Role of Transcription Factor BEND3 and Its Potential Effect on Cancer Progression
Abstract
Simple Summary
Abstract
1. Introduction
2. BEND3 Structure and Its Interaction with DNA
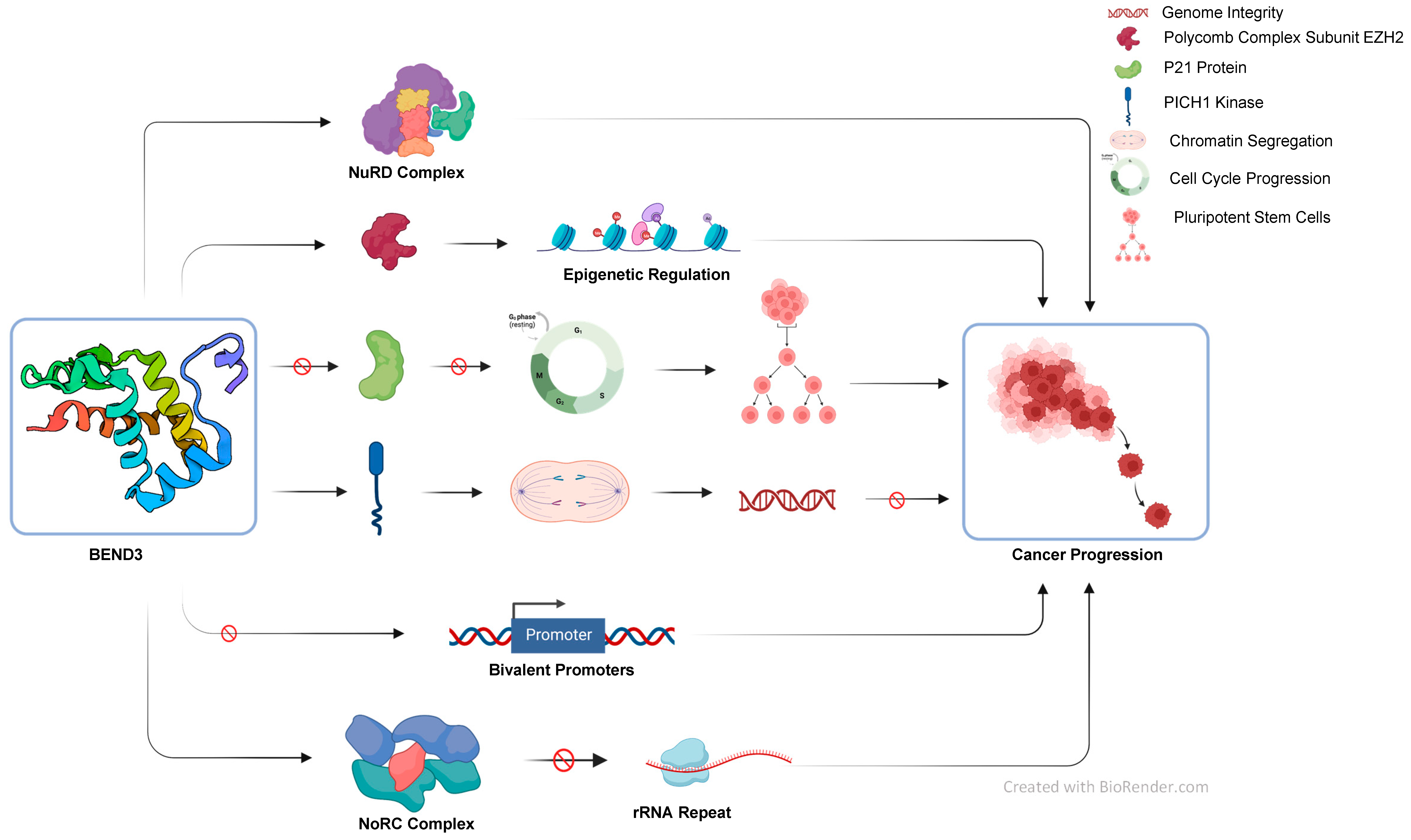
3. BEND3-Mediated Chromatin Regulation
4. Epigenetic Regulation by BEND3
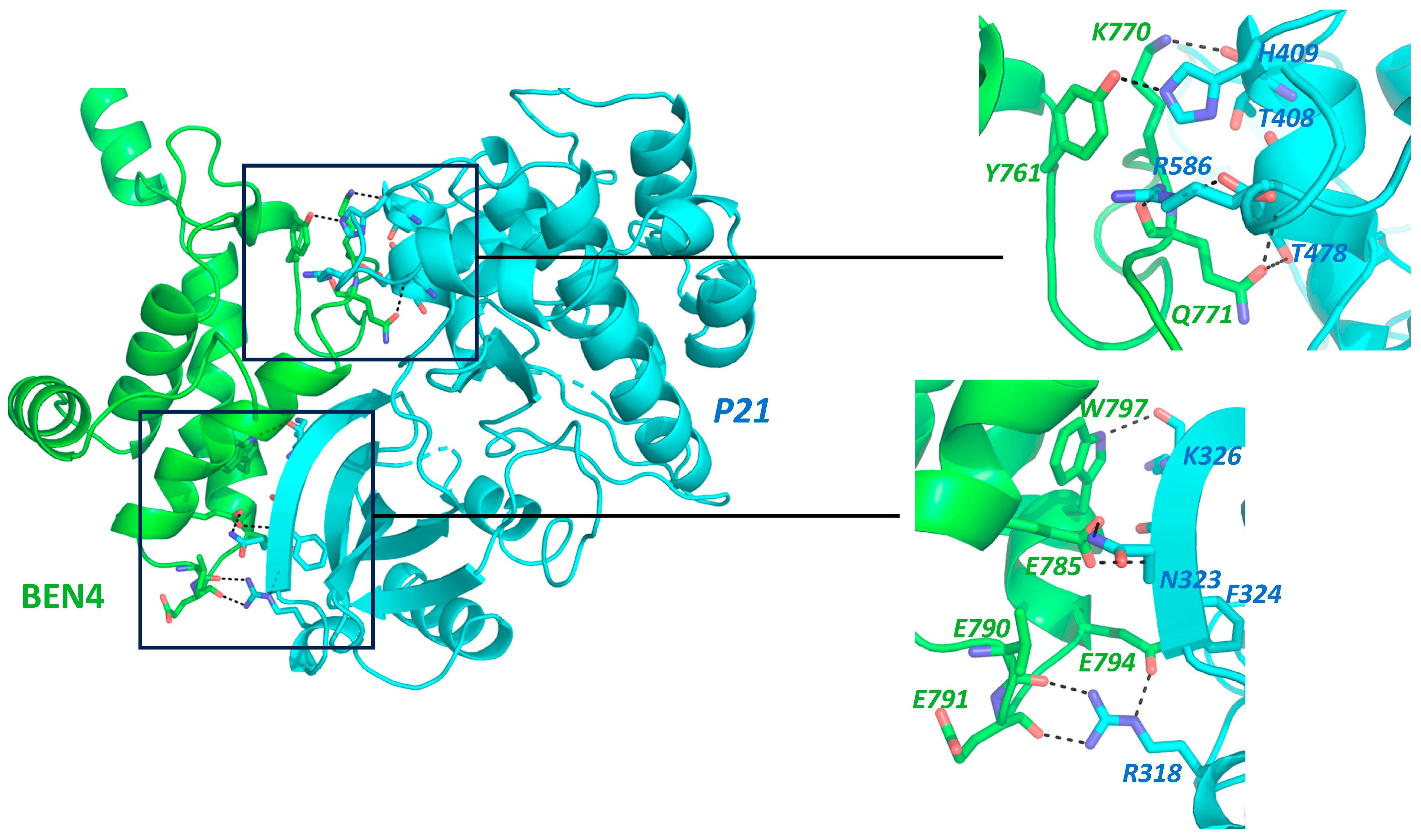
5. BEND3 in Cell-Cycle Regulation
6. Role of BEND3 in Pluripotency Maintenance
7. Discussion and Future Prospects
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ablett, M.P.; Singh, J.K.; Clarke, R.B. Stem Cells in Breast Tumours: Are They Ready for the Clinic? Eur. J. Cancer. 2012, 48, 2104–2116. [Google Scholar] [CrossRef] [PubMed]
- Witt, A.E.; Lee, C.-W.; Lee, T.I.; Azzam, D.J.; Wang, B.; Caslini, C.; Petrocca, F.; Grosso, J.; Jones, M.; Cohick, E.B.; et al. Identification of a Cancer Stem Cell-Specific Function for the Histone Deacetylases, HDAC1 and HDAC7, in Breast and Ovarian Cancer. Oncogene 2017, 36, 1707–1720. [Google Scholar] [CrossRef] [PubMed]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC Activation Is a Hallmark of Cancer Initiation and Maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-ΚB, an Active Player in Human Cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
- Kurniawan, F.; Chetlangia, N.; Kamran, M.; Redon, C.E.; Pongor, L.; Sun, Q.; Lin, Y.-C.; Mohan, V.; Shaqildi, O.; Asoudegi, D.; et al. BEND3 Safeguards Pluripotency by Repressing Differentiation-Associated Genes. Proc. Natl. Acad. Sci. USA 2022, 119, e2107406119. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; You, Q.; Huang, C.; Zhang, T.; Wang, M.; Zhang, T.; Yang, X.; Xiong, J.; Li, Y.; et al. Highly Enriched BEND3 Prevents the Premature Activation of Bivalent Genes during Differentiation. Science 2022, 375, 1053–1058. [Google Scholar] [CrossRef]
- Khan, A.; Giri, S.; Wang, Y.; Chakraborty, A.; Ghosh, A.K.; Anantharaman, A.; Aggarwal, V.; Sathyan, K.M.; Ha, T.; Prasanth, K.V.; et al. BEND3 Represses RDNA Transcription by Stabilizing a NoRC Component via USP21 Deubiquitinase. Proc. Natl. Acad. Sci. USA 2015, 112, 8338–8343. [Google Scholar] [CrossRef]
- Khan, A.; Prasanth, S.G. BEND3 Mediates Transcriptional Repression and Heterochromatin Organization. Transcription 2015, 6, 102–105. [Google Scholar] [CrossRef]
- Sathyan, K.M.; Shen, Z.; Tripathi, V.; Prasanth, K.V.; Prasanth, S.G. A BEN-Domain-Containing Protein Associates with Heterochromatin and Represses Transcription. J. Cell Sci. 2011, 124, 3149–3163. [Google Scholar] [CrossRef]
- Pitchai, G.P.; Kaulich, M.; Bizard, A.H.; Mesa, P.; Yao, Q.; Sarlos, K.; Streicher, W.W.; Nigg, E.A.; Montoya, G.; Hickson, I.D. A Novel TPR-BEN Domain Interaction Mediates PICH-BEND3 Association. Nucleic Acids Res. 2017, 45, 11413–11424. [Google Scholar] [CrossRef]
- Lai, A.Y.; Wade, P.A. Cancer Biology and NuRD: A Multifaceted Chromatin Remodelling Complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Barghout, S.H.; Aman, A.; Nouri, K.; Blatman, Z.; Arevalo, K.; Thomas, G.E.; MacLean, N.; Hurren, R.; Ketela, T.; Saini, M.; et al. A Genome-Wide CRISPR/Cas9 Screen in Acute Myeloid Leukemia Cells Identifies Regulators of TAK-243 Sensitivity. JCI Insight 2021, 6, e141518. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Prasanth, S. BENDing with Polycomb in Pluripotency and Cancer. BioEssays 2003, 2300046. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, J.; Niu, L.; Kamran, M.; Yang, A.W.H.; Jolma, A.; Dai, Q.; Hughes, T.R.; Patel, D.J.; Zhang, L.; et al. Distinct Structural Bases for Sequence-Specific DNA Binding by Mammalian BEN Domain Proteins. Genes Dev. 2022, 36, 225–240. [Google Scholar] [CrossRef]
- Abhiman, S.; Iyer, L.M.; Aravind, L. BEN: A Novel Domain in Chromatin Factors and DNA Viral Proteins. Bioinformatics 2008, 24, 458–461. [Google Scholar] [CrossRef]
- Shiheido, H.; Shimizu, J. Basic Amino Acid Residues Located in the N-Terminal Region of BEND3 Are Essential for Its Nuclear Localization. Biochem. Biophys. Res. Commun. 2015, 457, 589–594. [Google Scholar] [CrossRef]
- Xie, W.; Ling, T.; Zhou, Y.; Feng, W.; Zhu, Q.; Stunnenberg, H.G.; Grummt, I.; Tao, W. The Chromatin Remodeling Complex NuRD Establishes the Poised State of RRNA Genes Characterized by Bivalent Histone Modifications and Altered Nucleosome Positions. Proc. Natl. Acad. Sci. USA 2012, 109, 8161–8166. [Google Scholar] [CrossRef]
- Harikumar, A.; Meshorer, E. Chromatin Remodeling and Bivalent Histone Modifications in Embryonic Stem Cells. EMBO Rep. 2015, 16, 1609–1619. [Google Scholar] [CrossRef]
- dos Santos, R.L.; Tosti, L.; Radzisheuskaya, A.; Caballero, I.M.; Kaji, K.; Hendrich, B.; Silva, J.C.R. MBD3/NuRD Facilitates Induction of Pluripotency in a Context-Dependent Manner. Cell Stem Cell 2014, 15, 102–110. [Google Scholar] [CrossRef]
- Sims, J.K.; Wade, P.A. Mi-2/NuRD Complex Function Is Required for Normal S Phase Progression and Assembly of Pericentric Heterochromatin. Mol. Biol. Cell 2011, 22, 3094–3102. [Google Scholar] [CrossRef]
- Kaji, K.; Caballero, I.M.; MacLeod, R.; Nichols, J.; Wilson, V.A.; Hendrich, B. The NuRD Component Mbd3 Is Required for Pluripotency of Embryonic Stem Cells. Nat. Cell Biol. 2006, 8, 285–292. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, D.L.C.; Snoek, T.; Mullin, N.P.; Yates, A.; Bezstarosti, K.; Demmers, J.; Chambers, I.; Poot, R.A. An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell 2010, 6, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Fang, J.; Li, Y.; Zhang, J. Mbd3, a Component of NuRD/Mi-2 Complex, Helps Maintain Pluripotency of Mouse Embryonic Stem Cells by Repressing Trophectoderm Differentiation. PLoS ONE 2009, 4, e7684. [Google Scholar] [CrossRef] [PubMed]
- Denslow, S.A.; Wade, P.A. The Human Mi-2/NuRD Complex and Gene Regulation. Oncogene 2007, 26, 5433–5438. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wong, J.; Moreno, G.T.; Young, M.K.; Côté, J.; Wang, W. NURD, a Novel Complex with Both ATP-Dependent Chromatin-Remodeling and Histone Deacetylase Activities. Mol. Cell 1998, 2, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Cao, H.; Wang, Z.; Zhou, D.; Wu, C.; Wang, S.; Xia, D.; Zhang, D. CHD4/NuRD Complex Regulates Complement Gene Expression and Correlates with CD8 T Cell Infiltration in Human Hepatocellular Carcinoma. Clin. Epigenet. 2020, 12, 31. [Google Scholar] [CrossRef]
- Zhang, X.; DeSalle, L.M.; Patel, J.H.; Capobianco, A.J.; Yu, D.; Thomas-Tikhonenko, A.; McMahon, S.B. Metastasis-Associated Protein 1 (MTA1) Is an Essential Downstream Effector of the c-MYC Oncoprotein. Proc. Natl. Acad. Sci. USA 2005, 102, 13968–13973. [Google Scholar] [CrossRef]
- Fujita, N.; Jaye, D.L.; Geigerman, C.; Akyildiz, A.; Mooney, M.R.; Boss, J.M.; Wade, P.A. MTA3 and the Mi-2/NuRD Complex Regulate Cell Fate during B Lymphocyte Differentiation. Cell 2004, 119, 75–86. [Google Scholar] [CrossRef]
- Kusam, S.; Dent, A. Common Mechanisms for the Regulation of B Cell Differentiation and Transformation by the Transcriptional Repressor Protein BCL-6. Immunol. Res. 2007, 37, 177–186. [Google Scholar] [CrossRef]
- Morey, L.; Brenner, C.; Fazi, F.; Villa, R.; Gutierrez, A.; Buschbeck, M.; Nervi, C.; Minucci, S.; Fuks, F.; Di Croce, L. MBD3, a Component of the NuRD Complex, Facilitates Chromatin Alteration and Deposition of Epigenetic Marks. Mol. Cell. Biol. 2008, 28, 5912–5923. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Chen, Y.; Sun, Y.; Yang, F.; Yu, W.; Liang, J.; Sun, L.; Yang, X.; Shi, L.; et al. LSD1 Is a Subunit of the NuRD Complex and Targets the Metastasis Programs in Breast Cancer. Cell 2009, 138, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [PubMed]
- Tatetsu, H.; Kong, N.R.; Chong, G.; Amabile, G.; Tenen, D.G.; Chai, L. SALL4, the Missing Link between Stem Cells, Development and Cancer. Gene 2016, 584, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef] [PubMed]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Smirnov, E.; Chmúrčiaková, N.; Cmarko, D. Human RDNA and Cancer. Cells 2021, 10, 3452. [Google Scholar] [CrossRef]
- Stults, D.M.; Killen, M.W.; Williamson, E.P.; Hourigan, J.S.; Vargas, H.D.; Arnold, S.M.; Moscow, J.A.; Pierce, A.J. Human RRNA Gene Clusters Are Recombinational Hotspots in Cancer. Cancer Res. 2009, 69, 9096–9104. [Google Scholar] [CrossRef]
- Bywater, M.J.; Poortinga, G.; Sanij, E.; Hein, N.; Peck, A.; Cullinane, C.; Wall, M.; Cluse, L.; Drygin, D.; Anderes, K.; et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of P53. Cancer Cell 2012, 22, 51–65. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting Epigenetic Regulators for Cancer Therapy: Mechanisms and Advances in Clinical Trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Dawson, M.A. The Cancer Epigenome: Concepts, Challenges, and Therapeutic Opportunities. Science 2017, 355, 1147–1152. [Google Scholar] [CrossRef]
- Saksouk, N.; Barth, T.K.; Ziegler-Birling, C.; Olova, N.; Nowak, A.; Rey, E.; Mateos-Langerak, J.; Urbach, S.; Reik, W.; Torres-Padilla, M.-E.; et al. Redundant Mechanisms to Form Silent Chromatin at Pericentromeric Regions Rely on BEND3 and DNA Methylation. Mol. Cell 2014, 56, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Saurin, A.J.; Shiels, C.; Williamson, J.; Satijn, D.P.; Otte, A.P.; Sheer, D.; Freemont, P.S. The Human Polycomb Group Complex Associates with Pericentromeric Heterochromatin to Form a Novel Nuclear Domain. J. Cell Biol. 1998, 142, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Voncken, J.W.; Schweizer, D.; Aagaard, L.; Sattler, L.; Jantsch, M.F.; van Lohuizen, M. Chromatin-Association of the Polycomb Group Protein BMI1 Is Cell Cycle-Regulated and Correlates with Its Phosphorylation Status. J. Cell Sci. 1999, 112 Pt 24, 4627–4639. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.; Salmon-Divon, M.; Dvinge, H.; Hynes-Allen, A.; Balasooriya, G.; Leaford, D.; Behrens, A.; Bertone, P.; Hendrich, B. NuRD-Mediated Deacetylation of H3K27 Facilitates Recruitment of Polycomb Repressive Complex 2 to Direct Gene Repression. EMBO J. 2012, 31, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Dockerill, M.; Gregson, C.; O’ Donovan, D.H. Targeting PRC2 for the Treatment of Cancer: An Updated Patent Review (2016–2020). Expert Opin. Ther. Pat. 2021, 31, 119–135. [Google Scholar] [CrossRef]
- Kurniawan, F.; Prasanth, S.G. A BEN-Domain Protein and Polycomb Complex Work Coordinately to Regulate Transcription. Transcription 2022, 13, 82–87. [Google Scholar] [CrossRef]
- Liu, H.-H.; Tsai, Y.-S.; Lai, C.-L.; Tang, C.-H.; Lai, C.-H.; Wu, H.-C.; Hsieh, J.-T.; Yang, C.-R. Evolving Personalized Therapy for Castration-Resistant Prostate Cancer. BioMedicine 2014, 4, 2. [Google Scholar] [CrossRef]
- Huang, Y.; Li, W.; Yan, W.; Wu, J.; Chen, L.; Yao, X.; Gu, F.; Lv, L.; Zhao, J.; Zhao, M.; et al. Loss of PICH Promotes Chromosome Instability and Cell Death in Triple-Negative Breast Cancer. Cell Death Dis. 2019, 10, 428. [Google Scholar] [CrossRef]
- Pitchai, G.P.; Hickson, I.D.; Streicher, W.; Montoya, G.; Mesa, P. Characterization of the NTPR and BD1 Interacting Domains of the Human PICH-BEND3 Complex. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016, 72, 646–651. [Google Scholar] [CrossRef]
- Yao, Y.; Dai, W. Genomic Instability and Cancer. J. Carcinog. Mutagen. 2014, 5, 1000165. [Google Scholar] [CrossRef]
- Pradhan, J.; Noakes, P.G.; Bellingham, M.C. The Role of Altered BDNF/TrkB Signaling in Amyotrophic Lateral Sclerosis. Front. Cell. Neurosci. 2019, 13, 368. [Google Scholar] [CrossRef]
- Wang, C.S.; Kavalali, E.T.; Monteggia, L.M. BDNF Signaling in Context: From Synaptic Regulation to Psychiatric Disorders. Cell 2022, 185, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Guillot, C.R.; Kelly, M.E.; Phillips, N.B.; Su, M.-Y.; Douglas, M.E.; Poe, D.J.; Berman, M.E.; Liang, T. BDNF and Stress/Mood-Related Interactions on Emotional Disorder Symptoms, Executive Functioning, and Deliberate Self-Harm. J. Psychiatr. Res. 2023, 163, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Doyle, K.P.; Cekanaviciute, E.; Mamer, L.E.; Buckwalter, M.S. TGFβ Signaling in the Brain Increases with Aging and Signals to Astrocytes and Innate Immune Cells in the Weeks after Stroke. J. Neuroinflamm. 2010, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Meyers, E.A.; Kessler, J.A. TGF-β Family Signaling in Neural and Neuronal Differentiation, Development, and Function. Cold Spring Harb. Perspect. Biol. 2017, 9, a022244. [Google Scholar] [CrossRef]
- Malekan, M.; Nezamabadi, S.S.; Samami, E.; Mohebalizadeh, M.; Saghazadeh, A.; Rezaei, N. BDNF and Its Signaling in Cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 2621–2636. [Google Scholar] [CrossRef]
- Shi, X.; Yang, J.; Deng, S.; Xu, H.; Wu, D.; Zeng, Q.; Wang, S.; Hu, T.; Wu, F.; Zhou, H. TGF-β Signaling in the Tumor Metabolic Microenvironment and Targeted Therapies. J. Hematol. Oncol. 2022, 15, 135. [Google Scholar] [CrossRef]
- Aghajanirefah, A.; Nguyen, L.N.; Ohadi, M. BEND3 Is Involved in the Human-Specific Repression of Calreticulin: Implication for the Evolution of Higher Brain Functions in Human. Gene 2016, 576, 577–580. [Google Scholar] [CrossRef]
- Reid, K.M.; Kitchener, E.J.A.; Butler, C.A.; Cockram, T.O.J.; Brown, G.C. Brain Cells Release Calreticulin That Attracts and Activates Microglia, and Inhibits Amyloid Beta Aggregation and Neurotoxicity. Front. Immunol. 2022, 13, 859686. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Le, T.M.; Hattori, T.; Takarada-Iemata, M.; Ishii, H.; Roboon, J.; Tamatani, T.; Kannon, T.; Hosomichi, K.; Tajima, A.; et al. The ATF6β-Calreticulin Axis Promotes Neuronal Survival under Endoplasmic Reticulum Stress and Excitotoxicity. Sci. Rep. 2021, 11, 13086. [Google Scholar] [CrossRef]
- Dedhar, S.; Rennie, P.S.; Shago, M.; Hagesteijn, C.Y.; Yang, H.; Filmus, J.; Hawley, R.G.; Bruchovsky, N.; Cheng, H.; Matusik, R.J. Inhibition of Nuclear Hormone Receptor Activity by Calreticulin. Nature 1994, 367, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.H.; Veenstra, G.J.C. The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development. Epigenomes 2022, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.B.; Eichele, G.; Zhang, P.; Rawls, A.; Sands, A.T.; Bradley, A.; Olson, E.N.; Harper, J.W.; Elledge, S.J. P53-Independent Expression of P21Cip1 in Muscle and Other Terminally Differentiating Cells. Science 1995, 267, 1024–1027. [Google Scholar] [CrossRef]
- Savci-Heijink, C.D.; Halfwerk, H.; Koster, J.; Horlings, H.M.; van de Vijver, M.J. A Specific Gene Expression Signature for Visceral Organ Metastasis in Breast Cancer. BMC Cancer 2019, 19, 333. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Das, A. Peripheral Blood Mononuclear Cell Derived Biomarker Detection Using EXplainable Artificial Intelligence (XAI) Provides Better Diagnosis of Breast Cancer. Comput. Biol. Chem. 2023, 104, 107867. [Google Scholar] [CrossRef]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like Protein Activation by E1 Enzymes: The Apex for Downstream Signalling Pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef]
- Hyytinen, E.-R.; Saadut, R.; Chen, C.; Paull, L.; Koivisto, P.A.; Vessella, R.L.; Frierson, H.F.; Dong, J.-T. Defining the Region(s) of Deletion at 6q16-Q22 in Human Prostate Cancer. Genes Chromosom. Cancer 2002, 34, 306–312. [Google Scholar] [CrossRef]
- Morelli, C.; Karayianni, E.; Magnanini, C.; Mungall, A.J.; Thorland, E.; Negrini, M.; Smith, D.I.; Barbanti-Brodano, G. Cloning and Characterization of the Common Fragile Site FRA6F Harboring a Replicative Senescence Gene and Frequently Deleted in Human Tumors. Oncogene 2002, 21, 7266–7276. [Google Scholar] [CrossRef]
- Orphanos, V.; McGown, G.; Hey, Y.; Boyle, J.M.; Santibanez-Koref, M. Proximal 6q, a Region Showing Allele Loss in Primary Breast Cancer. Br. J. Cancer 1995, 71, 290–293. [Google Scholar] [CrossRef]
- Orphanos, V.; McGown, G.; Hey, Y.; Thorncroft, M.; Santibanez-Koref, M.; Russell, S.E.; Hickey, I.; Atkinson, R.J.; Boyle, J.M. Allelic Imbalance of Chromosome 6q in Ovarian Tumours. Br. J. Cancer 1995, 71, 666–669. [Google Scholar] [CrossRef]
- Zhang, Y.; Matthiesen, P.; Harder, S.; Siebert, R.; Castoldi, G.; Calasanz, M.J.; Wong, K.F.; Rosenwald, A.; Ott, G.; Atkin, N.B.; et al. A 3-CM Commonly Deleted Region in 6q21 in Leukemias and Lymphomas Delineated by Fluorescence in Situ Hybridization. Genes Chromosom. Cancer 2000, 27, 52–58. [Google Scholar] [CrossRef]
- Zhang, L.; Anglesio, M.S.; O’Sullivan, M.; Zhang, F.; Yang, G.; Sarao, R.; Mai, P.N.; Cronin, S.; Hara, H.; Melnyk, N.; et al. The E3 Ligase HACE1 Is a Critical Chromosome 6q21 Tumor Suppressor Involved in Multiple Cancers. Nat. Med. 2007, 13, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Roberts, C.W.M. Targeting EZH2 in Cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, B.V.S.K.; Nepal, S.; Varambally, S. Genomic and Epigenomic Alterations in Cancer. Am. J. Pathol. 2016, 186, 1724–1735. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naiyer, S.; Dwivedi, L.; Singh, N.; Phulera, S.; Mohan, V.; Kamran, M. Role of Transcription Factor BEND3 and Its Potential Effect on Cancer Progression. Cancers 2023, 15, 3685. https://doi.org/10.3390/cancers15143685
Naiyer S, Dwivedi L, Singh N, Phulera S, Mohan V, Kamran M. Role of Transcription Factor BEND3 and Its Potential Effect on Cancer Progression. Cancers. 2023; 15(14):3685. https://doi.org/10.3390/cancers15143685
Chicago/Turabian StyleNaiyer, Sarah, Lalita Dwivedi, Nishant Singh, Swastik Phulera, Vijay Mohan, and Mohammad Kamran. 2023. "Role of Transcription Factor BEND3 and Its Potential Effect on Cancer Progression" Cancers 15, no. 14: 3685. https://doi.org/10.3390/cancers15143685
APA StyleNaiyer, S., Dwivedi, L., Singh, N., Phulera, S., Mohan, V., & Kamran, M. (2023). Role of Transcription Factor BEND3 and Its Potential Effect on Cancer Progression. Cancers, 15(14), 3685. https://doi.org/10.3390/cancers15143685




