesterol and Cardiovascular Diseases
2.8. SQS Small-Molecule Inhibitors
| Compound | X-ray Diffraction (Å) | Potency | Potency Indicator | PDB Number/ PubChem CID |
|---|---|---|---|---|
| Zaragozic Acid A [70,71,72,73] | 1.89 | 10.1 | pKi in rat liver cells | 3VJC |
| Compound 21 [74] | 11.4 | pIC50 in rat microsomal SQS | 10591006 | |
| Compound 15 a [75] | 1.80 | 8.9 | pIC50 in Hep-G2 cells | 3V66 |
| NB 598 [76] | 7.7 | pIC50 in rat liver cells | 6443223 | |
| Compound 7 [77] | 2.00 | 5.19 | pIC50 in rat microsomal SQS | 3ASX |
| N-[(3R,5S)-7-Chloro-5-(2,3-dimethoxyphenyl)-1-neopentyl-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepine-3-acetyl]-L-aspartic acid [78] | 2.00 | 3Q2Z | ||
| SQ-109 [78] | 2.90 | 6.1 | pKi in Staphylococcus aureus | 3WSA |
| E5700 [79] | 2.32 | 8.8 | pIC50 in rat liver cells | 3WCC |
| ER-119884 [80] | 2.75 | 1.1 | pIC50 in rat liver cells | 3WCE |
| BPH1344 [49] | 2.80 | 6.59 | pIC50 in purified hSQS | 3WCG |
| BPH1218 [49] | 2.22 | 7.28 | pIC50 in purified hSQS | 3WCF |
| BPH1237 [49] | 2.50 | 7.05 | pIC50 in purified hSQS | 3WCH |
| BPH1325 [49] | 2.30 | 6.57 | pIC50 in purified hSQS | 3WCI |
| WC-9 [80] | 2.75 | 7.06 | pIC50 in T. cruzi | 3WCD |
| BPH652 [81] | 2.00 | 9.70 | pKi in rat microsomal SQS | 3LEE |
3. SQS in Ferroptosis
3.1. Overview of Ferroptosis
3.2. Molecular Mechanisms of Ferroptosis
3.3. Regulation of Ferroptosis through SQS
Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SQS | Squalene synthase |
| ROS | Reactive oxygen species |
| FPP | Farnesyl pyrophosphate |
| SQLE | Squalene epoxidase |
| FPS | Farnesyl-pyrophosphate synthase |
| Asp-RR | Aspartate-rich sequence motif regions |
| PSQPP | Presqualene pyrophosphate |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| PPi | inorganic phosphate |
| FDFT1 | Farnesyl-diphosphate farnesyltransferase 1 |
| hSQS | Human squalene synthase |
| PDB | Protein Data Bank |
| ER | Endoplasmatic reticulum |
| SQSD | Squalene synthase deficiency |
| (TNF-) | Tumor necrosis factor-α |
| (IL-1β) | Interleukin-1β |
| COAD | Colon adenocarcinoma |
| CVD | Cardiovascular disease |
| HMG-CoA | Methylglutaryl coenzyme A |
| CID | Compound identifier |
| ACD | Accidental cell death |
| RCD | Regulated cell death |
| ROS | Reactive oxygen species |
| PUFAs | Polyunsaturated fatty acids |
| GPX4 | Phospholipid peroxidase glutathione peroxidase 4 |
| CoQ10 | Coenzyme Q10 |
| GTP | Guanosine triphosphate |
| FINs | Ferroptosis inducers |
| ALCL | Anaplastic large cell lymphomas |
| shRNAs | Short hairpin RNAs |
References
- Park, J.; Matralis, A.N.; Berghuis, A.M.; Tsantrizos, Y.S. Human Isoprenoid Synthase Enzymes as Therapeutic Targets. Front. Chem. 2014, 2, 50. [Google Scholar] [CrossRef]
- Ha, N.T.; Lee, C.H. Roles of Farnesyl-Diphosphate Farnesyltransferase 1 in Tumour and Tumour Microenvironments. Cells 2020, 9, 2352. [Google Scholar] [CrossRef] [PubMed]
- Santana-Molina, C.; Rivas-Marin, E.; Rojas, A.M.; Devos, D.P. Origin and Evolution of Polycyclic Triterpene Synthesis. Mol. Biol. Evol. 2020, 37, 1925–1941. [Google Scholar] [CrossRef]
- Devarenne, T.P.; Ghosh, A.; Chappell, J. Regulation of Squalene Synthase, a Key Enzyme of Sterol Biosynthesis, in Tobacco. Plant Physiol. 2002, 129, 1095–1106. [Google Scholar] [CrossRef]
- Tansey, T.R.; Shechter, I. Squalene Synthase: Structure and Regulation. Prog. Nucleic Acid Res. Mol. Biol. 2000, 65, 157–195. [Google Scholar] [CrossRef]
- Biller, S.; Neuenschwander, K.; Ponpipom, M.; Poulter, D. Squalene synthase inhibitors. Curr. Pharm. Design 1996, 2, 1–40. [Google Scholar] [CrossRef]
- Fukunaga, K.; Arita, M.; Takahashi, M.; Morris, A.J.; Pfeffer, M.; Levy, B.D. Identification and Functional Characterization of a Presqualene Diphosphate Phosphatase. J. Biol. Chem. 2006, 281, 9490–9497. [Google Scholar] [CrossRef]
- Pandit, J.; Danley, D.E.; Schulte, G.K.; Mazzalupo, S.; Pauly, T.A.; Hayward, C.M.; Hamanaka, E.S.; Thompson, J.F.; Harwood, H.J. Crystal Structure of Human Squalene Synthase. J. Biol. Chem. 2000, 275, 30610–30617. [Google Scholar] [CrossRef]
- Tsujimoto, M. A Highly Unsaturated Hydrocarbon In Shark Liver Oil. J. Ind. Eng. Chem. 1916, 8, 889–896. [Google Scholar] [CrossRef]
- Popják, G.; Cornforth, J.W. Substrate Stereochemistry in Squalene Biosynthesis: The First Ciba Medal Lecture. Biochem. J. 1966, 101, 553.b4–568. [Google Scholar] [CrossRef]
- Ortiz de Montellano, P.R.; Wei, J.S.; Vinson, W.A.; Castillo, R.; Boparai, A.S. Substrate Selectivity of Squalene Synthetase. Biochemistry 1977, 16, 2680–2685. [Google Scholar] [CrossRef]
- Tanetoshi, K.; Kyozo, O.; Shuichi, S. Substrate Specificity of Squalene Synthetase. Biochim. Biophys. Acta BBA - Lipids Lipid Metab. 1980, 617, 218–224. [Google Scholar] [CrossRef]
- Sasiak, K.; Rilling, H. Purification to Homogeneity and Some Properties of Squalene Synthetase. Arch. Biochem. Biophys. 1988, 260, 622–627. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, T.L.; Jiang, G.; Straubhaar, J.R.; Conrad, D.G.; Shechter, I. Molecular Cloning, Expression, and Characterization of the CDNA for the Rat Hepatic Squalene Synthase. J. Biol. Chem. 1992, 267, 21368–21374. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; McKenzie, T.L.; Conrad, D.G.; Shechter, I. Transcriptional Regulation by Lovastatin and 25-Hydroxycholesterol in HepG2 Cells and Molecular Cloning and Expression of the CDNA for the Human Hepatic Squalene Synthase. J. Biol. Chem. 1993, 268, 12818–12824. [Google Scholar] [CrossRef]
- Chen, C.C.; Zhang, L.; Yu, X.; Ma, L.; Ko, T.P.; Guo, R.T. Versatile Cis-Isoprenyl Diphosphate Synthase Superfamily Members in Catalyzing Carbon-Carbon Bond Formation. ACS Catal. 2020, 10, 4717–4725. [Google Scholar] [CrossRef]
- Harwood, H.J.; Barbacci-Tobin, E.G.; Petras, S.F.; Lindsey, S.; Pellarin, L.D. 3-(4-Chlorophenyl)-2-(4-Diethylaminoethoxyphenyl)-a-Pentenonitrile Monohydrogen Citrate and Related Analogs. Biochem. Pharmacol. 1997, 53, 839–864. [Google Scholar] [CrossRef]
- Blagg, B.S.J.; Jarstfer, M.B.; Rogers, D.H.; Poulter, C.D. Recombinant Squalene Synthase. A Mechanism for the Rearrangement of Presqualene Diphosphate to Squalene. J. Am. Chem. Soc. 2002, 124, 8846–8853. [Google Scholar] [CrossRef]
- Sun, C.; Ding, Y.; Cheng, B.; Zong, Y. Using Engineered Escherichia Coli to Synthesize Squalene with Optimized Manipulation of Squalene Synthase and Mevalonate Pathway. bioRxiv 2020. [Google Scholar] [CrossRef]
- Koohang, A.; Coates, R.M.; Owen, D.R.; Poulter, C.D. Synthesis and Evaluation of Aziridine Analogues of Presqualene Diphosphate as Squalene Synthase Inhibitors. J. Org. Chem. 1999, 64, 6–7. [Google Scholar] [CrossRef]
- Liu, C.-I.; Jeng, W.-Y.; Chang, W.-J.; Shih, M.-F.; Ko, T.-P.; Wang, A.H.-J. Structural Insights into the Catalytic Mechanism of Human Squalene Synthase. Acta Crystallographica. Sect. D Biol. Crystallogr. 2014, 70 Pt 2, 231–241. [Google Scholar] [CrossRef]
- Shechter, I.; Conrad, D.G.; Hart, I.; Berger, R.C.; McKenzie, T.L.; Bleskan, J.; Patterson, D. Localization of the Squalene Synthase Gene (FDFT1) to Human Chromosome 8p22-P23.1. Genomics 1994, 20, 116–118. [Google Scholar] [CrossRef]
- Coman, D.; Vissers, L.E.L.M.; Riley, L.G.; Kwint, M.P.; Hauck, R.; Koster, J.; Geuer, S.; Hopkins, S.; Hallinan, B.; Sweetman, L.; et al. Squalene Synthase Deficiency: Clinical, Biochemical, and Molecular Characterization of a Defect in Cholesterol Biosynthesis. Am. J. Hum. Genet. 2018, 103, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Y.; Rong, Q.-X.; Chen, Y.; Yuan, Q.; Shen, Y.; Guo, J.; Yang, Y.; Zha, L.; Wu, H.; Huang, L.; et al. Molecular Cloning and Functional Analysis of Squalene Synthase (SS) in Panax Notoginseng. Int. J. Biol. Macromol. 2017, 95, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Jennings, S.M.; Tsay, Y.H.; Fisch, T.M.; Robinson, G.W. Molecular Cloning and Characterization of the Yeast Gene for Squalene Synthetase. Proc. Natl. Acad. Sci. USA 1991, 88, 6038–6042. [Google Scholar] [CrossRef]
- Gu, P.; Ishii, Y.; Spencer, T.A.; Shechter, I. Function-Structure Studies and Identification of Three Enzyme Domains Involved in the Catalytic Activity in Rat Hepatic Squalene Synthase. J. Biol. Chem. 1998, 273, 12515–12525. [Google Scholar] [CrossRef]
- Coman, D.; Vissers, L.; Waterham, H.; Christodoulou, J.; Wevers, R.A.; Pitt, J. Squalene Synthase Deficiency. 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK553533/ (accessed on 31 May 2023).
- Akamine, S.; Nakamori, K.; Chechetka, S.A.; Banba, M.; Umehara, Y.; Kouchi, H.; Izui, K.; Hata, S. CDNA Cloning, MRNA Expression, and Mutational Analysis of the Squalene Synthase Gene of Lotus Japonicus. Biochim. Biophys. Acta BBA - Gene Struct. Expr. 2003, 1626, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Do, R.; Paré, G.; Montpetit, A.; Hudson, T.J.; Gaudet, D.; Engert, J.C. K45R Variant of Squalene Synthase Increases Total Cholesterol Levels in Two Study Samples from a French Canadian Population. Hum. Mutat. 2008, 29, 689–694. [Google Scholar] [CrossRef]
- Faust, J.R.; Goldstein, J.L.; Brown, M.S. Synthesis of Ubiquinone and Cholesterol in Human Fibroblasts: Regulation of a Branched Pathway. Arch. Biochem. Biophys. 1979, 192, 86–99. [Google Scholar] [CrossRef]
- Guan, G.; Dai, P.-H.; Osborne, T.F.; Kim, J.B.; Shechter, I. Multiple Sequence Elements Are Involved in the Transcriptional Regulation of the Human Squalene Synthase Gene*. J. Biol. Chem. 1997, 272, 10295–10302. [Google Scholar] [CrossRef]
- Shimano, H.; Horton, J.D.; Hammer, R.E.; Shimomura, I.; Brown, M.S.; Goldstein, J.L. Overproduction of Cholesterol and Fatty Acids Causes Massive Liver Enlargement in Transgenic Mice Expressing Truncated SREBP-1a. J. Clin. Investig. 1996, 98, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Shimano, H.; Horton, J.D.; Shimomura, I.; Hammer, R.E.; Brown, M.S.; Goldstein, J.L. Isoform 1c of Sterol Regulatory Element Binding Protein Is Less Active than Isoform 1a in Livers of Transgenic Mice and in Cultured Cells. J. Clin. Investig. 1997, 99, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of Cholesterol Synthesis in Preference to Fatty Acid Synthesis in Liver and Adipose Tissue of Transgenic Mice Overproducing Sterol Regulatory Element-Binding Protein-2. J. Clin. Investig. 1998, 101, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Health & Human Services. Gram-Negative Bacteria Infections in Healthcare Settings CDC. 2011. Available online: https://www.cdc.gov/hai/organisms/gram-negative-bacteria.html#:~:text=Gram%2Dnegative%20bacteria%20cause%20infections (accessed on 14 May 2023).
- Hardardóttir, I.; Grunfeld, C.; Feingold, K.R. Effects of Endotoxin on Lipid Metabolism. Biochem. Soc. Trans. 1995, 23, 1013–1018. [Google Scholar] [CrossRef]
- Feingold, K.R.; Grunfeld, C. Tumor Necrosis Factor-Alpha Stimulates Hepatic Lipogenesis in the Rat in Vivo. J. Clin. Investig. 1987, 80, 184–190. [Google Scholar] [CrossRef]
- Kitagawa, S.; Yamaguchi, Y.; Kunitomo, M.; Imaizumi, N.; Fujiwara, M. Altered Vasoconstrictor Responsiveness in Vitamin D-Induced Arteriosclerotic Rat Aortas. JPN J. Pharmacol. 1993, 61, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, H.; Tazoe, F.; Okazaki, S.; Isoo, N.; Tsukamoto, K.; Sekiya, M.; Yahagi, N.; Iizuka, Y.; Ohashi, K.; Kitamine, T.; et al. Increased Cholesterol Biosynthesis and Hypercholesterolemia in Mice Overexpressing Squalene Synthase in the Liver. J. Lipid Res. 2006, 47, 1950–1958. [Google Scholar] [CrossRef]
- Robinson, G.W.; Tsay, Y.H.; Kienzle, B.K.; Smith-Monroy, C.A.; Bishop, R.W. Conservation between Human and Fungal Squalene Synthetases: Similarities in Structure, Function, and Regulation. Mol. Cell. Biol. 1993, 13, 2706–2717. [Google Scholar] [CrossRef]
- Popjak, G.; Agnew, W. Squalene Synthetase. Mol. Cell. Biochem. 1979, 27, 97–116. [Google Scholar] [CrossRef]
- Radisky, E.S.; Poulter, C.D. Squalene Synthase: Steady-State, Pre-Steady-State, and Isotope-Trapping Studies. Biochemistry 2000, 39, 1748–1760. [Google Scholar] [CrossRef]
- Yang, Y.-F.; Chang, Y.-C.; Jan, Y.-H.; Yang, C.-J.; Huang, M.-S.; Hsiao, M. Squalene Synthase Promotes the Invasion of Lung Cancer Cells via the Osteopontin/ERK Pathway. Oncogenesis 2020, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-F.; Jan, Y.-H.; Liu, Y.-P.; Yang, C.-J.; Su, C.-Y.; Chang, Y.-C.; Lai, T.-C.; Chiou, J.; Tsai, H.-Y.; Lu, J.; et al. Squalene Synthase Induces Tumor Necrosis Factor Receptor 1 Enrichment in Lipid Rafts to Promote Lung Cancer Metastasis. Am. J. Respir. Crit. Care Med. 2014, 190, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, I.; Gianfanti, F.; Desbats, M.A.; Orso, G.; Berretta, M.; Prayer-Galetti, T.; Ragazzi, E.; Cocetta, V. Cholesterol Metabolic Reprogramming in Cancer and Its Pharmacological Modulation as Therapeutic Strategy. Front. Oncol. 2021, 11, 682911. [Google Scholar] [CrossRef]
- Drost, F.-J.H.; Osses, D.F.; Nieboer, D.; Steyerberg, E.W.; Bangma, C.H.; Roobol, M.J.; Schoots, I.G. Prostate MRI, with or without MRI-Targeted Biopsy, and Systematic Biopsy for Detecting Prostate Cancer. Cochrane Database Syst. Rev. 2019, 2019, CD012663. [Google Scholar] [CrossRef]
- Fukuma, Y.; Matsui, H.; Koike, H.; Sekine, Y.; Shechter, I.; Ohtake, N.; Nakata, S.; Ito, K.; Suzuki, K. Role of Squalene Synthase in Prostate Cancer Risk and the Biological Aggressiveness of Human Prostate Cancer. Prostate Cancer Prostatic Dis. 2012, 15, 339–345. [Google Scholar] [CrossRef]
- Shang, N.; Li, Q.; Ko, T.-P.; Chan, H.-C.; Li, J.; Zheng, Y.; Huang, C.-H.; Ren, F.; Chen, C.-C.; Zhu, Z.; et al. Squalene Synthase as a Target for Chagas Disease Therapeutics. PLoS Pathog. 2014, 10, e1004114. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Shirasago, Y.; Suzuki, T.; Aizaki, H.; Hanada, K.; Wakita, T.; Nishijima, M.; Fukasawa, M. Targeting Cellular Squalene Synthase, an Enzyme Essential for Cholesterol Biosynthesis, Is a Potential Antiviral Strategy against Hepatitis c Virus. J. Virol. 2014, 89, 2220–2232. [Google Scholar] [CrossRef]
- Zheng, J.; Conrad, M. The Metabolic Underpinnings of Ferroptosis. Cell Metab. 2020, 32, 920–937. [Google Scholar] [CrossRef]
- Garcia-Bermudez, J.; Baudrier, L.; Bayraktar, E.C.; Shen, Y.; La, K.; Guarecuco, R.; Yucel, B.; Fiore, D.; Tavora, B.; Freinkman, E.; et al. Squalene Accumulation in Cholesterol Auxotrophic Lymphomas Prevents Oxidative Cell Death. Nature 2019, 567, 118–122. [Google Scholar] [CrossRef]
- CDC. High Cholesterol Facts|cdc.gov. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/cholesterol/facts.htm#:~:text=High%20total%20cholesterol%20in%20the%20United%20States&text=Slightly%20more%20than%20half%20of (accessed on 16 May 2023).
- American Cancer Society. Key Statistics for Lung Cancer. Available online: https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html#:~:text=Lung%20cancer%20is%20by%20far (accessed on 16 May 2023).
- CDC. What Is Lung Cancer?|CDC. 2022. Available online: https://www.cdc.gov/cancer/lung/basic_info/what-is-lung-cancer.htm#:~:text=When%20cancer%20starts%20in%20the (accessed on 15 May 2023).
- National Cancer Institute. Non-Small Cell Lung Cancer Treatment. National Cancer Institute. Available online: https://www.cancer.gov/types/lung/patient/non-small-cell-lung-treatment-pdq (accessed on 17 May 2023).
- Mayo Clinic. Colon Cancer—Symptoms and Causes. Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/colon-cancer/symptoms-causes/syc-20353669 (accessed on 20 May 2023).
- Li, C.; Wang, Y.; Liu, D.; Wong, C.C.; Coker, O.O.; Zhang, X.; Liu, C.; Zhou, Y.; Liu, Y.; Kang, W.; et al. Squalene Epoxidase Drives Cancer Cell Proliferation and Promotes Gut Dysbiosis to Accelerate Colorectal Carcinogenesis. Gut 2022, 71, 2253–2265. [Google Scholar] [CrossRef]
- Jiang, H.; Tang, E.; Chen, Y.; Liu, H.; Zhao, Y.; Lin, M.; He, L. Squalene Synthase Predicts Poor Prognosis in Stage I–III Colon Adenocarcinoma and Synergizes Squalene Epoxidase to Promote Tumor Progression. Cancer Sci. 2021, 113, 971–985. [Google Scholar] [CrossRef]
- Prostate Cancer|American Cancer Fund®. Available online: https://www.americancancerfund.org/cancer-types/prostate-cancer/?gclid=EAIaIQobChMI8qTksrWv_gIV_A-zAB0zoADOEAAYAiAAEgIlQfD_BwE (accessed on 22 May 2023).
- What is Prostate Cancer? Prostate Cancer Foundation. Available online: https://www.pcf.org/about-prostate-cancer/what-is-prostate-cancer/?utm_source=google_cpc&utm_medium=ad_grant&utm_campaign=awareness_patients_general&gclid=EAIaIQobChMItpnLpLWv_gIVHSuzAB0jgQOeEAAYASAAEgI_0PD_BwE (accessed on 31 May 2023).
- CDC—Chagas Disease—Epidemiology & Risk Factors. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/parasites/chagas/epi.html (accessed on 9 May 2023).
- CDC. Hepatitis C Information. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/hepatitis/hcv/index.htm (accessed on 7 May 2023).
- CDC. Heart Disease Facts. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/heartdisease/facts.htm#:~:text=Heart%20disease%20is%20the%20leading (accessed on 1 May 2023).
- Liao, J.K. Squalene Synthase Inhibitor Lapaquistat Acetate. Circulation 2011, 123, 1925–1928. [Google Scholar] [CrossRef]
- Jones, P.H.; Davidson, M.H.; Stein, E.A.; Bays, H.E.; McKenney, J.M.; Miller, E.; Cain, V.A.; Blasetto, J.W. Comparison of the Efficacy and Safety of Rosuvastatin versus Atorvastatin, Simvastatin, and Pravastatin across Doses (STELLAR**STELLAR = Statin Therapies for Elevated Lipid Levels Compared across Doses to Rosuvastatin. Trial). Am. J. Cardiol. 2003, 92, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, H. CCR5 in Multiple Sclerosis: Expression, Regulation and Modulation by Statins. Available online: https://scholarlypublications.universiteitleiden.nl/access/item%3A2866261/view (accessed on 31 May 2023).
- Oesterle, A.; Laufs, U.; Liao, J.K. Pleiotropic Effects of Statins on the Cardiovascular System. Circ. Res. 2017, 120, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Flint, O.P.; Masters, B.A.; Gregg, R.E.; Durham, S.K. Inhibition of Cholesterol Synthesis by Squalene Synthase Inhibitors Does Not Induce Myotoxicityin Vitro. Toxicol. Appl. Pharmacol. 1997, 145, 91–98. [Google Scholar] [CrossRef]
- Bansal, V.; Vaidya, S. Characterization of 2 Distinct Allyl Pyrophosphatase Activities from Rat-Liver Microsomes. Arch. Biochem. Biophys. 1994, 315, 393–399. [Google Scholar] [CrossRef]
- Baxter, A.; Fitzgerald, B.J.; Hutson, J.L.; McCarthy, A.D.; Motteram, J.M.; Ross, B.C.; Sapra, M.; Snowden, M.A.; Watson, N.S.; Williams, R.J. Squalestatin 1, a Potent Inhibitor of Squalene Synthase, Which Lowers Serum Cholesterol in Vivo. J. Biol. Chem. 1992, 267, 11705–11708. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, S.; Harwood, H.J. Inhibition of Mammalian Squalene Synthetase Activity by Zaragozic Acid a Is a Result of Competitive Inhibition Followed by Mechanism-Based Irreversible Inactivation. J. Biol. Chem. 1995, 270, 9083–9096. [Google Scholar] [CrossRef]
- Bergstrom, J.D.; Dufresne, C.; Bills, G.F.; Nallin-Omstead, M.; Byrne, K.P. Discovery, Biosynthesis, and Mechanism of Action of the Zaragozic Acids: Potent Inhibitors of Squalene Synthase. Annu. Rev. Microbiol. 1995, 49, 607–639. [Google Scholar] [CrossRef]
- Bergstrom, J.D.; Kurtz, M.M.; Rew, D.J.; Amend, A.M.; Karkas, J.D.; Bostedor, R.G.; Bansal, V.S.; Dufresne, C.; VanMiddlesworth, F.L.; Hensens, O.D. Zaragozic Acids: A Family of Fungal Metabolites That Are Picomolar Competitive Inhibitors of Squalene Synthase. Proc. Natl. Acad. Sci. USA 1993, 90, 80–84. [Google Scholar] [CrossRef]
- Brown, G.R.; Butlin, R.J.; Chapman, S.; Eakin, M.A.; Foubister, A.J.; Freeman, S.; Griffiths, D.; Harrison, P.J.; Johnson, M.C. Phenoxypropylamines: A New Series of Squalene Synthase Inhibitors. J. Med. Chem. 1995, 38, 4157–4160. [Google Scholar] [CrossRef]
- Ichikawa, M.; Ohtsuka, M.; Ohki, H.; Haginoya, N.; Itoh, M.; Sugita, K.; Usui, H.; Suzuki, M.; Terayama, K.; Kanda, A. Discovery of Novel Tricyclic Compounds as Squalene Synthase Inhibitors. Bioorganic Med. Chem. 2012, 20, 3072–3093. [Google Scholar] [CrossRef]
- Gotteland, J.-P.; Brunel, I.; Gendre, F.; Desire, J.; Delhon, A.; Junquero, D.; Oms, P.; Halazy, S. (Aryloxy)methylsilane Derivatives as New Cholesterol Biosynthesis Inhibitors: Synthesis and Hypocholesterolemic Activity of a New Class of Squalene Epoxidase Inhibitors. J. Med. Chem. 1995, 38, 3207–3216. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Yokomizo, A.; Itoh, M.; Sugita, K.; Usui, H.; Shimizu, H.; Suzuki, M.; Terayama, K.; Kanda, A. Discovery of a New 2-Aminobenzhydrol Template for Highly Potent Squalene Synthase Inhibitors. Bioorganic Med. Chem. 2011, 19, 1930–1949. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.-Y.; Liu, Y.-L.; Li, K.; Cao, R.; Zhu, W.; Axelson, J.; Pang, R.; Oldfield, E. Head-To-Head Prenyl Tranferases: Anti-Infective Drug Targets. J. Med. Chem. 2012, 55, 4367–4372. [Google Scholar] [CrossRef]
- Sealey-Cardona, M.; Cammerer, S.; Jones, S.; Ruiz-Pérez, L.M.; Brun, R.; Gilbert, I.H.; Urbina, J.A.; González-Pacanowska, D. Kinetic Characterization of Squalene Synthase from Trypanosoma Cruzi: Selective Inhibition by Quinuclidine Derivatives. Antimicrob. Agents Chemother. 2007, 51, 2123–2129. [Google Scholar] [CrossRef] [PubMed]
- Urbina, J.A.; Concepcion, J.L.; Montalvetti, A.; Rodriguez, J.B.; Docampo, R. Mechanism of Action of 4-Phenoxyphenoxyethyl Thiocyanate (WC-9) against Trypanosoma Cruzi, the Causative Agent of Chagas’ Disease. Antimicrob. Agents Chemother. 2003, 47, 2047–2050. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.-Y.; Liu, C.-I.; Liu, Y.-L.; Zhang, Y.; Wang, K.; Jeng, W.-Y.; Ko, T.-P.; Cao, R.; Jiang, A.; Oldfield, E. Mechanism of Action and Inhibition of Dehydrosqualene Synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 21337–21342. [Google Scholar] [CrossRef]
- Ponpipom, M.M.; Girotra, N.N.; Bugianesi, R.L.; Roberts, C.D.; Berger, G.D.; Burk, R.M.; Marquis, R.W.; Parsons, W.H.; Bartizal, K.F.; Bergstom, J.D. Structure-Activity Relationships of C1 and C6 Side Chains of Zaragozic Acid a Derivatives. J. Med. Chem. 1994, 37, 4031–4051. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Luo, G.; Zhang, X.; Lu, F.; Qiao, L.; He, W.; Li, G.; Zhang, Y. Discovery of Potential Inhibitors of Squalene Synthase from Traditional Chinese Medicine Based on Virtual Screening and in Vitro Evaluation of Lipid-Lowering Effect. Molecules 2018, 23, 1040. [Google Scholar] [CrossRef]
- Song, Y.; Lin, F.-Y.; Yin, F.; Hensler, M.; Poveda, C.A.R.; Mukkamala, D.; Cao, R.; Wang, H.; Morita, C.T.; Pacanowska, D.G.; et al. Phosphonosulfonates Are Potent, Selective Inhibitors of Dehydrosqualene Synthase and Staphyloxanthin Biosynthesis in Staphylococcus aureus. J. Med. Chem. 2009, 52, 976–988. [Google Scholar] [CrossRef]
- Magnin, D.R.; Biller, S.A.; Dickson, J.K.; Logan, J.V.; Lawrence, R.M.; Chen, Y.; Sulsky, R.B.; Ciosek, C.P.; Harrity, T.W.; Jolibois, K.G. 1,1-Bisphosphonate Squalene Synthase Inhibitors: Interplay between the Isoprenoid Subunit and the Diphosphate Surrogate. J. Med. Chem. 1995, 38, 2596–2605. [Google Scholar] [CrossRef]
- Amin, D.; Rutledge, R.Z.; Needle, S.J.; Neuenswander, K.; Bilder, G.E.; Perrone, M.H.; Hele, D.; Bush, R. RPR 101821, a new potent cholesterol-lowering agent: Inhibition of squalene synthase and 7-dehydrocholesterol reductase. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1996, 353, 233–240. [Google Scholar] [CrossRef]
- Brown, G.R.; Foubister, A.J.; Freeman, S.; McTaggart, F.; Mirrlees, D.J.; Reid, A.C.; Smith, G.J.; Taylor, M.J.; Thomason, D.A.; Whittamore, P.R.O. Novel Optimised Quinuclidine Squalene Synthase Inhibitors. Bioorganic Med. Chem. Lett. 1997, 7, 597–600. [Google Scholar] [CrossRef]
- Iwasawa, Y.; Shibata, J.; Mitsuya, M.; Masaki, H.; Hayashi, M.; Kanno, T.; Sawasaki, Y.; Hisaka, A.; Kamei, T.; Tomimoto, K. J-104,123, a Novel and Orally-Active Inhibitor of Squalene Synthase: Stereoselective Synthesis and Cholesterol Lowering Effects in Dogs. Bioorganic Med. Chem. Lett. 1996, 6, 463–466. [Google Scholar] [CrossRef]
- Brown, G.R.; Clarke, D.S.; Foubister, A.J.; Freeman, S.; Harrison, P.J.; Johnson, M.C.; Mallion, K.B.; McCormick, J.; McTaggart, F.; Reid, A.C.; et al. Synthesis and Activity of a Novel Series of 3-Biarylquinuclidine Squalene Synthase Inhibitors. J. Med. Chem. 1996, 39, 2971–2979. [Google Scholar] [CrossRef]
- Prashad, M.; Kathawala, F.G.; Scallen, T. N-(Arylalkyl)Farnesylamines: New Potent Squalene Synthetase Inhibitors. J. Med. Chem. 1993, 36, 1501–1504. [Google Scholar] [CrossRef]
- Rodrígues-Poveda, C.A.; González-Pacanowska, D.; Szajnman, S.H.; Rodríguez, J.B. 2-Alkylaminoethyl-1,1-Bisphosphonic Acids Are Potent Inhibitors of the Enzymatic Activity of Trypanosoma Cruzi Squalene Synthase. Antimicrob. Agents Chemother. 2012, 56, 4483–4486. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, J.A.; Damon, R.E.; Fell, J.B.; Perez, L.B.; Scallen, T.J.; Vedamanda, T.R. Squalene Synthase Inhibitors: Isosteric Replacements of the Farnesyl Chain of Benzyl Farnesyl Amine. Bioorganic Med. Chem. Lett. 1996, 6, 2491–2494. [Google Scholar] [CrossRef]
- Sharratt, P.J.; Hutson, J.L.; Inglis, G.G.A.; Lester, M.G.; Procopiou, P.A.; Watson, N.S. Structurally Simplified Squalestatins: Monocyclic 1,3-Dioxane Analogues. Bioorganic Med. Chem. Lett. 1994, 4, 661–666. [Google Scholar] [CrossRef]
- Fung, A.K.; Baker, W.R.; Fakhoury, S.; Stein, H.H.; Cohen, J.; Donner, B.G.; Garvey, D.S.; Spina, K.P.; Rosenberg, S.H. (1 Alpha, 2 Beta, 3 Beta, 4 Alpha)-1,2-Bis[N-Propyl-N-(4-Phenoxybenzyl) Amino]Carbonyl]Cyclobutane-3,4-Dicarboxylic Acid (A-87049): A Novel Potent Squalene Synthase Inhibitor. J. Med. Chem. 1997, 40, 2123–2125. [Google Scholar] [CrossRef]
- Orenes Lorente, S.; Gómez, R.; Jiménez, C.; Cammerer, S.; Yardley, V.; de Luca-Fradley, K.; Croft, S.L.; Ruiz Perez, L.M.; Urbina, J.; Gonzalez Pacanowska, D.; et al. Biphenylquinuclidines as Inhibitors of Squalene Synthase and Growth of Parasitic Protozoa. Bioorganic Med. Chem. 2005, 13, 3519–3529. [Google Scholar] [CrossRef]
- Magnin, D.R.; Biller, S.A.; Chen, Y.; Dickson, J.K.; Fryszman, O.M.; Lawrence, R.M.; Logan, J.V.; Sieber-McMaster, E.S.; Sulsky, R.B.; Traeger, S.C.; et al. Alpha-Phosphonosulfonic Acids: Potent and Selective Inhibitors of Squalene Synthase. J. Med. Chem. 1996, 39, 657–660. [Google Scholar] [CrossRef] [PubMed]
- Overhand, M.; Pieterman, E.; Cohen, L.H.; Valentijn, A.R.P.M.; van der Marel, G.A.; van Boom, J.H. Synthesis of Triphosphonate Analogues of Farnesyl Pyrophosphate. Inhibitors of Squalene Synthase and Protein:farnesyl Transferase. Bioorganic Med. Chem. Lett. 1997, 7, 2435–2440. [Google Scholar] [CrossRef]
- Biller, S.A.; Sofia, M.J.; DeLange, B.; Forster, C.; Gordon, E.M.; Harrity, T.; Rich, L.C.; Ciosek, C.P. The First Potent Inhibitor of Squalene Synthase: A Profound Contribution of an Ether Oxygen to Inhibitor-Enzyme Interaction. J. Am. Chem. Soc. 1991, 113, 8522–8524. [Google Scholar] [CrossRef]
- Lin, F.-Y.; Zhang, Y.; Hensler, M.; Liu, Y.-L.; Chow, O.A.; Zhu, W.; Wang, K.; Pang, R.; Thienphrapa, W.; Nizet, V.; et al. Dual Dehydrosqualene/Squalene Synthase Inhibitors: Leads for Innate Immune System-Based Therapeutics. ChemMedChem 2012, 7, 561–564. [Google Scholar] [CrossRef]
- Amin, D.; Cornell, S.A.; Gustafson, S.K.; Needle, S.J.; Ullrich, J.W.; Bilder, G.E.; Perrone, M.H. Bisphosphonates Used for the Treatment of Bone Disorders Inhibit Squalene Synthase and Cholesterol Biosynthesis. J. Lipid Res. 1992, 33, 1657–1663. [Google Scholar] [CrossRef]
- Ishihara, T.; Kakuta, H.; Moritani, H.; Ugawa, T.; Sakamoto, S.; Tsukamoto, S.; Yanagisawa, I. Syntheses of 3-Ethylidenequinuclidine Derivatives as Squalene Synthase Inhibitors. Part 2: Enzyme Inhibition and Effects on Plasma Lipid Levels. Bioorganic Med. Chem. 2003, 11, 3735–3745. [Google Scholar] [CrossRef]
- Choi, S.-W.; Hur, N.-Y.; Ahn, S.-C.; Kim, D.-S.; Lee, J.-K.; Kim, D.-O.; Park, S.-K.; Kim, B.-Y.; Baik, M.-Y. Isolation and Structural Determination of Squalene Synthase Inhibitor from Prunus Mume Fruit. J. Microbiol. Biotechnol. 2007, 17, 1970–1975. [Google Scholar]
- Rodrigues, J.C.F.; Urbina, J.A.; de Souza, W. Antiproliferative and Ultrastructural Effects of BPQ-OH, a Specific Inhibitor of Squalene Synthase, on Leishmania Amazonensis. Exp. Parasitol. 2005, 111, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Biller, S.A.; Abt, J.W.; Pudzianowski, A.T.; Rich, L.C.; Slusarchyk, D.A.; Ciosek, C.P. Aromatic Isosteres as Conformational Probes for an Isoprenyl Subunit: Application to Inhibitors of Squalene Synthase. Bioorganic Med. Chem. Lett. 1993, 3, 595–600. [Google Scholar] [CrossRef]
- Thompson, J.F.; Danley, D.E.; Mazzalupo, S.; Milos, P.M.; Lira, M.E.; Harwood, H. Truncation of Human Squalene Synthase Yields Active, Crystallizable Protein. Arch. Biochem. Biophys. 1998, 350, 283–290. [Google Scholar] [CrossRef]
- Nishimoto, T.; Amano, Y.; Tozawa, R.; Ishikawa, E.; Imura, Y.; Yukimasa, H.; Sugiyama, Y. Lipid-Lowering Properties of TAK-475, a Squalene Synthase Inhibitor, in Vivo and in Vitro. Br. J. Pharmacol. 2003, 139, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Garvey, D.S.; Cohen, J.; Stein, H.; Rosenberg, S.H. Cyclopentanedi- and Tricarboxylic Acids as Squalene Synthase Inhibitors: Syntheses and Evaluation. Bioorganic Med. Chem. Lett. 1998, 8, 891–896. [Google Scholar] [CrossRef]
- Song, Y.; Liu, C.-I.; Lin, F.-Y.; No, J.H.; Hensler, M.; Liu, Y.-L.; Jeng, W.-Y.; Low, J.; Liu, G.Y.; Nizet, V.; et al. Inhibition of Staphyloxanthin Virulence Factor Biosynthesis in Staphylococcus aureus: In Vitro, in Vivo, and Crystallographic Results. J. Med. Chem. 2009, 52, 3869–3880. [Google Scholar] [CrossRef] [PubMed]
- Pospiech, M.; Owens, S.E.; Miller, D.J.; Austin-Muttitt, K.; Mullins, J.G.L.; Cronin, J.G.; Allemann, R.K.; Sheldon, I.M. Bisphosphonate Inhibitors of Squalene Synthase Protect Cells against Cholesterol-Dependent Cytolysins. FASEB J. 2021, 35, e21640. [Google Scholar] [CrossRef] [PubMed]
- Cammerer, S.B.; Jimenez, C.; Jones, S.; Gros, L.; Lorente, S.O.; Rodrigues, C.; Rodrigues, J.C.F.; Caldera, A.; Ruiz Perez, L.M.; da Souza, W.; et al. Quinuclidine Derivatives as Potential Antiparasitics. Antimicrob. Agents Chemother. 2007, 51, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Tanimoto, T.; Onodera, K.; Hosoya, T.; Takamatsu, Y.; Kinoshita, T.; Tago, K.; Kogen, H.; Fujioka, T.; Hamano, K.; Tsujita, Y. Schizostatin, a Novel Squalene Synthase Inhibitor Produced by the Mushroom, Schizophyllum commune. I. Taxonomy, Fermentation, Isolation, Physico-chemical Properties and Biological Activities. J. Antibiot. 1996, 49, 617–623. [Google Scholar] [CrossRef]
- Kourounakis, A.P.; Charitos, C.; Rekka, E.A.; Kourounakis, P.N. Lipid-Lowering (Hetero)Aromatic Tetrahydro-1,4-Oxazine Derivatives with Antioxidant and Squalene Synthase Inhibitory Activity. J. Med. Chem. 2008, 51, 5861–5865. [Google Scholar] [CrossRef]
- Ichikawa, M.; Ohtsuka, M.; Ohki, H.; Ota, M.; Haginoya, N.; Itoh, M.; Shibata, Y.; Sugita, K.; Ishigai, Y.; Terayama, K.; et al. Discovery of DF-461, a Potent Squalene Synthase Inhibitor. ACS Med. Chem. Lett. 2013, 4, 932–936. [Google Scholar] [CrossRef]
- Watanabe, S.; Hirai, H.; Kambara, T.; Kojima, Y.; Nlshida, H.; Sugiura, A.; Yamauchi, Y.; Yoshikawa, N.; Harwood, H.J.; Huang, L.H.; et al. Cj-13,981 and CJ-13,982, New Squalene Synthase Inhibitors. J. Antibiot. 2001, 54, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Shechter, I.; Gu, P.; Jiang, G.; Onofrey, T.J.; Cann, R.O.; Castro, A.; Spencer, T.A. Sulfobetaine Zwitterionic Inhibitors of Squalene Synthase. Bioorganic Med. Chem. Lett. 1996, 6, 2585–2588. [Google Scholar] [CrossRef]
- Tavridou, A.; Kaklamanis, L.; Megaritis, G.; Kourounakis, A.P.; Papalois, A.; Roukounas, D.; Rekka, E.A.; Kourounakis, P.N.; Charalambous, A.; Manolopoulos, V.G. Pharmacological Characterization in Vitro of EP2306 and EP2302, Potent Inhibitors of Squalene Synthase and Lipid Biosynthesis. Eur. J. Pharmacol. 2006, 535, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Griebenow, N.; Flessner, T.; Buchmueller, A.; Raabe, M.; Bischoff, H.; Kolkhof, P. Identification and Optimization of Tetrahydro-2H-3-Benzazepin-2-Ones as Squalene Synthase Inhibitors. Bioorganic Med. Chem. Lett. 2011, 21, 2554–2558. [Google Scholar] [CrossRef] [PubMed]
- Lolli, M.L.; Rolando, B.; Tosco, P.; Chaurasia, S.; Stilo, A.D.; Lazzarato, L.; Gorassini, E.; Ferracini, R.; Oliaro-Bosso, S.; Fruttero, R.; et al. Synthesis and Preliminary Pharmacological Characterisation of a New Class of Nitrogen-Containing Bisphosphonates (N-BPs). Bioorganic Med. Chem. 2010, 18, 2428–2438. [Google Scholar] [CrossRef]
- Macías-Alonso, M.; Andrés, L.S.; Córdova-Guerrero, I.; Estolano-Cobián, A.; Díaz-Rubio, L.; Marrero, J.G. Inhibition of Squalene Synthase of Rat Liver by Abietane Diterpenes Derivatives. Nat. Prod. Res. 2021, 35, 2972–2976. [Google Scholar] [CrossRef]
- Hiyoshi, H.; Yanagimachi, M.; Ito, M.; Ohtsuka, I.; Yoshida, I.; Saeki, T.; Tanaka, H. Effect of ER-27856, a Novel Squalene Synthase Inhibitor, on Plasma Cholesterol in Rhesus Monkeys: Comparison with 3-Hydroxy-3-Methylglutaryl-Coa Reductase Inhibitors. J. Lipid Res. 2000, 41, 1136–1144. [Google Scholar] [CrossRef]
- Shiuan, D.; Chen, Y.-H.; Lin, H.-K.; Huang, K.-J.; Tai, D.-F.; Chang, D.-K. Discovering Peptide Inhibitors of Human Squalene Synthase through Screening the Phage-Displayed Cyclic Peptide C7c Library. Appl. Biochem. Biotechnol. 2016, 179, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Ugawa, T.; Kakuta, H.; Moritani, H.; Matsuda, K.; Ishihara, T.; Yamaguchi, M.; Naganuma, S.; Iizumi, Y.; Shikama, H. YM-53601, a Novel Squalene Synthase Inhibitor, Reduces Plasma Cholesterol and Triglyceride Levels in Several Animal Species. Br. J. Pharmacol. 2000, 131, 63–70. [Google Scholar] [CrossRef]
- Wasko, B.M.; Smits, J.P.; Shull, L.W.; Wiemer, D.F.; Hohl, R.J. A Novel Bisphosphonate Inhibitor of Squalene Synthase Combined with a Statin or a Nitrogenous Bisphosphonate in Vitro. J. Lipid Res. 2011, 52, 1957–1964. [Google Scholar] [CrossRef]
- Amin, D.; Rutledge, R.Z.; Needle, S.N.; Galczenski, H.F.; Neuenschwander, K.; Scotese, A.C.; Maguire, M.P.; Bush, R.C.; Hele, D.J.; Bilder, G.E.; et al. RPR 107393, a Potent Squalene Synthase Inhibitor and Orally Effective Cholesterol-Lowering Agent: Comparison with Inhibitors of HMG-CoA Reductase. J. Pharmacol. Exp. Ther. 1997, 281, 746–752. [Google Scholar] [PubMed]
- Wattanasin, S.; Boettcher, B.R.; Scallen, T. N-Hydroxyglycine Derivatives as Novel Inhibitors of Squalene Synthase. Bioorganic Med. Chem. Lett. 1997, 7, 3039–3044. [Google Scholar] [CrossRef]
- Iwasawa, Y.; Hayashi, M.; Nomoto, T.; Shibata, J.; Mitsuya, M.; Hirota, K.; Yonemoto, M.; Kamei, T.; Miura, K.; Tomimoto, K. Synthesis and Biological Activity of J-104,118, a Novel, Potent Inhibitor of Squalene Synthase. Bioorganic Med. Chem. Lett. 1995, 5, 1989–1994. [Google Scholar] [CrossRef]
- Ishihara, T.; Kakuta, H.; Moritani, H.; Ugawa, T.; Yanagisawa, I. Synthesis and Biological Evaluation of Novel Propylamine Derivatives as Orally Active Squalene Synthase Inhibitors. Bioorganic Med. Chem. 2004, 12, 5899–5908. [Google Scholar] [CrossRef]
- Prashad, M. Amidinium Cation as a Mimic of Allylic Carbocation: Synthesis and Squalene Synthetase Inhibitory Activity of an Amidinium Analog of a Carbocation Intermediate. J. Med. Chem. 1993, 36, 631–632. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The Molecular Machinery of Regulated Cell Death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Albini, S.; Zakharova, V.; Ait-Si-Ali, S. Chapter 3—Histone Modifications. ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/B9780128148792000030 (accessed on 22 February 2023).
- Tang, D.; Kroemer, G. Ferroptosis. Curr. Biol. 2020, 30, R1292–R1297. [Google Scholar] [CrossRef]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Wong, R.S. Apoptosis in Cancer: From Pathogenesis to Treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular Mechanisms and Health Implications. Cell Res. 2020, 31, 107–125. [Google Scholar] [CrossRef]
- D’Arcy, M.S. Cell Death: A Review of the Major Forms of Apoptosis, Necrosis and Autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef]
- Cookson, B.T.; Brennan, M.A. Pro-Inflammatory Programmed Cell Death. Trends Microbiol. 2001, 9, 113–114. [Google Scholar] [CrossRef] [PubMed]
- Eagle, H. Nutrition Needs of Mammalian Cells in Tissue Culture. Science 1955, 122, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Eagle, H.; Piez, K.A.; Oyama, V.I. The Biosynthesis of Cystine in Human Cell Cultures. J. Biol. Chem. 1961, 236, 1425–1428. [Google Scholar] [CrossRef]
- Bannai, S.; Kitamura, E. Transport Interaction of L-Cystine and L-Glutamate in Human Diploid Fibroblasts in Culture. J. Biol. Chem. 1980, 255, 2372–2376. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, A.M.; Chirillo, R.; Aversa, I.; Sacco, A.; Costanzo, F.; Biamonte, F. Ferroptosis and Cancer: Mitochondria Meet the “Iron Maiden” Cell Death. Cells 2020, 9, 1505. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Organelle-Specific Regulation of Ferroptosis. Cell Death Differ. 2021, 28, 2843–2856. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Cotto-Rios, X.M.; Gavathiotis, E. Unraveling Cell Death Mysteries. Nat. Chem. Biol. 2016, 12, 470–471. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.; Huang, Z.; Lin, Z.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, Present and Future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chakraborty, B.; Safi, R.; Kazmin, D.; Chang, C.; McDonnell, D.P. Dysregulated Cholesterol Homeostasis Results in Resistance to Ferroptosis Increasing Tumorigenicity and Metastasis in Cancer. Nat. Commun. 2021, 12, 5103. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.; Skouta, R. The Selenoprotein Glutathione Peroxidase 4: From Molecular Mechanisms to Novel Therapeutic Opportunities. Biomedicines 2022, 10, 891. [Google Scholar] [CrossRef]
- Yu, H.; Guo, P.; Xie, X.; Wang, Y.; Chen, G. Ferroptosis, a New Form of Cell Death, and Its Relationships with Tumourous Diseases. J. Cell. Mol. Med. 2016, 21, 648–657. [Google Scholar] [CrossRef]
- Gaschler, M.M.; Stockwell, B.R. Lipid Peroxidation in Cell Death. Biochem. Biophys. Res. Commun. 2017, 482, 419–425. [Google Scholar] [CrossRef]
- Lei, G.; Mao, C.; Yan, Y.; Zhuang, L.; Gan, B. Ferroptosis, Radiotherapy, and Combination Therapeutic Strategies. Protein Cell 2021, 12, 836–857. [Google Scholar] [CrossRef]
- Hassannia, B.; Vandenabeele, P.; Vanden Berghe, T. Targeting Ferroptosis to Iron out Cancer. Cancer Cell 2019, 35, 830–849. [Google Scholar] [CrossRef]
- Shimada, K.; Skouta, R.; Kaplan, A.; Yang, W.S.; Hayano, M.; Dixon, S.J.; Brown, L.M.; Valenzuela, C.A.; Wolpaw, A.J.; Stockwell, B.R. Global Survey of Cell Death Mechanisms Reveals Metabolic Regulation of Ferroptosis. Nat. Chem. Biol. 2016, 12, 497–503. [Google Scholar] [CrossRef]
- Rasheed, S.; Nelson-Rees, W.A.; Toth, E.M.; Arnstein, P.; Gardner, M.B. Characterization of a Newly Derived Human Sarcoma Cell Line (HT-1080). Cancer 1974, 33, 1027–1033. [Google Scholar] [CrossRef]

| Compound | Potency | Potency Indicator | PubChem CID |
|---|---|---|---|
| Squalestatin 2/Zaragozic Acid B [74] | 10.5 | pKi in Hep-G2 cells | 9940176 |
| Squalestatin 3/Zaragozic Acid C [74] | 10.4 | pKi in Hep-G2 cells | 11814656 |
| Compound 5d [82] | 10.4 | pIC50 in rat microsomal SQS | 44352892 |
| Compound 19 [75] | 10.3 | pIC50 in rat microsomal SQS | 10662370 |
| L735021 [83] | 9.9 | pIC50 in rat microsomal SQS | 9853075 |
| Compound 505374 [83] | 9.52 | pIC50 in Hep-G2 cells | |
| Compound 33b [76] | 9.29 | pIC50 in rat liver cells | |
| Compound 6 [84] | 9.0 | pIC50 in S. aureus | 56947056 |
| SQ34919 [85] | 9.0 | pIC50 in rat microsomal SQS | 10382597 |
| RPR 101821 [86] | 9 | pIC50 in rat microsomal SQS | |
| Compound 22a [78] | 8.82 | pIC50 in rat liver cells | |
| Compound 1e [87] | 8.7 | pKi in rat microsomal SQS | 44370557 |
| J104123 [88] | 8.6 | pIC50 in dogs | 9848748 |
| Compound 2d [86] | 8.6 | pIC50 in rat microsomal SQS | 10405846 |
| Compound 23 [89] | 8.5 | pIC50 in rat microsomal SQS | 19077552 |
| ER-28448 [80] | 8.44 | pIC50 in rat liver cells | 11540104 |
| Compound 7 [90] | 8.4 | pIC50 in rat liver cells | 10358175 |
| Compound 11 [91] | 8.30 | pIC50 in T. cruzi | |
| Compound 8 [92] | 8.1 | pIC50 in rat liver cells | 98110129 |
| Compound 16a [93] | 8.0 | pIC50 in rat microsomal SQS | 44373472 |
| Compound 5j [94] | 7.9 | pIC50 in rat liver cells | 10699948 |
| Compound 4ª [95] | 7.89 | pIC50 in rat microsomal SQS | |
| Compound 4 [96] | 7.8 | pIC50 in rat liver cells | 56947060 |
| Compound 1 [97] | 7.7 | pIC50 in rat microsomal SQS | 19956992 |
| CP-424677 [9] | 7.49 | pIC50 in rat microsomal SQS | |
| A-87049 [96] | 7.43 | pIC50 in rat liver cells | |
| Compound 6 [98] | 7.4 | pKi in rat microsomal SQS | 44370654 |
| CP-320473 [9] | 7.25 | pIC50 in rat microsomal SQS | |
| Compound 11 [99] | 7.22 | pKi in human liver cells | |
| Compound 4q [77] | 7.2 | pIC50 in rat microsomal SQS | 10411224 |
| YM 175 [100] | 7.19 | pIC50 in rat liver cells | 119188 |
| YM53579 [101] | 7.10 | pIC50 in rat microsomal SQS | 11372147 |
| Chlorogenic acid [102] | 7 | pIC50 in pig liver cells | 1794427 |
| BPQ-OH [103] | 6.96 | pIC50 in rat liver cells | 9817140 |
| Compound 15 [104] | 6.9 | pIC50 in rat microsomal SQS | 44370656 |
| CP294838 [105] | 6.9 | pIC50 in rat liver cells | 9889227 |
| TAK475 (Lapaquistat) [106] | 6.82 | pIC50 in Hep-G2 cells | 9960389 |
| Compound 23 [107] | 6.77 | pIC50 in rat liver cells | |
| P-3622 [18] | 6.70 | pIC50 in Hep-G2 cells | |
| EB 1053 [101] | 6.68 | pIC50 in rat liver cells | 130821 |
| Compound 14 [108] | 6.6 | pIC50 in rat liver cells | 44382842 |
| L731128 [73] | 6.6 | pIC50 in rat liver cells | 9931928 |
| PHPBP [101] | 6.51 | pIC50 in rat liver cells | |
| Compound 15 [108] | 6.5 | pKi in purified hSQS | 44185382 |
| MPEX098 [109] | 6.39 | pIC50 in purified hSQS | |
| BPH830 [109] | 6.3 | pKi in purified hSQS | 44182294 |
| Compound 4g [110] | 6.2 | pIC50 in L. donovani | 44584870 |
| Schizostatin [111] | 6.08 | pIC50 in rat liver cells | 9862523 |
| Compound 9 [112] | 6.0 | pIC50 in rat microsomal SQS | 25147760 |
| DF-461 [113] | 5.96 | pIC50 in rat liver cells | 57777744 |
| CJ-13,982 [114] | 5.96 | pIC50 in rat liver cells | 10428617 |
| 2R,3S diphosphate enantiomer [21] | 5.93 | pIC50 in yeast | |
| Compound 17 [115] | 5.7 | pIC50 in rat microsomal SQS | 10475079 |
| CJ-981 [115] | 5.55 | pIC50 in rat liver cells | |
| EP2302 [116] | 5.52 | pIC50 in Hep-G2 cells | |
| Compound 19 [117] | 5.48 | pIC50 in human liver | |
| Compound 20 [118] | 5.4 | pIC50 in rat microsomal SQS | 46866079 |
| BMS-188494 [69] | 5.39 | pIC50 in rat microsomal SQS | 154098 |
| BMS-187745 [69] | 5.16 | pIC50 in rat microsomal SQS | 153978 |
| EP2306 [117] | 4.88 | pIC50 in Hep-G2 cells | |
| Carnosol [119] | 4.75 | pIC50 in rat liver cells | 442009 |
| CP-458003 [9] | 4.52 | pIC50 in rat microsomal SQS | |
| ER-27856 [120] | 4.41 | pIC50 in rat liver cells | 9896881 |
| CLSPHSMFC [121] | 4.19 | pIC50 in Hep-G2 cells | |
| SMFC [122] | 4.12 | pIC50 in Hep-G2 cells | 5272743 |
| YM-53601 [122] | 4.10 | pIC50 in Hep-G2 cells | 9907532 |
| CKTE [122] | 4.06 | pIC50 in Hep-G2 cells | |
| WHQW [122] | 4.05 | pIC50 in Hep-G2 cells | |
| CJ-15,183 [115] | 4.01 | pIC50 in human liver cells | 9894585 |
| Compound 5 [123] | 8.14–8.24 | pIC50 in Hep-G2 cells | |
| RPR 107393 [124] | 6.05–6.22 | pIC50 in rat liver cells | 10314587 |
| Compound 12 [125] | 6.6–6.8 | pIC50 in rat liver cells | 56947012 |
| J104118 [126] | 9.1–9.3 | pIC50 in rat microsomal SQS | 10460101 |
| (3-{[1-(prop-2-en-1-yl)-9H-carbazol-2-yl]oxy}propyl)(propan-2-yl)amine [127] | 7.2–7.5 | pIC50 in Hep-G2 cells | 9949081 |
| Compound 1 [128] | 5.2–7.0 | pIC50 in rat liver cells | 10409462 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picón, D.F.; Skouta, R. Unveiling the Therapeutic Potential of Squalene Synthase: Deciphering Its Biochemical Mechanism, Disease Implications, and Intriguing Ties to Ferroptosis. Cancers 2023, 15, 3731. https://doi.org/10.3390/cancers15143731
Picón DF, Skouta R. Unveiling the Therapeutic Potential of Squalene Synthase: Deciphering Its Biochemical Mechanism, Disease Implications, and Intriguing Ties to Ferroptosis. Cancers. 2023; 15(14):3731. https://doi.org/10.3390/cancers15143731
Chicago/Turabian StylePicón, David Figueredo, and Rachid Skouta. 2023. "Unveiling the Therapeutic Potential of Squalene Synthase: Deciphering Its Biochemical Mechanism, Disease Implications, and Intriguing Ties to Ferroptosis" Cancers 15, no. 14: 3731. https://doi.org/10.3390/cancers15143731
APA StylePicón, D. F., & Skouta, R. (2023). Unveiling the Therapeutic Potential of Squalene Synthase: Deciphering Its Biochemical Mechanism, Disease Implications, and Intriguing Ties to Ferroptosis. Cancers, 15(14), 3731. https://doi.org/10.3390/cancers15143731







