Comprehensive Analysis and Drug Modulation of Human Endogenous Retrovirus in Hepatocellular Carcinomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Liver Samples and Clinical Data
2.2. RNA Extraction and RNA Sequencing (RNA-Seq)
2.3. Metatranscriptome Analysis
2.4. Retrotranscriptome and Transcriptome Quantification
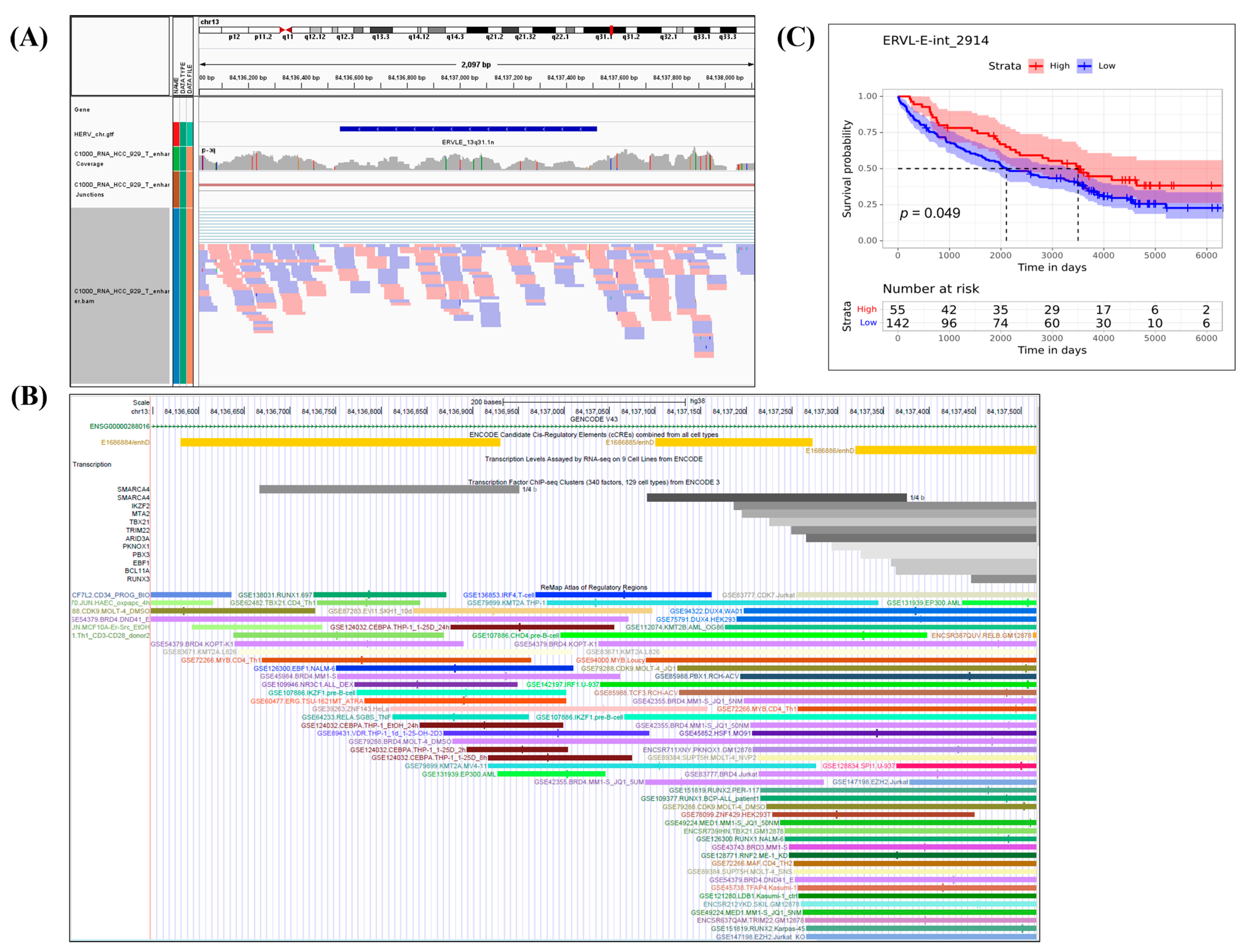
2.5. Structure Analysis of Survival-Related HERVs, Their Genomic Regions, and Nearby Related Genes
2.6. Molecular Classification
2.7. Correlation Analysis of HERV Subgroups and the Expression Levels of Genes Related to HERV Restriction, Viral Immunity, RNA Transport, and Stemness
2.8. Tumor Microenvironment Analysis
2.9. Drug Treatment of HCC Cell Line
2.10. Statistical Analyses
3. Results
3.1. Clinical Data and Metatranscriptomic Analysis
3.2. Retrotranscriptome and Transcriptome Quantification Analyses
3.3. Analysis of Nearby Genes of the 180 Survival-Related DE HERVs by Telescope
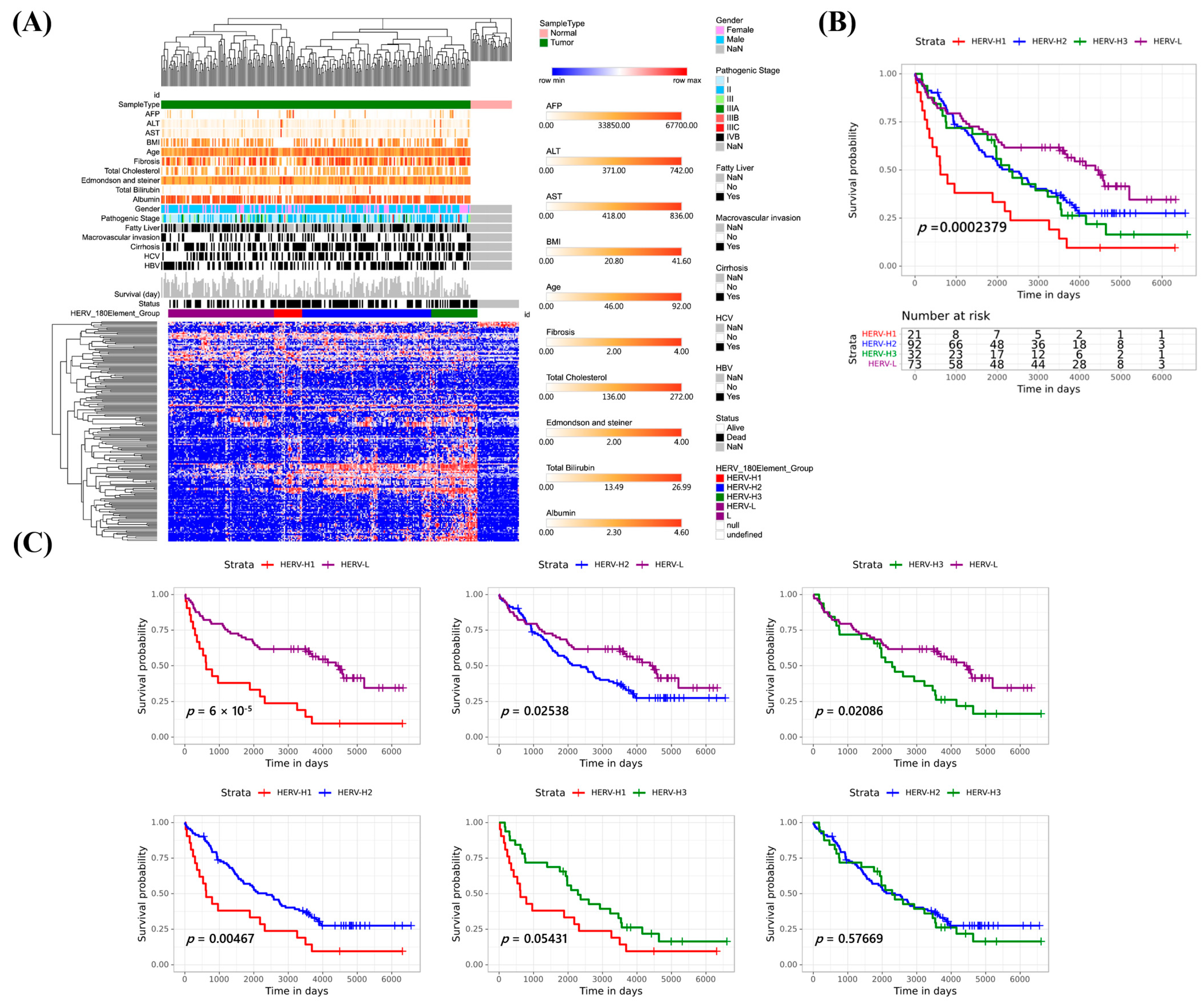
3.4. Molecular Classification of 254 Taiwanese HCCs Based on the Expression Profiles of 180 Survival-Related DE HERVs
3.5. Retrotranscriptome Quantification Analyses of the HCC Subgroups
3.6. Associations of HCC Subgroups with Survival-Related DE HERVs, Clinical Relevance, Mutations, Copy Number Aberrations, and Structural Variants (SVs)
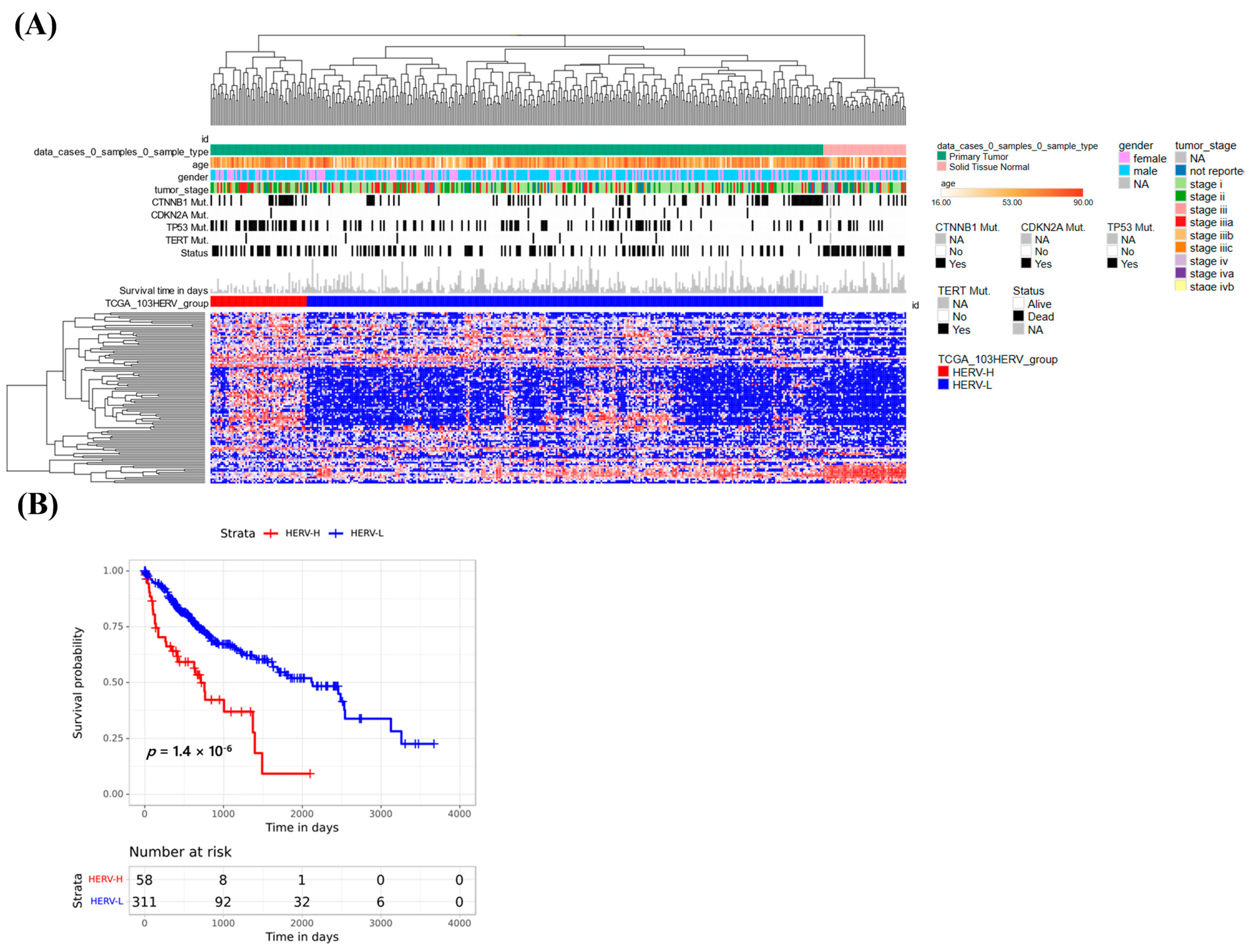
3.7. Classification Using HERV Expression in the Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC)
3.8. Analysis of Host Differential Gene Expression and Molecular Pathways between HERV-H and HERV-L
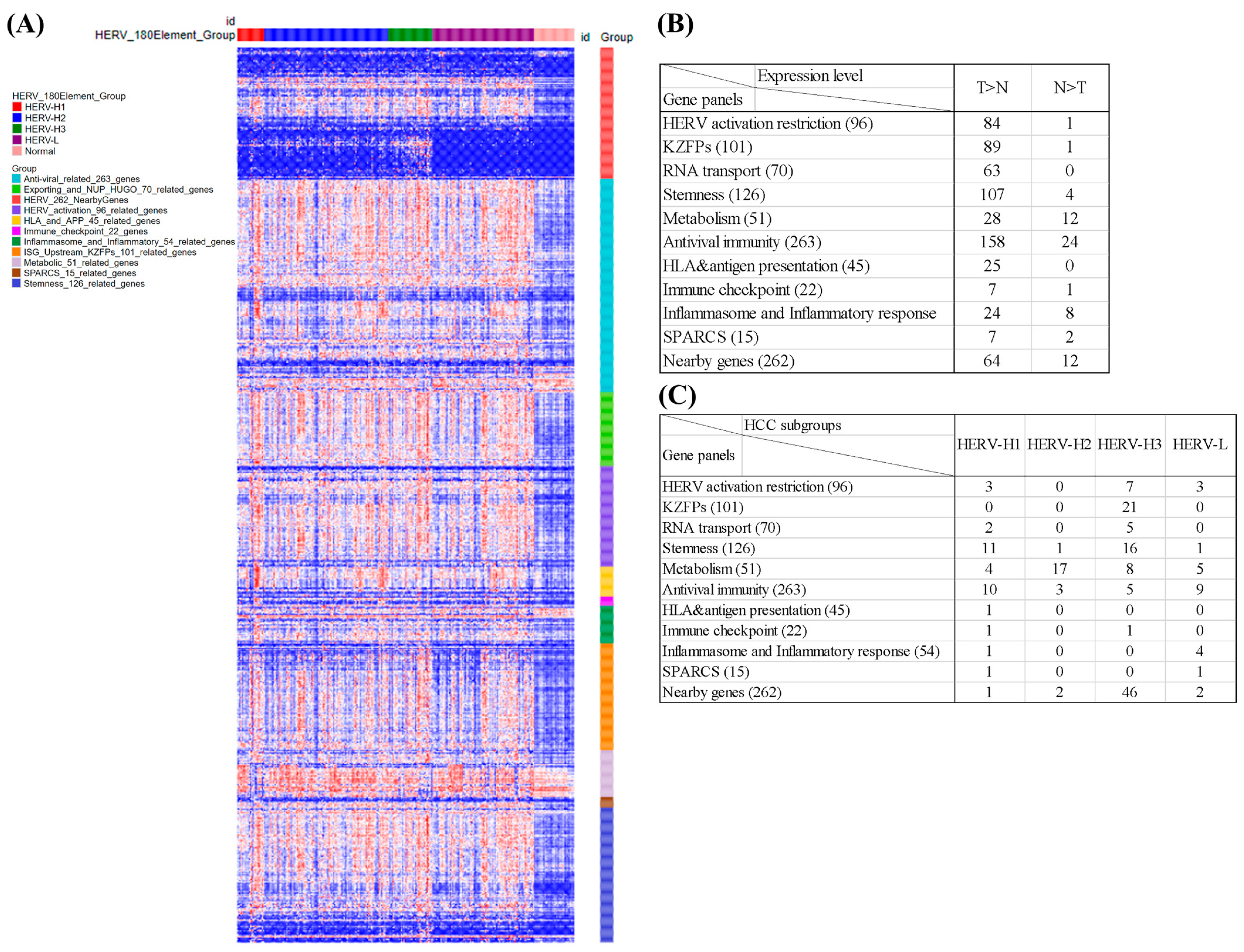
3.9. Analysis of 10 Gene Panels and Nearby Genes of the HCC Subgroups
3.10. Associations of the Tumor Microenvironment with Four HCC Subgroups
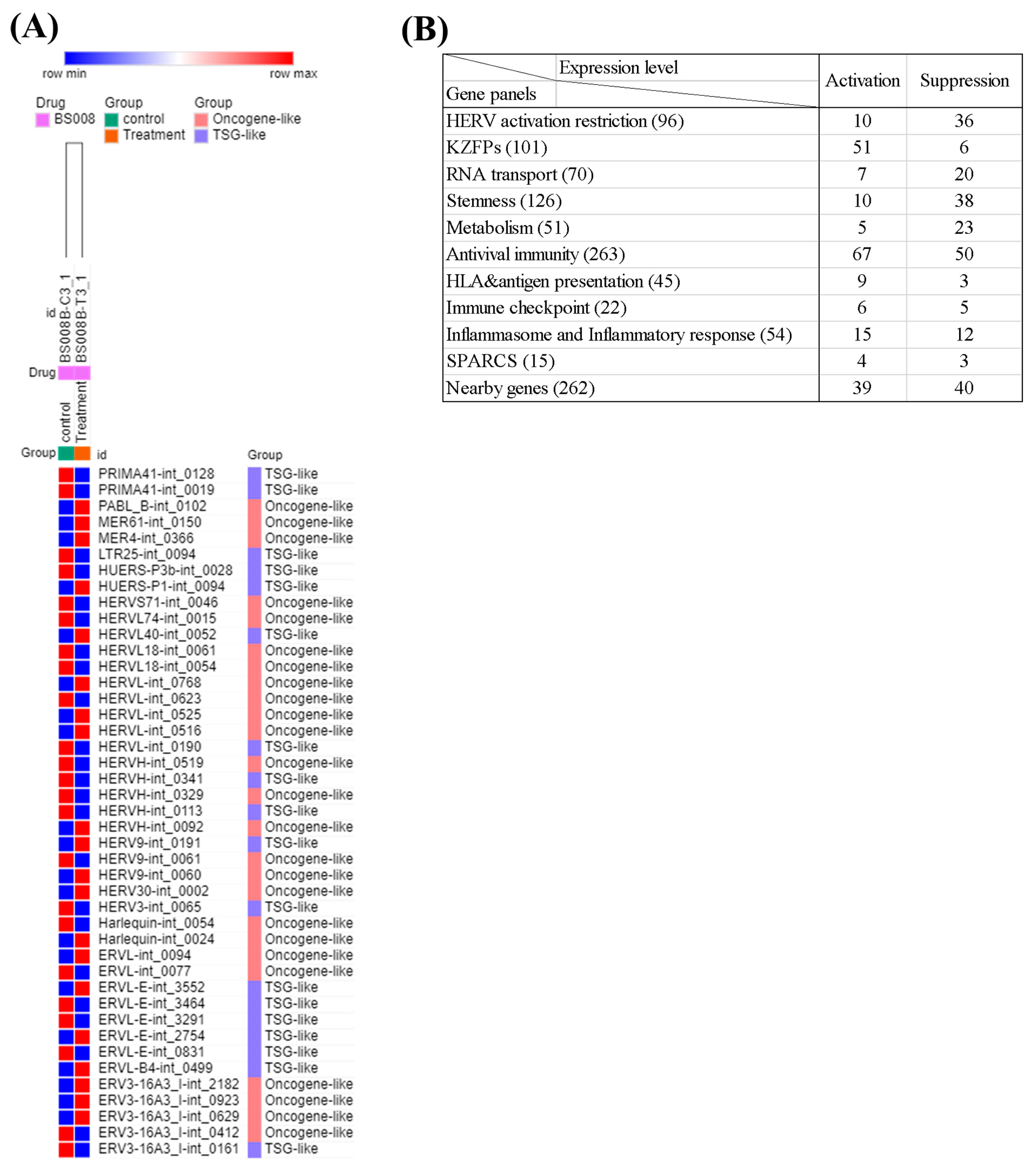
3.11. Analysis of the Effects of Splicing-Modulating Drugs on the Expression of HERV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of hepatocellular carcinoma. Hepatology 2021, 73, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef]
- Muller, M.; Bird, T.G.; Nault, J.C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J. Hepatol. 2020, 72, 990–1002. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Steiner, M.C.; Marston, J.L.; Iniguez, L.P.; Bendall, M.L.; Chiappinelli, K.B.; Nixon, D.F.; Crandall, K.A. Locus-Specific Characterization of Human Endogenous Retrovirus Expression in Prostate, Breast, and Colon Cancers. Cancer Res. 2021, 81, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Topham, J.T.; Titmuss, E.; Pleasance, E.D.; Williamson, L.M.; Karasinska, J.M.; Culibrk, L.; Lee, M.K.C.; Mendis, S.; Denroche, R.E.; Jang, G.H.; et al. Endogenous Retrovirus Transcript Levels Are Associated with Immunogenic Signatures in Multiple Metastatic Cancer Types. Mol. Cancer Ther. 2020, 19, 1889–1897. [Google Scholar] [CrossRef]
- Smith, C.C.; Beckermann, K.E.; Bortone, D.S.; De Cubas, A.A.; Bixby, L.M.; Lee, S.J.; Panda, A.; Ganesan, S.; Bhanot, G.; Wallen, E.M.; et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Investig. 2018, 128, 4804–4820. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Fueyo, R.; Judd, J.; Feschotte, C.; Wysocka, J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022, 23, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.F.; Xing, X.Y.; Kim, S.; et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef]
- Babaian, A.; Mager, D.L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 2016, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, C.; Tsang, J.; Bireau, C.; Heidmann, T.; Dewannieux, M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017, 13, e1006451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.W.; Liang, J.Q.; Zheng, S. Expressional activation and functional roles of human endogenous retroviruses in cancers. Rev. Med. Virol. 2019, 29, e2025. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Kimura, I.; Soper, A.; Coudray, A.; Koyanagi, Y.; Nakaoka, H.; Inoue, I.; Turelli, P.; Trono, D.; Sato, K. Endogenous retroviruses drive KRAB zinc-finger protein family expression for tumor suppression. Sci. Adv. 2020, 6, eabc3020. [Google Scholar] [CrossRef]
- Shukla, R.; Upton, K.R.; Munoz-Lopez, M.; Gerhardt, D.J.; Fisher, M.E.; Nguyen, T.; Brennan, P.M.; Baillie, J.K.; Collino, A.; Ghisletti, S.; et al. Endogenous Retrotransposition Activates Oncogenic Pathways in Hepatocellular Carcinoma. Cell 2013, 153, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Schauer, S.N.; Carreira, P.E.; Shukla, R.; Gerhardt, D.J.; Gerdes, P.; Sanchez-Luque, F.J.; Nicoli, P.; Kindlova, M.; Ghisletti, S.; Dos Santos, A.; et al. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res. 2018, 28, 639–653. [Google Scholar] [CrossRef]
- Petrizzo, A.; Ragone, C.; Cavalluzzo, B.; Mauriello, A.; Manolio, C.; Tagliamonte, M.; Buonaguro, L. Human Endogenous Retrovirus Reactivation: Implications for Cancer Immunotherapy. Cancers 2021, 13, 1999. [Google Scholar] [CrossRef]
- Kong, Y.; Rose, C.M.; Cass, A.A.; Williams, A.G.; Darwish, M.; Lianoglou, S.; Haverty, P.M.; Tong, A.J.; Blanchette, C.; Albert, M.L.; et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 2019, 10, 5228. [Google Scholar] [CrossRef] [PubMed]
- Canadas, I.; Thummalapalli, R.; Kim, J.W.; Kitajima, S.; Jenkins, R.W.; Christensen, C.L.; Campisi, M.; Kuang, Y.N.; Zhang, Y.X.; Gjini, E.; et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 2018, 24, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Au, L.; Hatipoglu, E.; de Massy, M.R.; Litchfield, K.; Beattie, G.; Rowan, A.; Schnidrig, D.; Thompson, R.; Byrne, F.; Horswell, S.; et al. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell 2021, 39, 1497–1518.e1411. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.L.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Roulois, D.; Yau, H.L.; Singhania, R.; Wang, Y.D.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.N.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Hsu, M.H.; Tu, S.J.; Yen, J.C.; Lee, Y.T.; Fang, H.Y.; Chang, J.G. Metatranscriptomic Analysis of Human Lung Metagenomes from Patients with Lung Cancer. Genes 2021, 12, 1458. [Google Scholar] [CrossRef] [PubMed]
- Bendall, M.L.; de Mulder, M.; Iniguez, L.P.; Lecanda-Sanchez, A.; Perez-Losada, M.; Ostrowski, M.A.; Jones, R.B.; Mulder, L.C.F.; Reyes-Teran, G.; Crandall, K.A.; et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput. Biol. 2019, 15, e1006453. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; pp. 65–90. [Google Scholar]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 24 August 2022).
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e1323. [Google Scholar] [CrossRef]
- Sheng, W.Q.; LaFleur, M.W.; Nguyen, T.H.; Chen, S.J.; Chakravarthy, A.; Conway, J.R.; Li, Y.; Chen, H.; Yang, H.; Hsu, P.H.; et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 2018, 174, 549–563.e519. [Google Scholar] [CrossRef]
- Geis, F.K.; Goff, S.P. Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview. Viruses 2020, 12, 884. [Google Scholar] [CrossRef] [PubMed]
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018, 28, 836–845. [Google Scholar] [CrossRef]
- Guo, E.S.; Xiao, R.R.; Wu, Y.F.; Lu, F.N.; Liu, C.; Yang, B.; Li, X.; Fu, Y.; Wang, Z.Z.; Li, Y.; et al. WEE1 inhibition induces anti-tumor immunity by activating ERV and the dsRNA pathway. J. Exp. Med. 2021, 219, e20210789. [Google Scholar] [CrossRef]
- Zhou, X.L.; Singh, M.; Santos, G.S.; Guerlavais, V.; Carvajal, L.A.; Aivado, M.; Zhan, Y.; Oliveira, M.M.S.; Westerberg, L.S.; Annis, D.A.; et al. Pharmacologic Activation of p53 Triggers Viral Mimicry Response Thereby Abolishing Tumor Immune Evasion and Promoting Antitumor Immunity. Cancer Discov. 2021, 11, 3090–3105. [Google Scholar] [CrossRef] [PubMed]
- Foroushani, A.K.; Chim, B.; Wong, M.; Rastegar, A.; Smith, P.T.; Wang, S.F.; Barbian, K.; Martens, C.; Hafner, M.; Muljo, S.A. Posttranscriptional regulation of human endogenous retroviruses by RNA-binding motif protein 4, RBM4. Proc. Natl. Acad. Sci. USA 2020, 117, 26520–26530. [Google Scholar] [CrossRef]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015, 522, 221–225. [Google Scholar] [CrossRef]
- Srour, N.; Villarreal, O.D.; Hardikar, S.; Yu, Z.B.; Preston, S.; Jr, W.H.M.; Szewczyk, M.M.; Barsyte-Lovejoy, D.; Xu, H.; Chen, T.P.; et al. PRMT7 ablation stimulates anti-tumor immunity and sensitizes melanoma to immune checkpoint blockade. Cell Rep. 2022, 38, 110582. [Google Scholar] [CrossRef]
- Berta, D.G.; Kuisma, H.; Valimaki, N.; Raisanen, M.; Jantti, M.; Pasanen, A.; Karhu, A.; Kaukomaa, J.; Taira, A.; Cajuso, T.; et al. Deficient H2A.Z deposition is associated with genesis of uterine leiomyoma. Nature 2021, 596, 398–403. [Google Scholar] [CrossRef]
- Ohtani, H.; Liu, M.M.; Zhou, W.D.; Liang, G.N.; Jones, P.A. Switching roles for DNA and histone methylation depend on evolutionary ages of human endogenous retroviruses. Genome Res. 2018, 28, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Groh, S.; Milton, A.V.; Marinelli, L.; Sickinger, C.V.; Russo, A.; Bollig, H.; de Almeida, G.P.; Schmidt, A.; Forne, I.; Imhof, A.; et al. Morc3 silences endogenous retroviruses by enabling Daxx-mediated histone H3.3 incorporation. Nat. Commun. 2021, 12, 5996. [Google Scholar] [CrossRef]
- Yu, C.H.; Lei, X.Y.; Chen, F.; Mao, S.; Lv, L.; Liu, H.L.; Hu, X.Y.; Wang, R.H.; Shen, L.C.; Zhang, N.; et al. ARID1A loss derepresses a group of human endogenous retrovirus-H loci to modulate BRD4-dependent transcription. Nat. Commun. 2022, 13, 3501. [Google Scholar] [CrossRef]
- Xu, W.Q.; Li, J.H.; He, C.X.; Wen, J.; Ma, H.H.; Rong, B.W.; Diao, J.B.; Wang, L.Y.; Wang, J.H.; Wu, F.Z.; et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 2021, 591, 317–321. [Google Scholar] [CrossRef]
- Chelmicki, T.; Roger, E.; Teissandier, A.; Dura, M.; Bonneville, L.; Rucli, S.; Dossin, F.; Fouassier, C.; Lameiras, S.; Bourc’his, D. m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591, 312–316. [Google Scholar] [CrossRef]
- Wu, Z.; Zhou, J.; Zhang, X.Y.; Zhang, Z.W.; Xie, Y.T.; Liu, J.B.; Ho, Z.V.; Panda, A.; Qiu, X.T.; Cejas, P.; et al. Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence. Nat. Genet. 2021, 53, 881–894. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.S. Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions. Viruses 2021, 13, 497. [Google Scholar] [CrossRef]
- Ku, Y.; Park, J.H.; Cho, R.; Lee, Y.; Park, H.M.; Kim, M.; Hur, K.; Byun, S.Y.; Liu, J.; Lee, Y.S.; et al. Noncanonical immune response to the inhibition of DNA methylation by Staufen1 via stabilization of endogenous retrovirus RNAs. Proc. Natl. Acad. Sci. USA 2021, 118, e2016289118. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.S.; Zhao, W.W.; Liu, Y.; Tan, X.T.; Li, X.; Zou, Q.; Xiao, Z.T.; Xu, H.; Wang, Y.T.; Yang, X.R. Function of HNRNPC in breast cancer cells by controlling the dsRNA-induced interferon response. EMBO J. 2018, 37, e99017. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Sachs, P.; Ding, D.; Bergmaier, P.; Lamp, B.; Schlagheck, C.; Finkernagel, F.; Nist, A.; Stiewe, T.; Mermoud, J.E. SMARCAD1 ATPase activity is required to silence endogenous retroviruses in embryonic stem cells. Nat. Commun. 2019, 10, 1335. [Google Scholar] [CrossRef]
- Yang, B.; Fang, L.; Gao, Q.Q.; Xu, C.; Xu, J.Q.; Chen, Z.X.; Wang, Y.X.; Yang, P. Species-specific KRAB-ZFPs function as repressors of retroviruses by targeting PBS regions. Proc. Natl. Acad. Sci. USA 2022, 119, e2119415119. [Google Scholar] [CrossRef] [PubMed]
- Imbeault, M.; Helleboid, P.Y.; Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 2017, 543, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Helleboid, P.Y.; Heusel, M.; Duc, J.; Piot, C.; Thorball, C.W.; Coluccio, A.; Pontis, J.; Imbeault, M.; Turelli, P.; Aebersold, R. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J. 2019, 38, e101220. [Google Scholar] [CrossRef]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Cho, U.H.; Hetzer, M.W. Nuclear Periphery Takes Center Stage: The Role of Nuclear Pore Complexes in Cell Identity and Aging. Neuron 2020, 106, 899–911. [Google Scholar] [CrossRef]
- Ding, B.J.; Sepehrimanesh, M. Nucleocytoplasmic Transport: Regulatory Mechanisms and the Implications in Neurodegeneration. Int. J. Mol. Sci. 2021, 22, 4165. [Google Scholar] [CrossRef]
- Vitiello, G.A.F.; Ferreira, W.A.S.; de Lima, V.C.C.; Medina, T.D. Antiviral Responses in Cancer: Boosting Antitumor Immunity Through Activation of Interferon Pathway in the Tumor Microenvironment. Front. Immunol. 2021, 12, 782852. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-stimulated genes: What do they all do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qian, C.; Cao, X. Post-translational modification control of innate immunity. Immunity 2016, 45, 15–30. [Google Scholar] [CrossRef]
- Dilthey, A.T. State-of-the-art genome inference in the human MHC. Int. J. Biochem. Cell B 2021, 131, 105882. [Google Scholar] [CrossRef] [PubMed]
- Curdy, N.; Lanvin, O.; Laurent, C.; Fournie, J.J.; Franchini, D.M. Regulatory Mechanisms of Inhibitory Immune Checkpoint Receptor Expression. Trends Cell Biol. 2019, 29, 777–790. [Google Scholar] [CrossRef]
- Zheng, D.P.; Liwinski, T.; Elinav, E. Inflammasome activation and regulation: Toward a better understanding of complex mechanisms. Cell Discov. 2020, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.W.K.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016, 540, 433–437. [Google Scholar] [CrossRef]
- Miranda, A.; Hamilton, P.T.; Zhang, A.W.; Pattnaik, S.; Becht, E.; Mezheyeuski, A.; Bruun, J.; Micke, P.; de Reynies, A.; Nelson, B.H. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. USA 2019, 116, 9020–9029. [Google Scholar] [CrossRef]
- Bidkhori, G.; Benfeitas, R.; Klevstig, M.; Zhang, C.; Nielsen, J.; Uhlen, M.; Boren, J.; Mardinoglu, A. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc. Natl. Acad. Sci. USA 2018, 115, E11874–E11883. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Chang, W.H.; Chang, Y.S.; Yang, J.M.; Chang, C.S.; Hsu, K.C.; Chen, Y.T.; Liu, T.Y.; Chen, Y.C.; Lin, S.Y.; et al. Alternative splicing in human cancer cells is modulated by the amiloride derivative 3,5-diamino-6-chloro-N-(N-(2,6-dichlorobenzoyl)carbamimidoyl)pyrazine-2-carboxide. Mol. Oncol. 2019, 13, 1744–1762. [Google Scholar] [CrossRef]
- Solovyov, A.; Vabret, N.; Arora, K.S.; Snyder, A.; Funt, S.A.; Bajorin, D.F.; Rosenberg, J.E.; Bhardwaj, N.; Ting, D.T.; Greenbaum, B.D. Global Cancer Transcriptome Quantifies Repeat Element Polarization between Immunotherapy Responsive and T Cell Suppressive Classes. Cell Rep. 2018, 23, 512–521. [Google Scholar] [CrossRef]
- Golkaram, M.; Salmans, M.L.; Kaplan, S.; Vijayaraghavan, R.; Martins, M.; Khan, N.; Garbutt, C.; Wise, A.; Yao, J.; Casimiro, S.; et al. HERVs establish a distinct molecular subtype in stage II/III colorectal cancer with poor outcome. Npj Genom. Med. 2021, 6, 13. [Google Scholar] [CrossRef]
- Alcazer, V.; Bonaventura, P.; Tonon, L.; Michel, E.; Mutez, V.; Fabres, C.; Chuvin, N.; Boulos, R.; Estornes, Y.; Maguer-Satta, V.; et al. HERVs characterize normal and leukemia stem cells and represent a source of shared epitopes for cancer immunotherapy. Am. J. Hematol. 2022, 97, 1200–1214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Q.; Fang, H.; Gladysz, K.; Barbour, J.A.; Wong, J.W.H. Original Research Overexpression of transposable elements is associated with immune evasion and poor outcome in colorectal cancer. Eur. J. Cancer 2021, 157, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Curty, G.; Menezes, A.N.; Brant, A.C.; Rougvie, M.D.; Moreira, M.A.M.; Soares, M.A. Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression. Cancers 2021, 13, 3513. [Google Scholar] [CrossRef] [PubMed]
- Bendall, M.L.; Francis, J.H.; Shoushtari, A.N.; Nixon, D.F. Specific human endogenous retroviruses predict metastatic potential in uveal melanoma. JCI Insight 2022, 7, e147172. [Google Scholar] [CrossRef]
- Natoli, M.; Gallon, J.; Lu, H.N.; Amgheib, A.; Pinato, D.J.; Mauri, F.A.; Marafioti, T.; Akarca, A.U.; Ullmo, I.; Ip, J.; et al. Transcriptional analysis of multiple ovarian cancer cohorts reveals prognostic and immunomodulatory consequences of ERV expression. J. Immunother. Cancer 2021, 9, e001519. [Google Scholar] [CrossRef]
- Kolbe, A.R.; Bendall, M.L.; Pearson, A.T.; Paul, D.; Nixon, D.F.; Perez-Losada, M.; Crandall, K.A. Human Endogenous Retrovirus Expression Is Associated with Head and Neck Cancer and Differential Survival. Viruses 2020, 12, 956. [Google Scholar] [CrossRef]
- Groh, S.; Schotta, G. Silencing of endogenous retroviruses by heterochromatin. Cell. Mol. Life Sci. 2017, 74, 2055–2065. [Google Scholar] [CrossRef]
- Chang, Y.S.; Tu, S.J.; Chen, H.D.; Chung, C.C.; Hsu, M.H.; Chou, Y.P.; Lee, Y.T.; Yen, J.C.; Jeng, L.B.; Chang, J.G. Whole genome and RNA sequencing analyses for 254 Taiwanese hepatocellular carcinomas. Biomark. Res. 2023, 11, 68. [Google Scholar] [CrossRef]
- Warkocki, Z. An update on post-transcriptional regulation of retrotransposons. FEBS Lett. 2022, 597, 380–406. [Google Scholar] [CrossRef]
- Bacon, C.W.; Challa, A.; Hyder, U.; Shukla, A.; Borkar, A.N.; Bayo, J.; Liu, J.; Wu, S.Y.; Chiang, C.M.; Kutateladze, T.G.; et al. KAP1 Is a Chromatin Reader that Couples Steps of RNA Polymerase II Transcription to Sustain Oncogenic Programs. Mol. Cell 2020, 78, 1133–1151.e1114. [Google Scholar] [CrossRef]
- Vibert, J.; Saulnier, O.; Collin, C.; Petit, F.; Borgman, K.J.E.; Vigneau, J.; Gautier, M.; Zaidi, S.; Pierron, G.; Watson, S.; et al. Oncogenic chimeric transcription factors drive tumor-specific transcription, processing, and translation of silent genomic regions. Mol. Cell 2022, 82, 2458–2471.e2459. [Google Scholar] [CrossRef] [PubMed]
- Randolph, K.; Hyder, U.; D’Orso, I. KAP1/TRIM28: Transcriptional Activator and/or Repressor of Viral and Cellular Programs? Front. Cell. Infect. Microbiol. 2022, 12, 834636. [Google Scholar] [CrossRef] [PubMed]
- Kogan, A.A.; Topper, M.J.; Dellomo, A.J.; Stojanovic, L.; McLaughlin, L.J.; Creed, T.M.; Eberly, C.L.; Kingsbury, T.J.; Baer, M.R.; Kessler, M.D.; et al. Activating STING1-dependent immune signaling in TP53 mutant and wild-type acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2022, 119, e2123227119. [Google Scholar] [CrossRef]
- Liu, M.; Jia, L.; Li, H.; Liu, Y.; Han, J.; Wang, X.; Li, T.; Li, J.; Zhang, B.; Zhai, X.; et al. p53 Binding Sites in Long Terminal Repeat 5Hs (LTR5Hs) of Human Endogenous Retrovirus K Family (HML-2 Subgroup) Play Important Roles in the Regulation of LTR5Hs Transcriptional Activity. Microbiol. Spectr. 2022, 10, e0048522. [Google Scholar] [CrossRef]
- Zhang, N.; Ashizawa, T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr. Opin. Genet. Dev. 2017, 44, 17–29. [Google Scholar] [CrossRef]
- Rivas, S.R.; Valdez, M.J.M.; Govindarajan, V.; Seetharam, D.; Doucet-O’Hare, T.T.; Heiss, J.D.; Shah, A.H. The Role of HERV-K in Cancer Stemness. Viruses 2022, 14, 2019. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, J.; Vincendeau, M. SnapShot: Human endogenous retroviruses. Cell 2022, 185, 400–400.e1. [Google Scholar] [CrossRef]
- Llovet, J.M.; Castet, F.; Heikenwalder, M.; Maini, M.K.; Mazzaferro, V.; Pinato, D.J.; Pikarsky, E.; Zhu, A.X.; Finn, R.S. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2022, 19, 151–172. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-S.; Hsu, M.-H.; Chung, C.-C.; Chen, H.-D.; Tu, S.-J.; Lee, Y.-T.; Yen, J.-C.; Liu, T.-C.; Chang, J.-G. Comprehensive Analysis and Drug Modulation of Human Endogenous Retrovirus in Hepatocellular Carcinomas. Cancers 2023, 15, 3664. https://doi.org/10.3390/cancers15143664
Chang Y-S, Hsu M-H, Chung C-C, Chen H-D, Tu S-J, Lee Y-T, Yen J-C, Liu T-C, Chang J-G. Comprehensive Analysis and Drug Modulation of Human Endogenous Retrovirus in Hepatocellular Carcinomas. Cancers. 2023; 15(14):3664. https://doi.org/10.3390/cancers15143664
Chicago/Turabian StyleChang, Ya-Sian, Ming-Hon Hsu, Chin-Chun Chung, Hong-Da Chen, Siang-Jyun Tu, Ya-Ting Lee, Ju-Chen Yen, Ta-Chih Liu, and Jan-Gowth Chang. 2023. "Comprehensive Analysis and Drug Modulation of Human Endogenous Retrovirus in Hepatocellular Carcinomas" Cancers 15, no. 14: 3664. https://doi.org/10.3390/cancers15143664
APA StyleChang, Y.-S., Hsu, M.-H., Chung, C.-C., Chen, H.-D., Tu, S.-J., Lee, Y.-T., Yen, J.-C., Liu, T.-C., & Chang, J.-G. (2023). Comprehensive Analysis and Drug Modulation of Human Endogenous Retrovirus in Hepatocellular Carcinomas. Cancers, 15(14), 3664. https://doi.org/10.3390/cancers15143664




