Decoding Oncofusions: Unveiling Mechanisms, Clinical Impact, and Prospects for Personalized Cancer Therapies
Abstract
Simple Summary
Abstract
1. Introduction
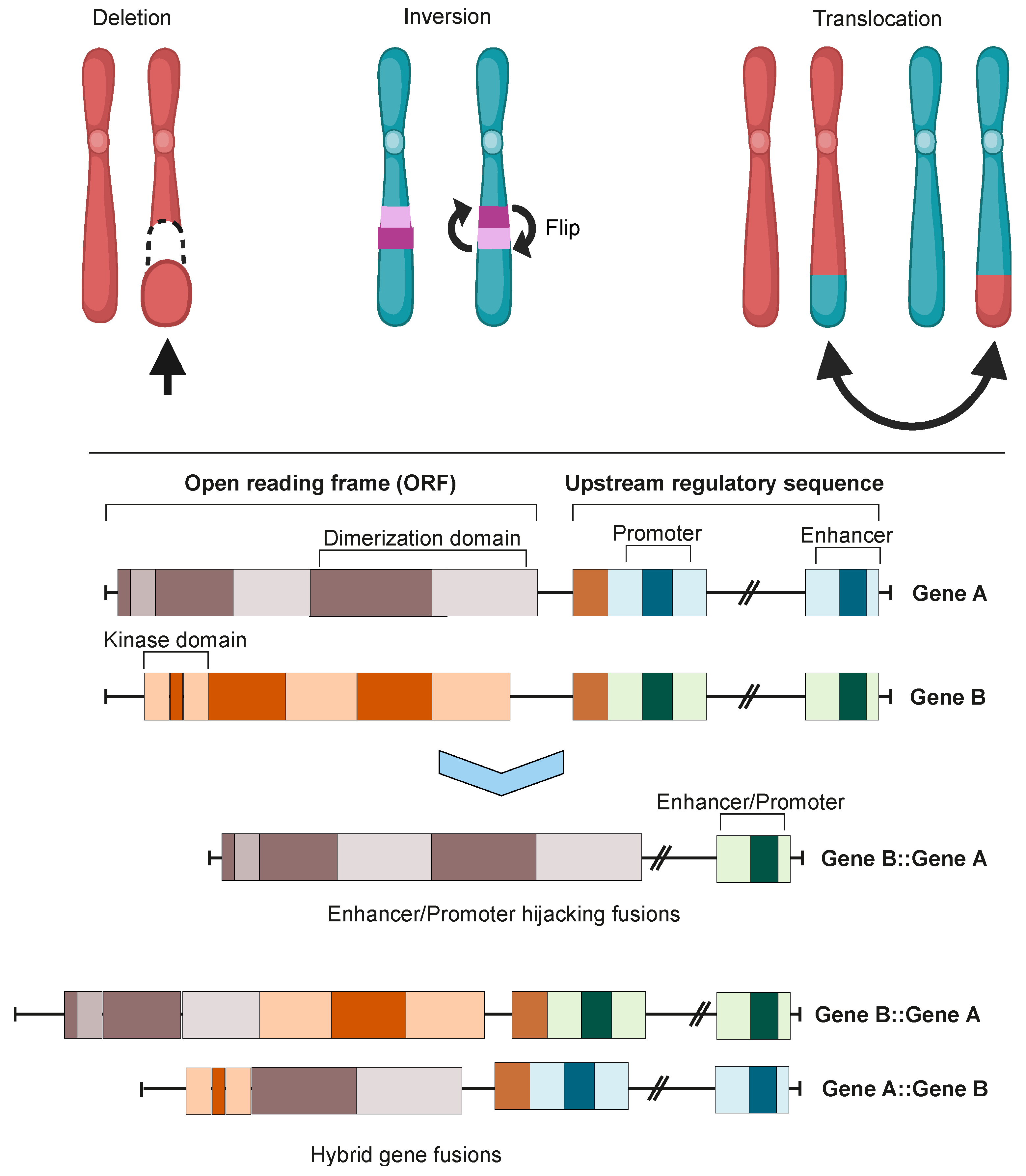
2. Oncofusion Formation
3. Oncofusions’ Role in Cancer Development
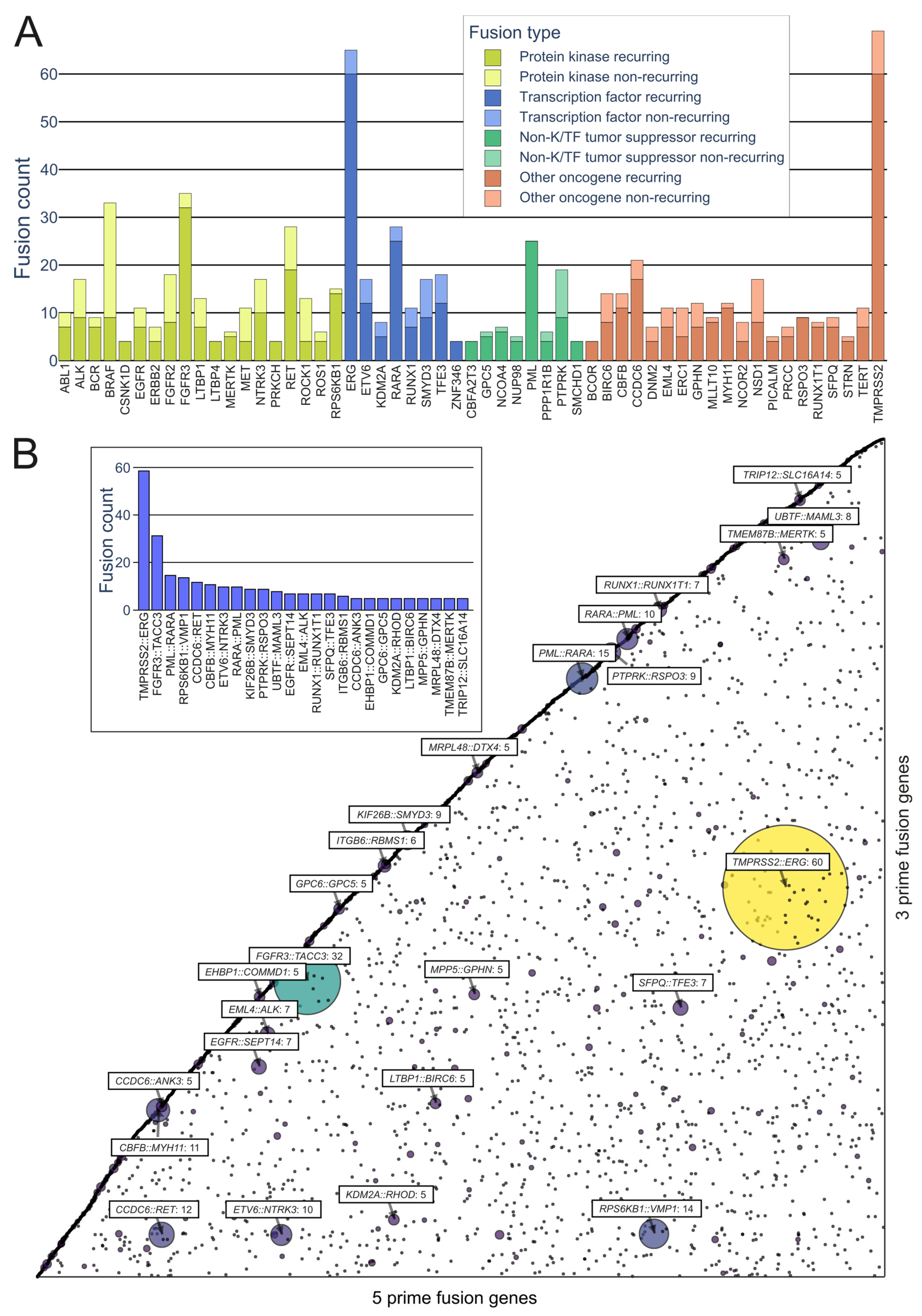
Prominent Oncofusions in Cancer
4. Fusion Identification
4.1. Fusion Callers
4.2. Protein-Level Analysis of Oncofusions
5. Functional Validation of Gene Fusions
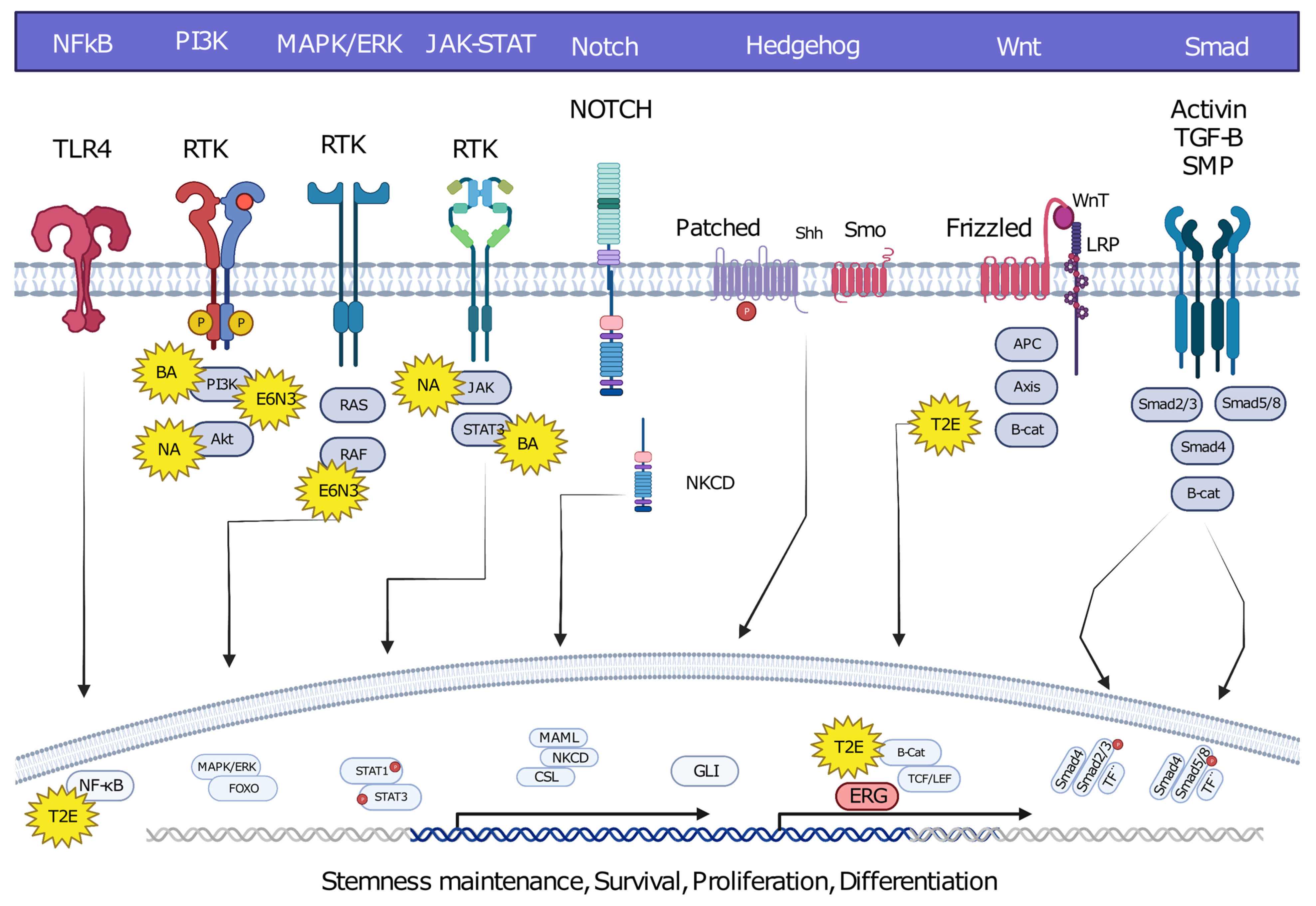
6. Oncofusion-Specific Molecular Mechanisms
7. The Importance of Oncofusion-Specific Molecular Mechanisms
7.1. BCR::ABL
7.2. ETV6::NTRK3
7.3. NPM::ALK
7.4. TMPRSS2::ERG
8. Gene Fusions as Neoantigens
8.1. Neoantigen Immunogenicity
8.2. Treatment Potential for T-Cell Therapies
9. Clinical Implications
10. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Latysheva, N.S.; Babu, M.M. Discovering and Understanding Oncogenic Gene Fusions through Data Intensive Computational Approaches. Nucleic Acids Res. 2016, 44, 4487–4503. [Google Scholar] [CrossRef]
- Nowell, P.; Hungerford, D. Chromosome Studies on Normal and Leukemic Human Leukocytes. J. Natl. Cancer Inst. 1960, 25, 85–109. [Google Scholar]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, T.; Yamaguchi, K.; Urakami, K.; Shimoda, Y.; Ohnami, S.; Ohshima, K.; Tanabe, T.; Naruoka, A.; Kamada, F.; Serizawa, M.; et al. Japanese Version of The Cancer Genome Atlas, JCGA, Established Using Fresh Frozen Tumors Obtained from 5143 Cancer Patients. Cancer Sci. 2020, 111, 687–699. [Google Scholar] [CrossRef]
- Salokas, K.; Weldatsadik, R.G.; Varjosalo, M. Human Transcription Factor and Protein Kinase Gene Fusions in Human Cancer. Sci. Rep. 2020, 10, 14169. [Google Scholar] [CrossRef] [PubMed]
- Erikson, J.; Nishikura, K.; ar-Rushdi, A.; Finan, J.; Emanuel, B.; Lenoir, G.; Nowell, P.C.; Croce, C.M. Translocation of an Immunoglobulin Kappa Locus to a Region 3′ of an Unrearranged c-Myc Oncogene Enhances c-Myc Transcription. Proc. Natl. Acad. Sci. USA 1983, 80, 7581–7585. [Google Scholar] [CrossRef]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human C-Myc Onc Gene Is Located on the Region of Chromosome 8 That Is Translocated in Burkitt Lymphoma Cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef]
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the C-Myc Gene into the Immunoglobulin Heavy Chain Locus in Human Burkitt Lymphoma and Murine Plasmacytoma Cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef]
- Heisterkamp, N.; Stam, K.; Groffen, J.; de Klein, A.; Grosveld, G. Structural Organization of the Bcr Gene and Its Role in the Ph’ Translocation. Nature 1985, 315, 758–761. [Google Scholar] [CrossRef]
- Shtivelman, E.; Lifshitz, B.; Gale, R.P.; Canaani, E. Fused Transcript of Abl and Bcr Genes in Chronic Myelogenous Leukaemia. Nature 1985, 315, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Ben-Neriah, Y.; Daley, G.Q.; Mes-Masson, A.M.; Witte, O.N.; Baltimore, D. The Chronic Myelogenous Leukemia-Specific P210 Protein Is the Product of the Bcr/Abl Hybrid Gene. Science 1986, 233, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Fainstein, E.; Marcelle, C.; Rosner, A.; Canaani, E.; Gale, R.P.; Dreazen, O.; Smith, S.D.; Croce, C.M. A New Fused Transcript in Philadelphia Chromosome Positive Acute Lymphocytic Leukaemia. Nature 1987, 330, 386–388. [Google Scholar] [CrossRef]
- Reckel, S.; Hamelin, R.; Georgeon, S.; Armand, F.; Jolliet, Q.; Chiappe, D.; Moniatte, M.; Hantschel, O. Differential Signaling Networks of Bcr-Abl P210 and P190 Kinases in Leukemia Cells Defined by Functional Proteomics. Leukemia 2017, 31, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Cutler, J.A.; Tahir, R.; Sreenivasamurthy, S.K.; Mitchell, C.; Renuse, S.; Nirujogi, R.S.; Patil, A.H.; Heydarian, M.; Wong, X.; Wu, X.; et al. Differential Signaling through P190 and P210 BCR-ABL Fusion Proteins Revealed by Interactome and Phosphoproteome Analysis. Leukemia 2017, 31, 1513–1524. [Google Scholar] [CrossRef]
- Efremov, G.D. Hemoglobins Lepore and Anti-Lepore. Hemoglobin 1978, 2, 197–233. [Google Scholar] [CrossRef] [PubMed]
- Mertens, F.; Johansson, B.; Fioretos, T.; Mitelman, F. The Emerging Complexity of Gene Fusions in Cancer. Nat. Rev. Cancer 2015, 15, 371–381. [Google Scholar] [CrossRef]
- Kumar-Sinha, C.; Kalyana-Sundaram, S.; Chinnaiyan, A.M. Landscape of Gene Fusions in Epithelial Cancers: Seq and Ye Shall Find. Genome Med. 2015, 7, 129. [Google Scholar] [CrossRef]
- Johansson, B.; Mertens, F.; Schyman, T.; Björk, J.; Mandahl, N.; Mitelman, F. Most Gene Fusions in Cancer Are Stochastic Events. Genes Chromosomes Cancer 2019, 58, 607–611. [Google Scholar] [CrossRef]
- Zhang, J.; Mardis, E.R.; Maher, C.A. INTEGRATE-Neo: A Pipeline for Personalized Gene Fusion Neoantigen Discovery. Bioinformatics 2017, 33, 555–557. [Google Scholar] [CrossRef]
- Yi-Mi, W.; Cieslik, M.; Lonigro, R.J.; Pankaj, V.; Reimers, M.A.; Xuhong, C.; Yu, N.; Lisha, W.; Kunju, L.P.; de Sarkar, N.; et al. Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 2018, 173, 1770–1782.e14. [Google Scholar] [CrossRef]
- So, A.; Le Guen, T.; Lopez, B.S.; Guirouilh-Barbat, J. Genomic Rearrangements Induced by Unscheduled DNA Double Strand Breaks in Somatic Mammalian Cells. FEBS J. 2017, 284, 2324–2344. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.W.; Yang, L.; Park, H.-Y.; Lee, C.-W.; Cha, H.; Shin, H.-T.; Noh, K.-W.; Choi, Y.-L.; Park, W.-Y.; Park, P.J. Dysregulation of Cancer Genes by Recurrent Intergenic Fusions. Genome Biol. 2020, 21, 166. [Google Scholar] [CrossRef] [PubMed]
- Namba, S.; Ueno, T.; Kojima, S.; Kobayashi, K.; Kawase, K.; Tanaka, Y.; Inoue, S.; Kishigami, F.; Kawashima, S.; Maeda, N.; et al. Transcript-Targeted Analysis Reveals Isoform Alterations and Double-Hop Fusions in Breast Cancer. Commun. Biol. 2021, 4, 1320. [Google Scholar] [CrossRef]
- Ferrad, M.; Ghazzaui, N.; Issaoui, H.; Cook-Moreau, J.; Denizot, Y. Mouse Models of C-Myc Deregulation Driven by IgH Locus Enhancers as Models of B-Cell Lymphomagenesis. Front. Immunol. 2020, 11, 1564. [Google Scholar] [CrossRef]
- Panagopoulos, I.; Heim, S. Neoplasia-Associated Chromosome Translocations Resulting in Gene Truncation. Cancer Genom. Proteom. 2022, 19, 647–672. [Google Scholar] [CrossRef]
- Wang, T.; Wei, L.; Lu, Q.; Shao, Y.; You, S.; Yin, J.C.; Wang, S.; Shao, Y.; Chen, Z.; Wang, Z. Landscape of Potentially Targetable Receptor Tyrosine Kinase Fusions in Diverse Cancers by DNA-Based Profiling. NPJ Precis. Oncol. 2022, 6, 84. [Google Scholar] [CrossRef]
- Gagos, S.; Irminger-Finger, I. Chromosome Instability in Neoplasia: Chaotic Roots to Continuous Growth. Int. J. Biochem. Cell Biol. 2005, 37, 1014–1033. [Google Scholar] [CrossRef]
- Takeuchi, K. Discovery Stories of RET Fusions in Lung Cancer: A Mini-Review. Front. Physiol. 2019, 10, 216. [Google Scholar] [CrossRef]
- Long, M.; Langley, C.H. Natural Selection and the Origin of Jingwei, a Chimeric Processed Functional Gene in Drosophila. Science 1993, 260, 91–95. [Google Scholar] [CrossRef]
- Mitelman, F.; Johansson, B.; Mertens, F. The Impact of Translocations and Gene Fusions on Cancer Causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lee, J.-K.; Choi, Y.-L.; Kwon, M.; Park, P.J. Mechanisms and Consequences of Cancer Genome Instability: Lessons from Genome Sequencing Studies. Annu. Rev. Pathol. 2016, 11, 283–312. [Google Scholar] [CrossRef] [PubMed]
- Lobato, M.N.; Metzler, M.; Drynan, L.; Forster, A.; Pannell, R.; Rabbitts, T.H. Modeling Chromosomal Translocations Using Conditional Alleles to Recapitulate Initiating Events in Human Leukemias. J. Natl. Cancer Inst. Monogr. 2008, 2008, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Mertens, F.; Antonescu, C.R.; Mitelman, F. Gene Fusions in Soft Tissue Tumors: Recurrent and Overlapping Pathogenetic Themes. Genes Chromosomes Cancer 2016, 55, 291–310. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liang, W.-W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.-W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018, 23, 227–238.e3. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Z.; Jeselsohn, R. ESR1 Fusions and Therapeutic Resistance in Metastatic Breast Cancer. Front. Oncol. 2022, 12, 1037531. [Google Scholar] [CrossRef]
- Yu, L.; Davis, I.J.; Liu, P. Regulation of EWSR1-FLI1 Function by Post-Transcriptional and Post-Translational Modifications. Cancers 2023, 15, 382. [Google Scholar] [CrossRef]
- Apfelbaum, A.A.; Wrenn, E.D.; Lawlor, E.R. The Importance of Fusion Protein Activity in Ewing Sarcoma and the Cell Intrinsic and Extrinsic Factors That Regulate It: A Review. Front. Oncol. 2022, 12, 1044707. [Google Scholar] [CrossRef]
- Bowling, G.C.; Rands, M.G.; Dobi, A.; Eldhose, B. Emerging Developments in ETS-Positive Prostate Cancer Therapy. Mol. Cancer Ther. 2023, 22, 168–178. [Google Scholar] [CrossRef]
- Shen, Z.; Qiu, B.; Li, L.; Yang, B.; Li, G. Targeted Therapy of RET Fusion-Positive Non-Small Cell Lung Cancer. Front. Oncol. 2022, 12, 1033484. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.; Rabushko, E.; Rozenberg, J.M.; Mohammad, T.; Seryakov, A.; Sekacheva, M.; Buzdin, A. Clinically Relevant Fusion Oncogenes: Detection and Practical Implications. Ther. Adv. Med. Oncol. 2022, 14, 17588359221144108. [Google Scholar] [CrossRef]
- Hagstrom, M.; Fumero-Velázquez, M.; Dhillon, S.; Olivares, S.; Gerami, P. An Update on Genomic Aberrations in Spitz Naevi and Tumours. Pathology 2023, 55, 196–205. [Google Scholar] [CrossRef]
- Chu, Y.-H. This Is Your Thyroid on Drugs: Targetable Mutations and Fusions in Thyroid Carcinoma. Surg. Pathol. Clin. 2023, 16, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Peiris, M.N.; Meyer, A.N.; Nelson, K.N.; Donoghue, D.J. Oncogenic Driver FGFR3-TACC3 Requires Five Coiled-Coil Heptads for Activation and Disulfide Bond Formation for Stability. Oncotarget 2023, 14, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Kastenhuber, E.R.; Lalazar, G.; Houlihan, S.L.; Tschaharganeh, D.F.; Baslan, T.; Chen, C.-C.; Requena, D.; Tian, S.; Bosbach, B.; Wilkinson, J.E.; et al. DNAJB1-PRKACA Fusion Kinase Interacts with β-Catenin and the Liver Regenerative Response to Drive Fibrolamellar Hepatocellular Carcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 13076–13084. [Google Scholar] [CrossRef]
- Matissek, K.J.; Onozato, M.L.; Sun, S.; Zheng, Z.; Schultz, A.; Lee, J.; Patel, K.; Jerevall, P.-L.; Saladi, S.V.; Macleay, A.; et al. Expressed Gene Fusions as Frequent Drivers of Poor Outcomes in Hormone Receptor-Positive Breast Cancer. Cancer Discov. 2018, 8, 336–353. [Google Scholar] [CrossRef]
- Lei, J.T.; Shao, J.; Zhang, J.; Iglesia, M.; Chan, D.W.; Cao, J.; Anurag, M.; Singh, P.; He, X.; Kosaka, Y.; et al. Functional Annotation of ESR1 Gene Fusions in Estrogen Receptor-Positive Breast Cancer. Cell Rep. 2018, 24, 1434–1444.e7. [Google Scholar] [CrossRef]
- Kim, J.; Kim, S.; Ko, S.; In, Y.; Moon, H.-G.; Ahn, S.K.; Kim, M.K.; Lee, M.; Hwang, J.-H.; Ju, Y.S.; et al. Recurrent Fusion Transcripts Detected by Whole-Transcriptome Sequencing of 120 Primary Breast Cancer Samples. Genes Chromosomes Cancer 2015, 54, 681–691. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Finger, L.R.; Yunis, J.; Nowell, P.C.; Croce, C.M. Cloning of the Chromosome Breakpoint of Neoplastic B Cells with the t(14;18) Chromosome Translocation. Science 1984, 226, 1097–1099. [Google Scholar] [CrossRef]
- Vaux, D.L.; Cory, S.; Adams, J.M. Bcl-2 Gene Promotes Haemopoietic Cell Survival and Cooperates with c-Myc to Immortalize Pre-B Cells. Nature 1988, 335, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-P.; Liu, P.; Nelson, J.; Hamilton, R.L.; Bhargava, R.; Michalopoulos, G.; Chen, Q.; Zhang, J.; Ma, D.; Pennathur, A.; et al. Identification of Recurrent Fusion Genes across Multiple Cancer Types. Sci. Rep. 2019, 9, 1074. [Google Scholar] [CrossRef]
- Yu, Y.P.; Ding, Y.; Chen, Z.; Liu, S.; Michalopoulos, A.; Chen, R.; Gulzar, Z.G.; Yang, B.; Cieply, K.M.; Luvison, A.; et al. Novel Fusion Transcripts Associate with Progressive Prostate Cancer. Am. J. Pathol. 2014, 184, 2840–2849. [Google Scholar] [CrossRef]
- Yu, J.; Yu, J.; Mani, R.-S.; Cao, Q.; Brenner, C.J.; Cao, X.; Wang, G.X.; Wu, L.; Li, J.; Hu, M.; et al. An Integrated Network of Androgen Receptor, Polycomb, and TMPRSS2-ERG Gene Fusions in Prostate Cancer Progression. Cancer Cell 2010, 17, 443–454. [Google Scholar] [CrossRef]
- Andersson, A.K.; Ma, J.; Wang, J.; Chen, X.; Gedman, A.L.; Dang, J.; Nakitandwe, J.; Holmfeldt, L.; Parker, M.; Easton, J.; et al. The Landscape of Somatic Mutations in Infant MLL-Rearranged Acute Lymphoblastic Leukemias. Nat. Genet. 2015, 47, 330–337. [Google Scholar] [CrossRef]
- Milne, T.A. Mouse Models of MLL Leukemia: Recapitulating the Human Disease. Blood 2017, 129, 2217–2223. [Google Scholar] [CrossRef] [PubMed]
- Babiceanu, M.; Qin, F.; Xie, Z.; Jia, Y.; Lopez, K.; Janus, N.; Facemire, L.; Kumar, S.; Pang, Y.; Qi, Y.; et al. Recurrent Chimeric Fusion RNAs in Non-Cancer Tissues and Cells. Nucleic Acids Res. 2016, 44, 2859–2872. [Google Scholar] [CrossRef]
- Forsberg, L.A.; Rasi, C.; Razzaghian, H.R.; Pakalapati, G.; Waite, L.; Thilbeault, K.S.; Ronowicz, A.; Wineinger, N.E.; Tiwari, H.K.; Boomsma, D.; et al. Age-Related Somatic Structural Changes in the Nuclear Genome of Human Blood Cells. Am. J. Hum. Genet. 2012, 90, 217–228. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Tsukamoto, T.; Chinen, Y.; Shimura, Y.; Sasaki, N.; Nagoshi, H.; Sato, R.; Adachi, H.; Nakano, M.; Horiike, S.; et al. Detection of Novel and Recurrent Conjoined Genes in Non-Hodgkin B-Cell Lymphoma. J. Clin. Exp. Hematop. JCEH 2021, 61, 71–77. [Google Scholar] [CrossRef]
- Mukherjee, S.; Frenkel-Morgenstern, M. Evolutionary Impact of Chimeric RNAs on Generating Phenotypic Plasticity in Human Cells. Trends Genet. TIG 2022, 38, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. RNA Sequencing (RNA-Seq) and Its Application in Ovarian Cancer. Gynecol. Oncol. 2019, 152, 194–201. [Google Scholar] [CrossRef]
- Abeshouse, A.; Ahn, J.; Akbani, R.; Ally, A.; Amin, S.; Andry, C.D.; Annala, M.; Aprikian, A.; Armenia, J.; Arora, A.; et al. Cancer Genome Atlas Research Network The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.; Zhou, X. FusionGDB: Fusion Gene Annotation DataBase. Nucleic Acids Res. 2019, 47, D994–D1004. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Palsgrove, D.N.; Anders, R.A.; Smith, S.C.; Saoud, C.; Kwon, R.; Voltaggio, L.; Assarzadegan, N.; Oshima, K.; Rooper, L.; et al. A Novel NIPBL-NACC1 Gene Fusion Is Characteristic of the Cholangioblastic Variant of Intrahepatic Cholangiocarcinoma. Am. J. Surg. Pathol. 2021, 45, 1550–1560. [Google Scholar] [CrossRef]
- Haas, B.J.; Dobin, A.; Li, B.; Stransky, N.; Pochet, N.; Regev, A. Accuracy Assessment of Fusion Transcript Detection via Read-Mapping and de Novo Fusion Transcript Assembly-Based Methods. Genome Biol. 2019, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tsai, W.-H.; Ding, Y.; Chen, R.; Fang, Z.; Huo, Z.; Kim, S.; Ma, T.; Chang, T.-Y.; Priedigkeit, N.M.; et al. Comprehensive Evaluation of Fusion Transcript Detection Algorithms and a Meta-Caller to Combine Top Performing Methods in Paired-End RNA-Seq Data. Nucleic Acids Res. 2016, 44, e47. [Google Scholar] [CrossRef] [PubMed]
- Dorney, R.; Dhungel, B.P.; Rasko, J.E.J.; Hebbard, L.; Schmitz, U. Recent Advances in Cancer Fusion Transcript Detection. Brief. Bioinform. 2022, 24, bbac519. [Google Scholar] [CrossRef]
- Nattestad, M.; Goodwin, S.; Ng, K.; Baslan, T.; Sedlazeck, F.J.; Rescheneder, P.; Garvin, T.; Fang, H.; Gurtowski, J.; Hutton, E.; et al. Complex Rearrangements and Oncogene Amplifications Revealed by Long-Read DNA and RNA Sequencing of a Breast Cancer Cell Line. Genome Res. 2018, 28, 1126–1135. [Google Scholar] [CrossRef]
- Apostolides, M.; Jiang, Y.; Husić, M.; Siddaway, R.; Hawkins, C.; Turinsky, A.L.; Brudno, M.; Ramani, A.K. MetaFusion: A High-Confidence Metacaller for Filtering and Prioritizing RNA-Seq Gene Fusion Candidates. Bioinformatics 2021, 37, 3144–3151. [Google Scholar] [CrossRef]
- Thomas, B.B.; Mou, Y.; Keeler, L.; Magnan, C.; Funari, V.; Weiss, L.; Brown, S.; Agersborg, S. A Highly Sensitive and Specific Gene Fusion Algorithm Based on Multiple Fusion Callers and an Ensemble Machine Learning Approach. Blood 2020, 136, 12–13. [Google Scholar] [CrossRef]
- Hedges, D.J. RNA-Seq Fusion Detection in Clinical Oncology. Adv. Exp. Med. Biol. 2022, 1361, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Kuksin, M.; Morel, D.; Aglave, M.; Danlos, F.-X.; Marabelle, A.; Zinovyev, A.; Gautheret, D.; Verlingue, L. Applications of Single-Cell and Bulk RNA Sequencing in Onco-Immunology. Eur. J. Cancer 2021, 149, 193–210. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Bahleda, R.; Hierro, C.; Sanson, M.; Bridgewater, J.; Arkenau, H.-T.; Tran, B.; Kelley, R.K.; Park, J.O.; Javle, M.; et al. Futibatinib, an Irreversible FGFR1-4 Inhibitor, in Patients with Advanced Solid Tumors Harboring FGF/FGFR Aberrations: A Phase I Dose-Expansion Study. Cancer Discov. 2022, 12, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.K.; Yates, M.E.; Yang, S.; Oesterreich, S.; Lee, A.V.; Wang, X.-S. Fusion-Associated Carcinomas of the Breast: Diagnostic, Prognostic, and Therapeutic Significance. Genes Chromosomes Cancer 2022, 61, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Guo, L.; Zhao, R.; Teng, H.; Wang, Y.; Xiong, L.; Han, Y. Identification and Validation of Noncanonical RET Fusions in Non-Small-Cell Lung Cancer through DNA and RNA Sequencing. J. Mol. Diagn. JMD 2022, 24, 374–385. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Li, W.; Chen, L.; Ying, J. Intergenic Breakpoints Identified by DNA Sequencing Confound Targetable Kinase Fusion Detection in NSCLC. J. Thorac. Oncol. 2020, 15, 1223–1231. [Google Scholar] [CrossRef]
- Rathe, S.K.; Popescu, F.E.; Johnson, J.E.; Watson, A.L.; Marko, T.A.; Moriarity, B.S.; Ohlfest, J.R.; Largaespada, D.A. Identification of Candidate Neoantigens Produced by Fusion Transcripts in Human Osteosarcomas. Sci. Rep. 2019, 9, 358. [Google Scholar] [CrossRef]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; de Jong, P.; Rouleau, G. Gene Fusion with an ETS DNA-Binding Domain Caused by Chromosome Translocation in Human Tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef]
- Sand, L.G.L.; Szuhai, K.; Hogendoorn, P.C.W. Sequencing Overview of Ewing Sarcoma: A Journey across Genomic, Epigenomic and Transcriptomic Landscapes. Int. J. Mol. Sci. 2015, 16, 16176–16215. [Google Scholar] [CrossRef]
- Robinson, D.R.; Kalyana-Sundaram, S.; Wu, Y.-M.; Shankar, S.; Cao, X.; Ateeq, B.; Asangani, I.A.; Iyer, M.; Maher, C.A.; Grasso, C.S.; et al. Functionally Recurrent Rearrangements of the MAST Kinase and Notch Gene Families in Breast Cancer. Nat. Med. 2011, 17, 1646–1651. [Google Scholar] [CrossRef] [PubMed]
- Li, A.Y.; McCusker, M.G.; Russo, A.; Scilla, K.A.; Gittens, A.; Arensmeyer, K.; Mehra, R.; Adamo, V.; Rolfo, C. RET Fusions in Solid Tumors. Cancer Treat. Rev. 2019, 81, 101911. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Moccia, M.; Federico, G.; Carlomagno, F. RET Gene Fusions in Malignancies of the Thyroid and Other Tissues. Genes 2020, 11, 424. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, Z.; Sun, Y.; Wang, S.; Wu, M.; Ou, Q.; Xu, Y.; Chen, Z.; Shao, Y.; Liu, H.; et al. RET Fusions as Primary Oncogenic Drivers and Secondary Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in Patients with Non-Small-Cell Lung Cancer. J. Transl. Med. 2022, 20, 390. [Google Scholar] [CrossRef]
- Yau, D.T.-W.; Lacambra, M.D.; Chow, C.; To, K.-F. The Novel Finding of an FGFR1::TACC1 Fusion in an Undifferentiated Spindle Cell Sarcoma of Soft Tissue with Aggressive Clinical Course. Genes Chromosomes Cancer 2022, 61, 206–211. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Q.; Wang, Y.; Dai, X.; Chen, P.; Chen, A.; Zhou, S.; Dai, C.; Zhao, S.; Xiao, S.; et al. Case Report: A Novel LHFPL3::NTRK2 Fusion in Dysembryoplastic Neuroepithelial Tumor. Front. Oncol. 2022, 12, 1064817. [Google Scholar] [CrossRef]
- Deland, L.; Keane, S.; Bontell, T.O.; Fagman, H.; Sjögren, H.; Lind, A.E.; Carén, H.; Tisell, M.; Nilsson, J.A.; Ejeskär, K.; et al. Novel TPR::ROS1 Fusion Gene Activates MAPK, PI3K and JAK/STAT Signaling in an Infant-Type Pediatric Glioma. Cancer Genom. Proteom. 2022, 19, 711–726. [Google Scholar] [CrossRef]
- Hiwatari, M.; Seki, M.; Matsuno, R.; Yoshida, K.; Nagasawa, T.; Sato-Otsubo, A.; Yamamoto, S.; Kato, M.; Watanabe, K.; Sekiguchi, M.; et al. Novel TENM3-ALK Fusion Is an Alternate Mechanism for ALK Activation in Neuroblastoma. Oncogene 2022, 41, 2789–2797. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK Fusion-Positive Cancers and TRK Inhibitor Therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Amatu, A.; Sartore-Bianchi, A.; Bencardino, K.; Pizzutilo, E.G.; Tosi, F.; Siena, S. Tropomyosin Receptor Kinase (TRK) Biology and the Role of NTRK Gene Fusions in Cancer. Ann. Oncol. 2019, 30, viii5–viii15. [Google Scholar] [CrossRef]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK Gene Fusions as Novel Targets of Cancer Therapy across Multiple Tumour Types. ESMO Open 2016, 1, e000023. [Google Scholar] [CrossRef]
- Ardini, E.; Bosotti, R.; Borgia, A.L.; De Ponti, C.; Somaschini, A.; Cammarota, R.; Amboldi, N.; Raddrizzani, L.; Milani, A.; Magnaghi, P.; et al. The TPM3-NTRK1 Rearrangement Is a Recurring Event in Colorectal Carcinoma and Is Associated with Tumor Sensitivity to TRKA Kinase Inhibition. Mol. Oncol. 2014, 8, 1495–1507. [Google Scholar] [CrossRef] [PubMed]
- Vaishnavi, A.; Capelletti, M.; Le, A.T.; Kako, S.; Butaney, M.; Ercan, D.; Mahale, S.; Davies, K.D.; Aisner, D.L.; Pilling, A.B.; et al. Oncogenic and Drug-Sensitive NTRK1 Rearrangements in Lung Cancer. Nat. Med. 2013, 19, 1469–1472. [Google Scholar] [CrossRef]
- Dermawan, J.K.; Vanderbilt, C.M.; Chang, J.C.; Untch, B.R.; Singer, S.; Chi, P.; Tap, W.D.; Antonescu, C.R. FGFR2::TACC2 Fusion as a Novel KIT-Independent Mechanism of Targeted Therapy Failure in a Multidrug-Resistant Gastrointestinal Stromal Tumor. Genes Chromosomes Cancer 2022, 61, 412–419. [Google Scholar] [CrossRef]
- Wu, Y.-M.; Su, F.; Kalyana-Sundaram, S.; Khazanov, N.; Ateeq, B.; Cao, X.; Lonigro, R.J.; Vats, P.; Wang, R.; Lin, S.-F.; et al. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discov. 2013, 3, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.-M.; Arnau, G.M.; Ryland, G.L.; Huang, S.; Lira, M.E.; Emmanuel, Y.; Perez, O.D.; Irwin, D.; Fellowes, A.P.; Wong, S.Q.; et al. Multiplexed Transcriptome Analysis to Detect ALK, ROS1 and RET Rearrangements in Lung Cancer. Sci. Rep. 2017, 7, 42259. [Google Scholar] [CrossRef]
- Chuang, T.-P.; Lai, W.-Y.; Gabre, J.L.; Lind, D.E.; Umapathy, G.; Bokhari, A.A.; Bergman, B.; Kristenson, L.; Thorén, F.B.; Le, A.; et al. ALK Fusion NSCLC Oncogenes Promote Survival and Inhibit NK Cell Responses via SERPINB4 Expression. Proc. Natl. Acad. Sci. USA 2023, 120, e2216479120. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-P.; Tsung, A.; Liu, S.; Nalesnick, M.; Geller, D.; Michalopoulos, G.; Luo, J.-H. Detection of Fusion Transcripts in the Serum Samples of Patients with Hepatocellular Carcinoma. Oncotarget 2019, 10, 3352–3360. [Google Scholar] [CrossRef]
- Yu, Y.-P.; Liu, S.; Nelson, J.; Luo, J.-H. Detection of Fusion Gene Transcripts in the Blood Samples of Prostate Cancer Patients. Sci. Rep. 2021, 11, 16995. [Google Scholar] [CrossRef]
- Chen, Z.-H.; Yu, Y.P.; Tao, J.; Liu, S.; Tseng, G.; Nalesnik, M.; Hamilton, R.; Bhargava, R.; Nelson, J.B.; Pennathur, A.; et al. MAN2A1–FER Fusion Gene Is Expressed by Human Liver and Other Tumor Types and Has Oncogenic Activity in Mice. Gastroenterology 2017, 153, 1120–1132.e15. [Google Scholar] [CrossRef]
- Glenfield, C.; Innan, H. Gene Duplication and Gene Fusion Are Important Drivers of Tumourigenesis during Cancer Evolution. Genes 2021, 12, 1376. [Google Scholar] [CrossRef] [PubMed]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri, S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-Spondin Fusions in Colon Cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Slape, C.; Aplan, P.D. The Role of NUP98 Gene Fusions in Hematologic Malignancy. Leuk. Lymphoma 2004, 45, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 Gene Fusions and Hematopoietic Malignancies: Common Themes and New Biologic Insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef]
- Lanic, M.-D.; Guérin, R.; Sater, V.; Durdilly, P.; Ruminy, P.; Skálová, A.; Laé, M. A Novel SMARCA2-CREM Fusion Expending the Molecular Spectrum of Salivary Gland Hyalinazing Clear Cell Carcinoma beyond the FET Genes. Genes Chromosomes Cancer 2022, 62, 231–236. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Gao, X.; Zhao, W.; Zhou, F.; Liu, H.; Wang, W. Identification of Novel PIEZO1::CBFA2T3 and INO80C::SETBP1 Fusion Genes in an Acute Myeloid Leukemia Patient by RNA-Seq. Mol. Biol. Rep. 2022, 50, 1961–1966. [Google Scholar] [CrossRef]
- Ryzhova, M.V.; Shaikhaev, E.G.; Snigireva, G.P.; Gorelyshev, S.K.; Zheludkova, O.G.; Golanov, A.V. Novel BRAF::EPB41L2 gene fusion in posterior fossa pilocytic astrocytoma. Brief communication. Arkh. Patol. 2022, 84, 40–42. [Google Scholar] [CrossRef]
- Trubini, S.; Ubiali, A.; Paties, C.T.; Cavanna, L. Novel BRAF Mutation in Melanoma: A Case Report. Mol. Clin. Oncol. 2018, 8, 460–462. [Google Scholar] [CrossRef]
- Gong, L.-H.; Liu, W.-F.; Niu, X.-H.; Ding, Y. Two Cases of Spindle Cell Tumors with S100 and CD34 Co-Expression Showing Novel RAF1 Fusions. Diagn. Pathol. 2022, 17, 80. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Wang, T.; Wang, H.; Chen, X.; Cao, P.; Ma, X.; Liu, M.; Xu, P.; Bi, H.; et al. Competitive Evolved Sub-Clonal BCR::ABL1 and Novel MSI2::PC Fusion Genes in Myelodysplastic Syndrome with Isolated Del(5q). Hematol. Oncol. 2022, 41, 178–181. [Google Scholar] [CrossRef]
- Jain, P.; Iyer, S.; Straka, J.; Surrey, L.F.; Pogoriler, J.; Han, H.; Smith, T.; Busch, C.; Fox, E.; Li, M.; et al. Discovery and Functional Characterization of the Oncogenicity and Targetability of a Novel NOTCH1-ROS1 Gene Fusion in Pediatric Angiosarcoma. Cold Spring Harb. Mol. Case Stud. 2022, 8, a006222. [Google Scholar] [CrossRef]
- Zong, X.; Kang, Z.; Huang, D.; Zhang, X.; Gao, Y.; Wang, H.; Li, W.; Yan, J. One Novel ACOT7-NPHP4 Fusion Gene Identified in One Patient with Acute Lymphoblastic Leukemia: A Case Report. BMC Med. Genom. 2022, 15, 226. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.-G.; Xia, T.-L.; Fu, J.-C.; Li, T.; Zhong, Q.; Han, F. BCL6-SPECC1L: A Novel Fusion Gene in Nasopharyngeal Carcinoma. Technol. Cancer Res. Treat. 2022, 21, 15330338221139980. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.-H.; Zhang, F.-B.; Yan, H.; Yu, W.-Y.; Chen, M.; Guan, Y.-T. A Novel CDKN1A-JAZF1 Gene Fusion in Low-Grade Endometrial Stromal Sarcoma Arising from Endometriosis in Abdominal Wall Cesarean Section Scar: A Case Report and Literature Review. Taiwan. J. Obstet. Gynecol. 2022, 61, 1082–1085. [Google Scholar] [CrossRef] [PubMed]
- Patton, A.; Speeckaert, A.; Zeltman, M.; Cui, X.; Oghumu, S.; Iwenofu, O.H. A Novel IRF2BP2::CDX2 Gene Fusion in Digital Intravascular Myoepithelioma of Soft Tissue: An Enigma! Genes Chromosomes Cancer 2023, 62, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Guo, S.; Liu, D.; Chu, J.; Li, Y.; Wang, X.; Zhang, X.; Song, C.; Huang, Q. Pediatric Meningioma with a Novel MAML2-YAP1 Fusion Variant: A Case Report and Literature Review. BMC Pediatr. 2022, 22, 694. [Google Scholar] [CrossRef]
- Rai, S.; Singh, M.P.; Srivastava, S. Integrated Analysis Identifies Novel Fusion Transcripts in Laterally Spreading Tumors Suggestive of Distinct Etiology Than Colorectal Cancers. J. Gastrointest. Cancer 2022. [Google Scholar] [CrossRef]
- Georgantzoglou, N.; Shen, G.; Jour, G.; Linos, K. A Case of FN1-Fused Calcified Chondroid Mesenchymal Neoplasm of the Hand with Novel FGFR3 Partner Gene. Genes Chromosomes Cancer 2022, 62, 237–241. [Google Scholar] [CrossRef]
- Lipplaa, A.; Meijer, D.; van de Sande, M.A.J.; Gelderblom, H.; Bovée, J.V.M.G.; Mei, H.; Szuhai, K. A Novel CSF1 Translocation Involving Human Endogenous Retroviral Element (ERV) in a Tenosynovial Giant Cell Tumour. Genes Chromosomes Cancer 2023, 62, 223–230. [Google Scholar] [CrossRef]
- Goto, H.; Koga, Y.; Kohashi, K.; Ono, H.; Takemoto, J.; Matsuura, T.; Tajiri, T.; Ihara, K.; Oda, Y.; Ohga, S. Pancreatoblastoma with a Novel Fusion Gene of IQSEC1-RAF1. Pediatr. Blood Cancer 2023, 70, e30155. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Q.; Tang, M.; Barthel, F.; Amin, S.; Yoshihara, K.; Lang, F.M.; Martinez-Ledesma, E.; Lee, S.H.; Zheng, S.; et al. TumorFusions: An Integrative Resource for Cancer-Associated Transcript Fusions. Nucleic Acids Res. 2018, 46, D1144–D1149. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.E.; Jang, I.; Kim, S.; Cho, S.; Kim, D.; Kim, K.; Kim, J.; Hwang, J.; Kim, S.; Kim, J.; et al. ChimerDB 4.0: An Updated and Expanded Database of Fusion Genes. Nucleic Acids Res. 2020, 48, D817–D824. [Google Scholar] [CrossRef]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct Molecular Interaction Database in 2012. Nucleic Acids Res. 2011, 40, D841–D846. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-Spectrometric Exploration of Proteome Structure and Function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Meissner, F.; Geddes-McAlister, J.; Mann, M.; Bantscheff, M. The Emerging Role of Mass Spectrometry-Based Proteomics in Drug Discovery. Nat. Rev. Drug Discov. 2022, 21, 637–654. [Google Scholar] [CrossRef]
- Sharifi Tabar, M.; Francis, H.; Yeo, D.; Bailey, C.G.; Rasko, J.E.J. Mapping Oncogenic Protein Interactions for Precision Medicine. Int. J. Cancer 2022, 151, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Stangl, C.; Post, J.B.; van Roosmalen, M.J.; Hami, N.; Verlaan-Klink, I.; Vos, H.R.; van Es, R.M.; Koudijs, M.J.; Voest, E.E.; Snippert, H.J.G.; et al. Diverse BRAF Gene Fusions Confer Resistance to EGFR-Targeted Therapy via Differential Modulation of BRAF Activity. Mol. Cancer Res. MCR 2020, 18, 537–548. [Google Scholar] [CrossRef]
- McCoach, C.E.; Le, A.T.; Gowan, K.; Jones, K.; Schubert, L.; Doak, A.; Estrada-Bernal, A.; Davies, K.D.; Merrick, D.T.; Bunn, P.A.; et al. Resistance Mechanisms to Targeted Therapies in ROS1+ and ALK+ Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 3334–3347. [Google Scholar] [CrossRef]
- Szulzewsky, F.; Holland, E.C.; Vasioukhin, V. YAP1 and Its Fusion Proteins in Cancer Initiation, Progression and Therapeutic Resistance. Dev. Biol. 2021, 475, 205–221. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Yang, B.; Shao, X.; Ying, M. Driving the Degradation of Oncofusion Proteins for Targeted Cancer Therapy. Drug Discov. Today 2023, 28, 103584. [Google Scholar] [CrossRef] [PubMed]
- Brien, G.L.; Stegmaier, K.; Armstrong, S.A. Targeting Chromatin Complexes in Fusion Protein-Driven Malignancies. Nat. Rev. Cancer 2019, 19, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Gröschel, S.; Sanders, M.A.; Hoogenboezem, R.; de Wit, E.; Bouwman, B.A.M.; Erpelinck, C.; van der Velden, V.H.J.; Havermans, M.; Avellino, R.; van Lom, K.; et al. A Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell 2014, 157, 369–381. [Google Scholar] [CrossRef]
- Miller, P.J.; Hollenbach, A.D. The Oncogenic Fusion Protein Pax3-FKHR Has a Greater Post-Translational Stability Relative to Pax3 during Early Myogenesis. Biochim. Biophys. Acta 2007, 1770, 1450–1458. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tokheim, C.; Lee, J.D.; Gan, W.; North, B.J.; Liu, X.S.; Pandolfi, P.P.; Wei, W. Genetic Fusions Favor Tumorigenesis through Degron Loss in Oncogenes. Nat. Commun. 2021, 12, 6704. [Google Scholar] [CrossRef]
- Tan, Y.; Wang, X.; Song, H.; Zhang, Y.; Zhang, R.; Li, S.; Jin, W.; Chen, S.; Fang, H.; Chen, Z.; et al. A PML/RARα Direct Target Atlas Redefines Transcriptional Deregulation in Acute Promyelocytic Leukemia. Blood 2021, 137, 1503–1516. [Google Scholar] [CrossRef]
- Kim, P.; Jia, P.; Zhao, Z. Kinase Impact Assessment in the Landscape of Fusion Genes That Retain Kinase Domains: A Pan-Cancer Study. Brief. Bioinform. 2016, 19, 450–460. [Google Scholar] [CrossRef]
- An, J.; Ren, S.; Murphy, S.J.; Dalangood, S.; Chang, C.; Pang, X.; Cui, Y.; Wang, L.; Pan, Y.; Zhang, X.; et al. Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Mol. Cell 2015, 59, 904–916. [Google Scholar] [CrossRef]
- Li, K.; Wang, F.; Cao, W.-B.; Lv, X.-X.; Hua, F.; Cui, B.; Yu, J.-J.; Zhang, X.-W.; Shang, S.; Liu, S.-S.; et al. TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of P53-Mediated Senescence. Cancer Cell 2017, 31, 697–710.e7. [Google Scholar] [CrossRef]
- Yang, W.-C.; Shih, H.-M. The Deubiquitinating Enzyme USP37 Regulates the Oncogenic Fusion Protein PLZF/RARA Stability. Oncogene 2013, 32, 5167–5175. [Google Scholar] [CrossRef]
- Hong, Z.; Zhang, W.; Ding, D.; Huang, Z.; Yan, Y.; Cao, W.; Pan, Y.; Hou, X.; Weroha, S.J.; Karnes, R.J.; et al. DNA Damage Promotes TMPRSS2-ERG Oncoprotein Destruction and Prostate Cancer Suppression via Signaling Converged by GSK3β and WEE1. Mol. Cell 2020, 79, 1008–1023.e4. [Google Scholar] [CrossRef] [PubMed]
- Touriol, C.; Greenland, C.; Lamant, L.; Pulford, K.; Bernard, F.; Rousset, T.; Mason, D.Y.; Delsol, G. Further Demonstration of the Diversity of Chromosomal Changes Involving 2p23 in ALK-Positive Lymphoma: 2 Cases Expressing ALK Kinase Fused to CLTCL (Clathrin Chain Polypeptide-Like). Blood 2000, 95, 3204–3207. [Google Scholar] [CrossRef]
- Schneider, J.L.; Lin, J.J.; Shaw, A.T. ALK-Positive Lung Cancer: A Moving Target. Nat. Cancer 2023, 4, 330–343. [Google Scholar] [CrossRef]
- Qin, Z.; Sun, H.; Yue, M.; Pan, X.; Chen, L.; Feng, X.; Yan, X.; Zhu, X.; Ji, H. Phase Separation of EML4–ALK in Firing Downstream Signaling and Promoting Lung Tumorigenesis. Cell Discov. 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Tulpule, A.; Guan, J.; Neel, D.S.; Allegakoen, H.R.; Lin, Y.P.; Brown, D.; Chou, Y.-T.; Heslin, A.; Chatterjee, N.; Perati, S.; et al. Kinase-Mediated RAS Signaling via Membraneless Cytoplasmic Protein Granules. Cell 2021, 184, 2649–2664.e18. [Google Scholar] [CrossRef]
- Sampson, J.; Richards, M.W.; Choi, J.; Fry, A.M.; Bayliss, R. Phase-separated Foci of EML4-ALK Facilitate Signalling and Depend upon an Active Kinase Conformation. EMBO Rep. 2021, 22, e53693. [Google Scholar] [CrossRef]
- Okamura, R.; Boichard, A.; Kato, S.; Sicklick, J.K.; Bazhenova, L.; Kurzrock, R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis. Oncol. 2018, 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cremolini, C.; Morano, F.; Moretto, R.; Berenato, R.; Tamborini, E.; Perrone, F.; Rossini, D.; Gloghini, A.; Busico, A.; Zucchelli, G.; et al. Negative Hyper-Selection of Metastatic Colorectal Cancer Patients for Anti-EGFR Monoclonal Antibodies: The PRESSING Case–Control Study. Ann. Oncol. 2017, 28, 3009–3014. [Google Scholar] [CrossRef]
- Rolle, A.-F.L.; Klempner, S.J.; Garrett, C.R.; Seery, T.; Sanford, E.M.; Balasubramanian, S.; Ross, J.S.; Stephens, P.J.; Miller, V.A.; Ali, S.M.; et al. Identification and Characterization of RET Fusions in Advanced Colorectal Cancer. Oncotarget 2015, 6, 28929–28937. [Google Scholar] [CrossRef]
- Declercq, J.; Van Dyck, F.; Van Damme, B.; Van de Ven, W.J.M. Upregulation of Igf and Wnt Signalling Associated Genes in Pleomorphic Adenomas of the Salivary Glands in PLAG1 Transgenic Mice. Int. J. Oncol. 2008, 32, 1041–1047. [Google Scholar] [CrossRef]
- Schram, A.M.; Chang, M.T.; Jonsson, P.; Drilon, A. Fusions in Solid Tumours: Diagnostic Strategies, Targeted Therapy, and Acquired Resistance. Nat. Rev. Clin. Oncol. 2017, 14, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The Landscape of Kinase Fusions in Cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef] [PubMed]
- Amarante-Mendes, G.P.; Rana, A.; Datoguia, T.S.; Hamerschlak, N.; Brumatti, G. BCR-ABL1 Tyrosine Kinase Complex Signaling Transduction: Challenges to Overcome Resistance in Chronic Myeloid Leukemia. Pharmaceutics 2022, 14, 215. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.M.; Hediyeh-zadeh, S.; Sadras, T.; Huckstep, H.; Sandow, J.J.; Bartolo, R.C.; Kosasih, H.J.; Davidson, N.M.; Schmidt, B.; Bjelosevic, S.; et al. SFPQ-ABL1 and BCR-ABL1 Use Different Signaling Networks to Drive B-Cell Acute Lymphoblastic Leukemia. Blood Adv. 2022, 6, 2373–2387. [Google Scholar] [CrossRef]
- Latysheva, N.S.; Oates, M.E.; Maddox, L.; Flock, T.; Gough, J.; Buljan, M.; Weatheritt, R.J.; Babu, M.M. Molecular Principles of Gene Fusion Mediated Rewiring of Protein Interaction Networks in Cancer. Mol. Cell 2016, 63, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Baserga, A.; Gastoldi, G.L. The Philadelptia Chromosome. Biomedicine 1973, 18, 89–94. [Google Scholar]
- Stam, K.; Heisterkamp, N.; Grosveld, G.; de Klein, A.; Verma, R.S.; Coleman, M.; Dosik, H.; Groffen, J. Evidence of a New Chimeric Bcr/c-Abl MRNA in Patients with Chronic Myelocytic Leukemia and the Philadelphia Chromosome. N. Engl. J. Med. 1985, 313, 1429–1433. [Google Scholar] [CrossRef]
- Naldini, L.; Stacchini, A.; Cirillo, D.M.; Aglietta, M.; Gavosto, F.; Comoglio, P.M. Phosphotyrosine Antibodies Identify the P210c-Abl Tyrosine Kinase and Proteins Phosphorylated on Tyrosine in Human Chronic Myelogenous Leukemia Cells. Mol. Cell. Biol. 1986, 6, 1803–1811. [Google Scholar]
- Adnan-Awad, S.; Kim, D.; Hohtari, H.; Javarappa, K.K.; Brandstoetter, T.; Mayer, I.; Potdar, S.; Heckman, C.A.; Kytölä, S.; Porkka, K.; et al. Characterization of P190-Bcr-Abl Chronic Myeloid Leukemia Reveals Specific Signaling Pathways and Therapeutic Targets. Leukemia 2021, 35, 1964–1975. [Google Scholar] [CrossRef]
- McWhirter, J.R.; Wang, J.Y. An Actin-Binding Function Contributes to Transformation by the Bcr-Abl Oncoprotein of Philadelphia Chromosome-Positive Human Leukemias. EMBO J. 1993, 12, 1533–1546. [Google Scholar] [CrossRef]
- Aloisi, A.; Di Gregorio, S.; Stagno, F.; Guglielmo, P.; Mannino, F.; Sormani, M.P.; Bruzzi, P.; Gambacorti-Passerini, C.; Saglio, G.; Venuta, S.; et al. BCR-ABL Nuclear Entrapment Kills Human CML Cells: Ex Vivo Study on 35 Patients with the Combination of Imatinib Mesylate and Leptomycin B. Blood 2006, 107, 1591–1598. [Google Scholar] [CrossRef] [PubMed]
- Carrà, G.; Russo, I.; Guerrasio, A.; Morotti, A. Nuclear-Cytoplasmic Shuttling in Chronic Myeloid Leukemia: Implications in Leukemia Maintenance and Therapy. Cells 2019, 8, 1248. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Saglio, G. Molecular Pathways: BCR-ABL. Clin. Cancer Res. 2012, 18, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Gong, D.; He, C.; Xiao, M.; Zhang, M.; Huang, W. Targeted Therapy Using Larotrectinib and Venetoclax for the Relapsed/Refractory T-Cell Acute Lymphoblastic Leukemia Harboring a Cryptic ETV6-NTRK3 Fusion. Mol. Carcinog. 2023, 62, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhang, Z.; He, X.; Chen, M.; Pang, X.; Chen, C.; Du, T.; Zhang, H. Primary Inflammatory Myofibroblastic Tumour of the Liver: A Clinicopathological and Genetic Study Including a Subset with ETV6::NTRK3 Fusion. Histopathology 2023, 82, 925–936. [Google Scholar] [CrossRef]
- Jiang, H. A Novel ETV6-NTRK3 Gene Fusion in Primary Renal Fibrosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 4705–4708. [Google Scholar] [CrossRef]
- Knezevich, S.R.; McFadden, D.E.; Tao, W.; Lim, J.F.; Sorensen, P.H. A Novel ETV6-NTRK3 Gene Fusion in Congenital Fibrosarcoma. Nat. Genet. 1998, 18, 184–187. [Google Scholar] [CrossRef]
- Lannon, C.L.; Sorensen, P.H.B. ETV6-NTRK3: A Chimeric Protein Tyrosine Kinase with Transformation Activity in Multiple Cell Lineages. Semin. Cancer Biol. 2005, 15, 215–223. [Google Scholar] [CrossRef]
- Tognon, C.; Knezevich, S.R.; Huntsman, D.; Roskelley, C.D.; Melnyk, N.; Mathers, J.A.; Becker, L.; Carneiro, F.; MacPherson, N.; Horsman, D.; et al. Expression of the ETV6-NTRK3 Gene Fusion as a Primary Event in Human Secretory Breast Carcinoma. Cancer Cell 2002, 2, 367–376. [Google Scholar] [CrossRef]
- ETV6-NTRK3 Transformation Requires Insulin-like Growth Factor 1 Receptor Signaling and Is Associated with Constitutive IRS-1 Tyrosine Phosphorylation—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/12173038/ (accessed on 3 May 2023).
- Tognon, C.E.; Somasiri, A.M.; Evdokimova, V.E.; Trigo, G.; Uy, E.E.; Melnyk, N.; Carboni, J.M.; Gottardis, M.M.; Roskelley, C.D.; Pollak, M.; et al. ETV6-NTRK3-Mediated Breast Epithelial Cell Transformation Is Blocked by Targeting the IGF1R Signaling Pathway. Cancer Res. 2011, 71, 1060–1070. [Google Scholar] [CrossRef]
- Bai, R.Y.; Dieter, P.; Peschel, C.; Morris, S.W.; Duyster, J. Nucleophosmin-Anaplastic Lymphoma Kinase of Large-Cell Anaplastic Lymphoma Is a Constitutively Active Tyrosine Kinase That Utilizes Phospholipase C-Gamma to Mediate Its Mitogenicity. Mol. Cell. Biol. 1998, 18, 6951–6961. [Google Scholar] [CrossRef] [PubMed]
- Duyster, J.; Bai, R.Y.; Morris, S.W. Translocations Involving Anaplastic Lymphoma Kinase (ALK). Oncogene 2001, 20, 5623–5637. [Google Scholar] [CrossRef] [PubMed]
- Buetti-Dinh, A.; O’Hare, T.; Friedman, R. Sensitivity Analysis of the NPM-ALK Signalling Network Reveals Important Pathways for Anaplastic Large Cell Lymphoma Combination Therapy. PLoS ONE 2016, 11, e0163011. [Google Scholar] [CrossRef]
- Chiarle, R.; Simmons, W.J.; Cai, H.; Dhall, G.; Zamo, A.; Raz, R.; Karras, J.G.; Levy, D.E.; Inghirami, G. Stat3 Is Required for ALK-Mediated Lymphomagenesis and Provides a Possible Therapeutic Target. Nat. Med. 2005, 11, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Cussac, D.; Greenland, C.; Roche, S.; Bai, R.-Y.; Duyster, J.; Morris, S.W.; Delsol, G.; Allouche, M.; Payrastre, B. Nucleophosmin-Anaplastic Lymphoma Kinase of Anaplastic Large-Cell Lymphoma Recruits, Activates, and Uses Pp60c-Src to Mediate Its Mitogenicity. Blood 2004, 103, 1464–1471. [Google Scholar] [CrossRef]
- Khosh Kish, E.; Choudhry, M.; Gamallat, Y.; Buharideen, S.M.; Bismar, T.A. The Expression of Proto-Oncogene ETS-Related Gene (ERG) Plays a Central Role in the Oncogenic Mechanism Involved in the Development and Progression of Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 4772. [Google Scholar] [CrossRef]
- Zoma, M.; Curti, L.; Shinde, D.; Albino, D.; Mitra, A.; Sgrignani, J.; Mapelli, S.N.; Sandrini, G.; Civenni, G.; Merulla, J.; et al. EZH2-Induced Lysine K362 Methylation Enhances TMPRSS2-ERG Oncogenic Activity in Prostate Cancer. Nat. Commun. 2021, 12, 4147. [Google Scholar] [CrossRef]
- Wu, L.; Zhao, J.C.; Kim, J.; Jin, H.-J.; Wang, C.-Y.; Yu, J. ERG Is a Critical Regulator of Wnt/LEF1 Signaling in Prostate Cancer. Cancer Res. 2013, 73, 6068–6079. [Google Scholar] [CrossRef]
- Kaplan, Z.; Zielske, S.P.; Ibrahim, K.G.; Cackowski, F.C. Wnt and β-Catenin Signaling in the Bone Metastasis of Prostate Cancer. Life 2021, 11, 1099. [Google Scholar] [CrossRef]
- Abou-Ouf, H.; Assem, H.; Ghosh, S.; Karnes, R.J.; Stoletov, K.; Palanisamy, N.; Lewis, J.D.; Bismar, T.A. High Serine-Arginine Protein Kinase 1 Expression with PTEN Loss Defines Aggressive Phenotype of Prostate Cancer Associated with Lethal Outcome and Decreased Overall Survival. Eur. Urol. Open Sci. 2021, 23, 1–8. [Google Scholar] [CrossRef]
- Ruiz Moreno, J.M.; Medrano López, M.; Rodriguez Prats, J.L. Cavernous angioma of the retina: Therapeutic approach. J. Fr. Ophtalmol. 1987, 10, 731–733. [Google Scholar] [PubMed]
- Meisel Sharon, S.; Pozniak, Y.; Geiger, T.; Werner, H. TMPRSS2-ERG Fusion Protein Regulates Insulin-like Growth Factor-1 Receptor (IGF1R) Gene Expression in Prostate Cancer: Involvement of Transcription Factor Sp1. Oncotarget 2016, 7, 51375–51392. [Google Scholar] [CrossRef] [PubMed]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate Cancer. Nat. Rev. Dis. Primer 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, M.; Qin, Y.; Gao, W.; Tao, L.; Su, W.; Zhong, J. Neoantigen: A New Breakthrough in Tumor Immunotherapy. Front. Immunol. 2021, 12, 672356. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising Targets for Cancer Therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wu, J.; Chen, S.; Zhou, Z. Shared Neoantigens: Ideal Targets for off-the-Shelf Cancer Immunotherapy. Pharmacogenomics 2020, 21, 637–645. [Google Scholar] [CrossRef]
- Smith, B.; Kennedy, J.W. Thrombolysis in the Treatment of Acute Transmural Myocardial Infarction. Ann. Intern. Med. 1987, 106, 414–420. [Google Scholar] [CrossRef]
- Liu, Y.; Klein, J.; Bajpai, R.; Dong, L.; Tran, Q.; Kolekar, P.; Smith, J.L.; Ries, R.E.; Huang, B.J.; Wang, Y.-C.; et al. Etiology of Oncogenic Fusions in 5,190 Childhood Cancers and Its Clinical and Therapeutic Implication. Nat. Commun. 2023, 14, 1739. [Google Scholar] [CrossRef]
- Wei, Z.; Zhou, C.; Zhang, Z.; Guan, M.; Zhang, C.; Liu, Z.; Liu, Q. The Landscape of Tumor Fusion Neoantigens: A Pan-Cancer Analysis. iScience 2019, 21, 249–260. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, T.; Song, X.; Liu, B.; Wei, J. Gene Fusion Neoantigens: Emerging Targets for Cancer Immunotherapy. Cancer Lett. 2021, 506, 45–54. [Google Scholar] [CrossRef]
- Lin, Y.-Y.; Gawronski, A.; Hach, F.; Li, S.; Numanagić, I.; Sarrafi, I.; Mishra, S.; McPherson, A.; Collins, C.C.; Radovich, M.; et al. Computational Identification of Micro-Structural Variations and Their Proteogenomic Consequences in Cancer. Bioinformatics 2018, 34, 1672–1681. [Google Scholar] [CrossRef] [PubMed]
- Howarth, K.D.; Mirza, T.; Cooke, S.L.; Chin, S.-F.; Pole, J.C.; Turro, E.; Eldridge, M.D.; Garcia, R.M.; Rueda, O.M.; Boursnell, C.; et al. NRG1 Fusions in Breast Cancer. Breast Cancer Res. BCR 2021, 23, 3. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Koh, H.H.; Chang, E.S.; Choi, J.; Song, J.-Y.; Lee, M.-S.; Choi, Y.-L. Unearthing Novel Fusions as Therapeutic Targets in Solid Tumors Using Targeted RNA Sequencing. Front. Oncol. 2022, 12, 892918. [Google Scholar] [CrossRef] [PubMed]
- Mackall, C.L.; Rhee, E.H.; Read, E.J.; Khuu, H.M.; Leitman, S.F.; Bernstein, D.; Tesso, M.; Long, L.M.; Grindler, D.; Merino, M.; et al. A Pilot Study of Consolidative Immunotherapy in Patients with High-Risk Pediatric Sarcomas. Clin. Cancer Res. 2008, 14, 4850–4858. [Google Scholar] [CrossRef]
- Biernacki, M.A.; Foster, K.A.; Woodward, K.B.; Coon, M.E.; Cummings, C.; Cunningham, T.M.; Dossa, R.G.; Brault, M.; Stokke, J.; Olsen, T.M.; et al. CBFB-MYH11 Fusion Neoantigen Enables T Cell Recognition and Killing of Acute Myeloid Leukemia. J. Clin. Investig. 2020, 130, 5127–5141. [Google Scholar] [CrossRef]
- Yang, W.; Lee, K.-W.; Srivastava, R.M.; Kuo, F.; Krishna, C.; Chowell, D.; Makarov, V.; Hoen, D.; Dalin, M.G.; Wexler, L.; et al. Immunogenic Neoantigens Derived from Gene Fusions Stimulate T Cell Responses. Nat. Med. 2019, 25, 767–775. [Google Scholar] [CrossRef]
- Kalina, J.L.; Neilson, D.S.; Lin, Y.-Y.; Hamilton, P.T.; Comber, A.P.; Loy, E.M.H.; Sahinalp, S.C.; Collins, C.C.; Hach, F.; Lum, J.J. Mutational Analysis of Gene Fusions Predicts Novel MHC Class I-Restricted T-Cell Epitopes and Immune Signatures in a Subset of Prostate Cancer. Clin. Cancer Res. 2017, 23, 7596–7607. [Google Scholar] [CrossRef]
- Pavet, V.; Portal, M.M.; Moulin, J.C.; Herbrecht, R.; Gronemeyer, H. Towards Novel Paradigms for Cancer Therapy. Oncogene 2011, 30, 1–20. [Google Scholar] [CrossRef]
- Powers, M.P. The Ever-Changing World of Gene Fusions in Cancer: A Secondary Gene Fusion and Progression. Oncogene 2019, 38, 7197–7199. [Google Scholar] [CrossRef]
- Wong, S.; Witte, O.N. The BCR-ABL Story: Bench to Bedside and Back. Annu. Rev. Immunol. 2004, 22, 247–306. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Aubin, S.M.J.; Siddiqui, J.; Lonigro, R.J.; Sefton-Miller, L.; Miick, S.; Williamsen, S.; Hodge, P.; Meinke, J.; Blase, A.; et al. Urine TMPRSS2:ERG Fusion Transcript Stratifies Prostate Cancer Risk in Men with Elevated Serum PSA. Sci. Transl. Med. 2011, 3, 94ra72. [Google Scholar] [CrossRef] [PubMed]
- Villani, A.; Davidson, S.; Kanwar, N.; Lo, W.W.; Li, Y.; Cohen-Gogo, S.; Fuligni, F.; Edward, L.-M.; Light, N.; Layeghifard, M.; et al. The Clinical Utility of Integrative Genomics in Childhood Cancer Extends beyond Targetable Mutations. Nat. Cancer 2022, 4, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Ibn-Salem, J.; Sorn, P.; Suchan, M.; Holtsträter, C.; Lahrmann, U.; Vogler, I.; Schmoldt, K.; Lang, F.; Schrörs, B.; et al. Accurate Detection of Tumor-Specific Gene Fusions Reveals Strongly Immunogenic Personal Neo-Antigens. Nat. Biotechnol. 2022, 40, 1276–1284. [Google Scholar] [CrossRef]
- Larkin, R.; Hermsen, M.A.; London, N.R., Jr. Translocations and Gene Fusions in Sinonasal Malignancies. Curr. Oncol. Rep. 2023, 25, 269–278. [Google Scholar] [CrossRef]
- Nikanjam, M.; Okamura, R.; Barkauskas, D.A.; Kurzrock, R. Targeting Fusions for Improved Outcomes in Oncology Treatment. Cancer 2020, 126, 1315. [Google Scholar] [CrossRef] [PubMed]
- Rudzinski, E.R.; Drilon, A.; Moore, A.; Spinosa, S.; Willi, M.; Laetsch, T.W. Testing Methods to Diagnose TRK Fusion Cancer: A Plain Language Summary and Patient Perspective. Future Oncol. Lond. Engl. 2023, 18, 4141–4151. [Google Scholar] [CrossRef]
- Li, W.; Guo, L.; Liu, Y.; Dong, L.; Yang, L.; Chen, L.; Liu, K.; Shao, Y.; Ying, J. Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC. J. Thorac. Oncol. 2021, 16, 404–418. [Google Scholar] [CrossRef]
- Dankner, M.; Rose, A.A.N.; Rajkumar, S.; Siegel, P.M.; Watson, I.R. Classifying BRAF Alterations in Cancer: New Rational Therapeutic Strategies for Actionable Mutations. Oncogene 2018, 37, 3183–3199. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, L.; Yang, Q.; Sun, J. The Evolution of BRAF Activation in Non-Small-Cell Lung Cancer. Front. Oncol. 2022, 12, 882940. [Google Scholar] [CrossRef]
- Vojnic, M.; Kubota, D.; Kurzatkowski, C.; Offin, M.; Suzawa, K.; Benayed, R.; Schoenfeld, A.J.; Plodkowski, A.J.; Poirier, J.T.; Rudin, C.M.; et al. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR Therapy in EGFR-Mutated Lung Cancers. J. Thorac. Oncol. 2019, 14, 802–815. [Google Scholar] [CrossRef]
- Servetto, A.; Esposito, D.; Ferrara, R.; Signorelli, D.; Belli, S.; Napolitano, F.; Santaniello, A.; Ciciola, P.; Formisano, L.; Bianco, R. RET Rearrangements in Non-Small Cell Lung Cancer: Evolving Treatment Landscape and Future Challenges. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188810. [Google Scholar] [CrossRef] [PubMed]
- Barnes, E.J.; Eide, C.A.; Kaempf, A.; Bottomly, D.; Romine, K.A.; Wilmot, B.; Saunders, D.; McWeeney, S.K.; Tognon, C.E.; Druker, B.J. Secondary Fusion Proteins as a Mechanism of BCR::ABL1 Kinase-Independent Resistance in Chronic Myeloid Leukaemia. Br. J. Haematol. 2023, 200, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Liu, Y.; Liang, Z.; Wang, W.; Qin, T.; Liu, S.V.; Um, S.-W.; Luo, F.; Liu, J. Classical ALK G1202R Resistance Mutation Was Identified in a Lung Adenocarcinoma Patient with Rare LOC388942-ALK Fusion after Sequential Treatment with ALK-TKIs and Anlotinib: A Case Report. Ann. Transl. Med. 2022, 10, 1180. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Oxnard, G.R.; Cohen, E.F.; Mahadevan, N.R.; Alessi, J.V.; Hung, Y.P.; Bertram, A.A.; Heppner, D.E.; Ribeiro, M.F.; Sacardo, K.P.; et al. Genomic and Biological Study of Fusion Genes as Resistance Mechanisms to EGFR Inhibitors. Nat. Commun. 2022, 13, 5614. [Google Scholar] [CrossRef] [PubMed]
- Jonna, S.; Feldman, R.A.; Swensen, J.; Gatalica, Z.; Korn, W.M.; Borghaei, H.; Ma, P.C.; Nieva, J.J.; Spira, A.I.; Vanderwalde, A.M.; et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin. Cancer Res. 2019, 25, 4966–4972. [Google Scholar] [CrossRef]
- Shin, D.H.; Lee, D.; Hong, D.W.; Hong, S.H.; Hwang, J.-A.; Lee, B.I.; You, H.J.; Lee, G.K.; Kim, I.-H.; Lee, Y.-S.; et al. Oncogenic Function and Clinical Implications of SLC3A2-NRG1 Fusion in Invasive Mucinous Adenocarcinoma of the Lung. Oncotarget 2016, 7, 69450–69465. [Google Scholar] [CrossRef]
- Nam, R.K.; Sugar, L.; Wang, Z.; Yang, W.; Kitching, R.; Klotz, L.H.; Venkateswaran, V.; Narod, S.A.; Seth, A. Expression of TMPRSS2:ERG Gene Fusion in Prostate Cancer Cells Is an Important Prognostic Factor for Cancer Progression. Cancer Biol. Ther. 2007, 6, 40–45. [Google Scholar] [CrossRef]
- Kumar-Sinha, C.; Tomlins, S.A.; Chinnaiyan, A.M. Recurrent Gene Fusions in Prostate Cancer. Nat. Rev. Cancer 2008, 8, 497–511. [Google Scholar] [CrossRef]
- Demichelis, F.; Fall, K.; Perner, S.; Andrén, O.; Schmidt, F.; Setlur, S.R.; Hoshida, Y.; Mosquera, J.-M.; Pawitan, Y.; Lee, C.; et al. TMPRSS2:ERG Gene Fusion Associated with Lethal Prostate Cancer in a Watchful Waiting Cohort. Oncogene 2007, 26, 4596–4599. [Google Scholar] [CrossRef]
- Hägglöf, C.; Hammarsten, P.; Strömvall, K.; Egevad, L.; Josefsson, A.; Stattin, P.; Granfors, T.; Bergh, A. TMPRSS2-ERG Expression Predicts Prostate Cancer Survival and Associates with Stromal Biomarkers. PLoS ONE 2014, 9, e86824. [Google Scholar] [CrossRef]
- Song, C.; Chen, H. Predictive Significance of TMRPSS2-ERG Fusion in Prostate Cancer: A Meta-Analysis. Cancer Cell Int. 2018, 18, 177. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wan, R.; Guo, L.; Chang, G.; Jiang, D.; Meng, L.; Ying, J. Reliability Analysis of Exonic-Breakpoint Fusions Identified by DNA Sequencing for Predicting the Efficacy of Targeted Therapy in Non-Small Cell Lung Cancer. BMC Med. 2022, 20, 160. [Google Scholar] [CrossRef]
- Pagani, F.; Randon, G.; Guarini, V.; Raimondi, A.; Prisciandaro, M.; Lobefaro, R.; Di Bartolomeo, M.; Sozzi, G.; de Braud, F.; Gasparini, P.; et al. The Landscape of Actionable Gene Fusions in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 5319. [Google Scholar] [CrossRef]
- Zhou, Y.; El-Bahrawy, M. Gene Fusions in Tumourigenesis with Particular Reference to Ovarian Cancer. J. Med. Genet. 2021, 58, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Heyer, E.E.; Deveson, I.W.; Wooi, D.; Selinger, C.I.; Lyons, R.J.; Hayes, V.M.; O’Toole, S.A.; Ballinger, M.L.; Gill, D.; Thomas, D.M.; et al. Diagnosis of Fusion Genes Using Targeted RNA Sequencing. Nat. Commun. 2019, 10, 1388. [Google Scholar] [CrossRef] [PubMed]
- Heydt, C.; Wölwer, C.B.; Velazquez Camacho, O.; Wagener-Ryczek, S.; Pappesch, R.; Siemanowski, J.; Rehker, J.; Haller, F.; Agaimy, A.; Worm, K.; et al. Detection of Gene Fusions Using Targeted Next-Generation Sequencing: A Comparative Evaluation. BMC Med. Genom. 2021, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Carter, T.C.; He, M.M. Challenges of Identifying Clinically Actionable Genetic Variants for Precision Medicine. J. Healthc. Eng. 2016, 2016, 3617572. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Kirchner, M.; Kluck, K.; Kazdal, D.; Glade, J.; Allgäuer, M.; Kriegsmann, M.; Heußel, C.-P.; Herth, F.J.; Winter, H.; et al. Deciphering the Immunosuppressive Tumor Microenvironment in ALK- and EGFR-Positive Lung Adenocarcinoma. Cancer Immunol. Immunother. 2022, 71, 251–265. [Google Scholar] [CrossRef]
- Galkin, A.V.; Melnick, J.S.; Kim, S.; Hood, T.L.; Li, N.; Li, L.; Xia, G.; Steensma, R.; Chopiuk, G.; Jiang, J.; et al. Identification of NVP-TAE684, a Potent, Selective, and Efficacious Inhibitor of NPM-ALK. Proc. Natl. Acad. Sci. USA 2007, 104, 270–275. [Google Scholar] [CrossRef]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Baek, J.H.; Sun, J.-M.; Min, Y.J.; Cho, E.K.; Cho, B.C.; Kim, J.-H.; Ahn, M.-J.; Park, K. Efficacy of EGFR Tyrosine Kinase Inhibitors in Patients with EGFR-Mutated Non-Small Cell Lung Cancer except Both Exon 19 Deletion and Exon 21 L858R: A Retrospective Analysis in Korea. Lung Cancer 2015, 87, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Druker, B.J.; Talpaz, M.; Resta, D.J.; Peng, B.; Buchdunger, E.; Ford, J.M.; Lydon, N.B.; Kantarjian, H.; Capdeville, R.; Ohno-Jones, S.; et al. Efficacy and Safety of a Specific Inhibitor of the BCR-ABL Tyrosine Kinase in Chronic Myeloid Leukemia. N. Engl. J. Med. 2001, 344, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Pemovska, T.; Johnson, E.; Kontro, M.; Repasky, G.A.; Chen, J.; Wells, P.; Cronin, C.N.; McTigue, M.; Kallioniemi, O.; Porkka, K.; et al. Axitinib Effectively Inhibits BCR-ABL1(T315I) with a Distinct Binding Conformation. Nature 2015, 519, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.; Maley, C.C. Clonal Evolution in Cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase Drug Discovery 20 Years after Imatinib: Progress and Future Directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef]
- Meador, C.B.; Hata, A.N. Acquired Resistance to Targeted Therapies in NSCLC: Updates and Evolving Insights. Pharmacol. Ther. 2020, 210, 107522. [Google Scholar] [CrossRef]
- Viossat, Y.; Noble, R. A Theoretical Analysis of Tumour Containment. Nat. Ecol. Evol. 2021, 5, 826–835. [Google Scholar] [CrossRef]
- Naik, R.R.; Shakya, A.K. Exploring the Chemotherapeutic Potential of Currently Used Kinase Inhibitors: An Update. Front. Pharmacol. 2023, 13, 1064472. [Google Scholar] [CrossRef]
- Qin, S.; Li, A.; Yi, M.; Yu, S.; Zhang, M.; Wu, K. Recent Advances on Anti-Angiogenesis Receptor Tyrosine Kinase Inhibitors in Cancer Therapy. J. Hematol. Oncol. 2019, 12, 27. [Google Scholar] [CrossRef]
- Pottier, C.; Fresnais, M.; Gilon, M.; Jérusalem, G.; Longuespée, R.; Sounni, N.E. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers 2020, 12, 731. [Google Scholar] [CrossRef]
- Murumägi, A.; Ungureanu, D.; Arjama, M.; Bützow, R.; Lohi, J.; Sariola, H.; Kanerva, J.; Koskenvuo, M.; Kallioniemi, O. STRN-ALK Rearranged Pediatric Malignant Peritoneal Mesothelioma—Functional Testing of 527 Cancer Drugs in Patient-Derived Cancer Cells. Transl. Oncol. 2021, 14, 101027. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 Signalling Axis in Cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- A Phase I Clinical Trial of Ruxolitinib in Combination with Nilotinib in Chronic Myeloid Leukemia Patients with Molecular Evidence of Disease—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/30340199/ (accessed on 3 May 2023).
- Chirnomas, D.; Hornberger, K.R.; Crews, C.M. Protein Degraders Enter the Clinic—A New Approach to Cancer Therapy. Nat. Rev. Clin. Oncol. 2023, 20, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhou, Y.; Sun, D.; Yang, Y.; Liu, Y.; Li, X.; Li, H.; Chen, L. PROTACs: New Method to Degrade Transcription Regulating Proteins. Eur. J. Med. Chem. 2020, 207, 112698. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted Protein Degradation: Mechanisms, Strategies and Application. Signal Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef]
- Lai, A.C.; Toure, M.; Hellerschmied, D.; Salami, J.; Jaime-Figueroa, S.; Ko, E.; Hines, J.; Crews, C.M. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew. Chem. Int. Ed. 2016, 55, 807–810. [Google Scholar] [CrossRef]
- Burslem, G.M.; Schultz, A.R.; Bondeson, D.P.; Eide, C.A.; Savage Stevens, S.L.; Druker, B.J.; Crews, C.M. Targeting BCR-ABL1 in Chronic Myeloid Leukemia by PROTAC-Mediated Targeted Protein Degradation. Cancer Res. 2019, 79, 4744–4753. [Google Scholar] [CrossRef]
- Kang, C.H.; Lee, D.H.; Lee, C.O.; Du Ha, J.; Park, C.H.; Hwang, J.Y. Induced Protein Degradation of Anaplastic Lymphoma Kinase (ALK) by Proteolysis Targeting Chimera (PROTAC). Biochem. Biophys. Res. Commun. 2018, 505, 542–547. [Google Scholar] [CrossRef]
- Powell, C.E.; Gao, Y.; Tan, L.; Donovan, K.A.; Nowak, R.P.; Loehr, A.; Bahcall, M.; Fischer, E.S.; Jänne, P.A.; George, R.E.; et al. Chemically Induced Degradation of Anaplastic Lymphoma Kinase (ALK). J. Med. Chem. 2018, 61, 4249–4255. [Google Scholar] [CrossRef]
- Song, X.; Zhong, H.; Qu, X.; Yang, L.; Jiang, B. Two Novel Strategies to Overcome the Resistance to ALK Tyrosine Kinase Inhibitor Drugs: Macrocyclic Inhibitors and Proteolysis-Targeting Chimeras. MedComm 2021, 2, 341–350. [Google Scholar] [CrossRef]
- Sun, N.; Ren, C.; Kong, Y.; Zhong, H.; Chen, J.; Li, Y.; Zhang, J.; Zhou, Y.; Qiu, X.; Lin, H.; et al. Development of a Brigatinib Degrader (SIAIS117) as a Potential Treatment for ALK Positive Cancer Resistance. Eur. J. Med. Chem. 2020, 193, 112190. [Google Scholar] [CrossRef] [PubMed]
- Mayor-Ruiz, C.; Bauer, S.; Brand, M.; Kozicka, Z.; Siklos, M.; Imrichova, H.; Kaltheuner, I.H.; Hahn, E.; Seiler, K.; Koren, A.; et al. Rational Discovery of Molecular Glue Degraders via Scalable Chemical Profiling. Nat. Chem. Biol. 2020, 16, 1199–1207. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salokas, K.; Dashi, G.; Varjosalo, M. Decoding Oncofusions: Unveiling Mechanisms, Clinical Impact, and Prospects for Personalized Cancer Therapies. Cancers 2023, 15, 3678. https://doi.org/10.3390/cancers15143678
Salokas K, Dashi G, Varjosalo M. Decoding Oncofusions: Unveiling Mechanisms, Clinical Impact, and Prospects for Personalized Cancer Therapies. Cancers. 2023; 15(14):3678. https://doi.org/10.3390/cancers15143678
Chicago/Turabian StyleSalokas, Kari, Giovanna Dashi, and Markku Varjosalo. 2023. "Decoding Oncofusions: Unveiling Mechanisms, Clinical Impact, and Prospects for Personalized Cancer Therapies" Cancers 15, no. 14: 3678. https://doi.org/10.3390/cancers15143678
APA StyleSalokas, K., Dashi, G., & Varjosalo, M. (2023). Decoding Oncofusions: Unveiling Mechanisms, Clinical Impact, and Prospects for Personalized Cancer Therapies. Cancers, 15(14), 3678. https://doi.org/10.3390/cancers15143678





