The Prevalence and Molecular Landscape of Lynch Syndrome in the Affected and General Population
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
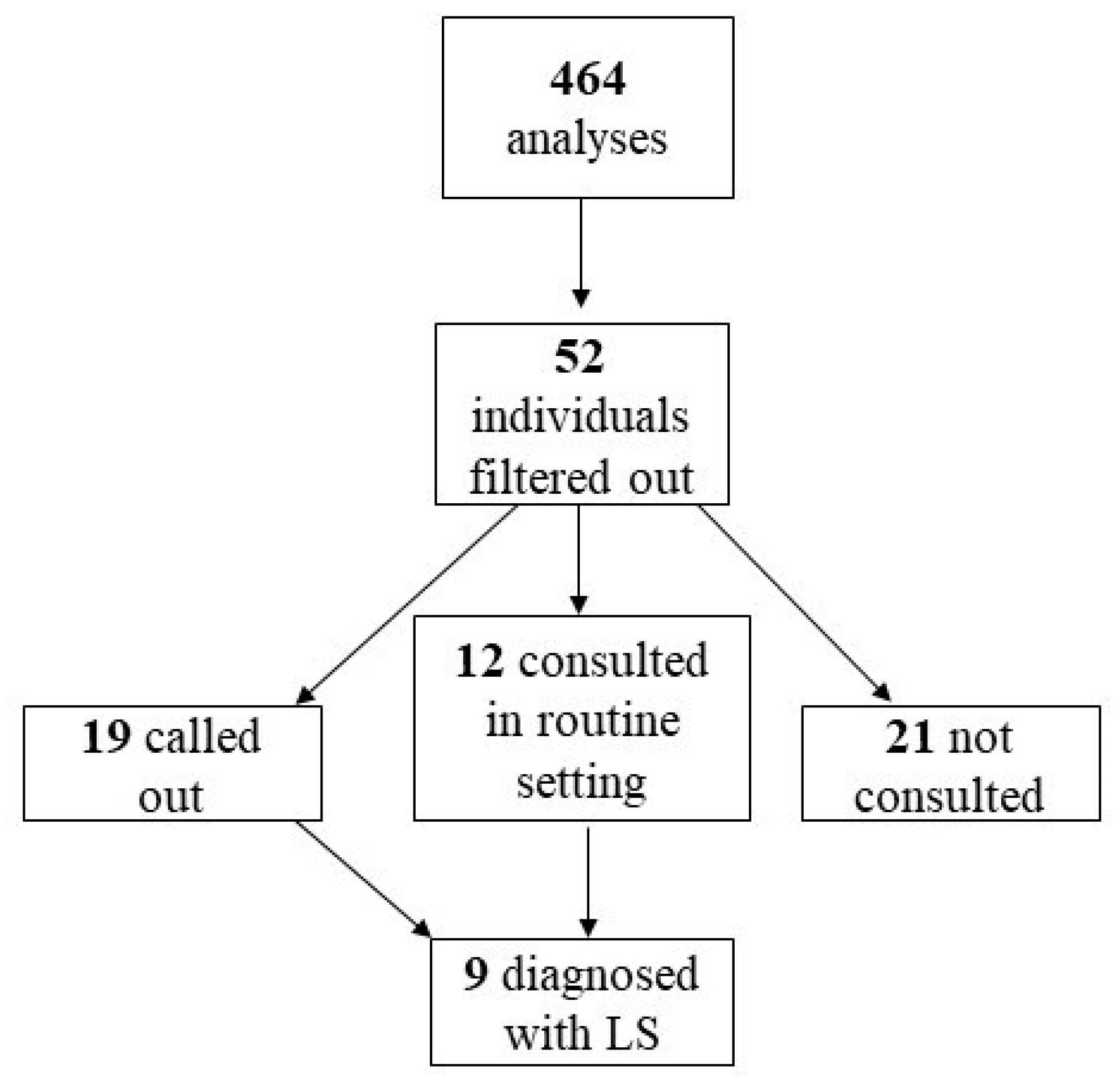
2.1. Study Group
2.2. Diagnostic Cohort
2.3. EstBB Cohort
2.4. MMR IHC Pilot Study
3. Methods
3.1. Molecular Methods
3.2. Statistical Methods
3.3. MMR IHC
4. Results
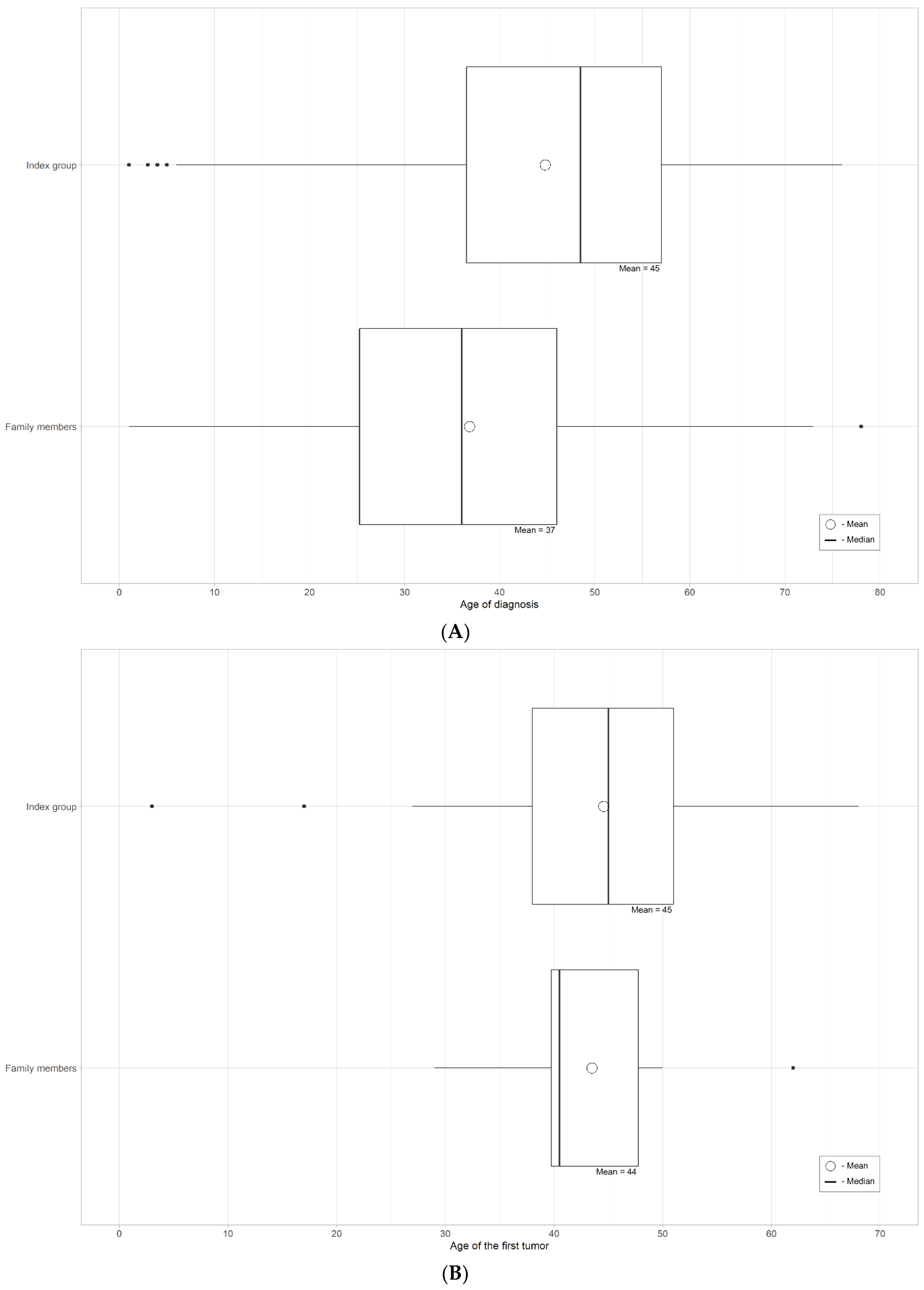
4.1. Results of the Detailed Clinical Information
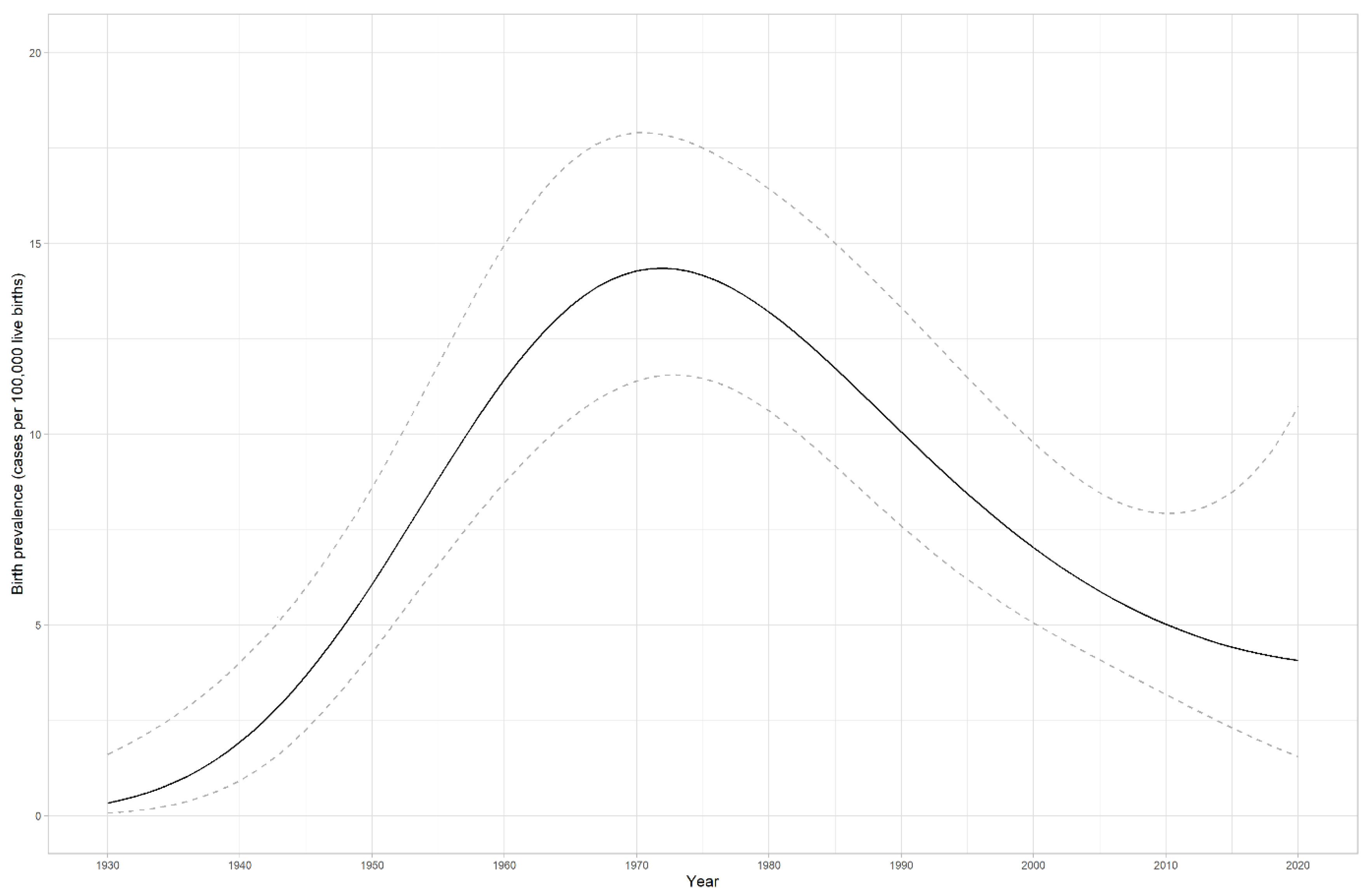
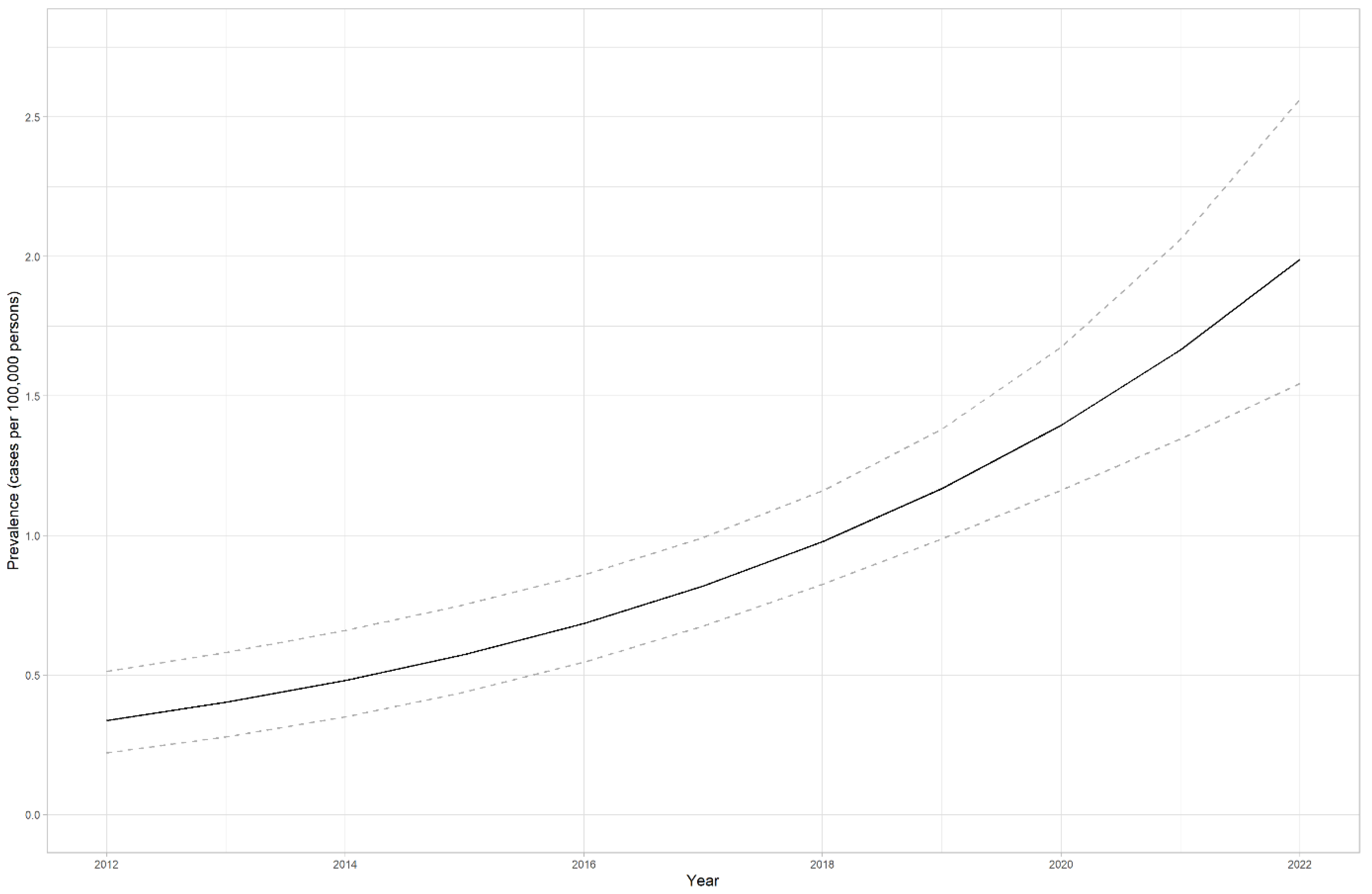
4.2. LS prevalence in Estonia
4.3. Clinical Aspects
4.4. MMR Genes Pathogenic Variants in the Estonian Population
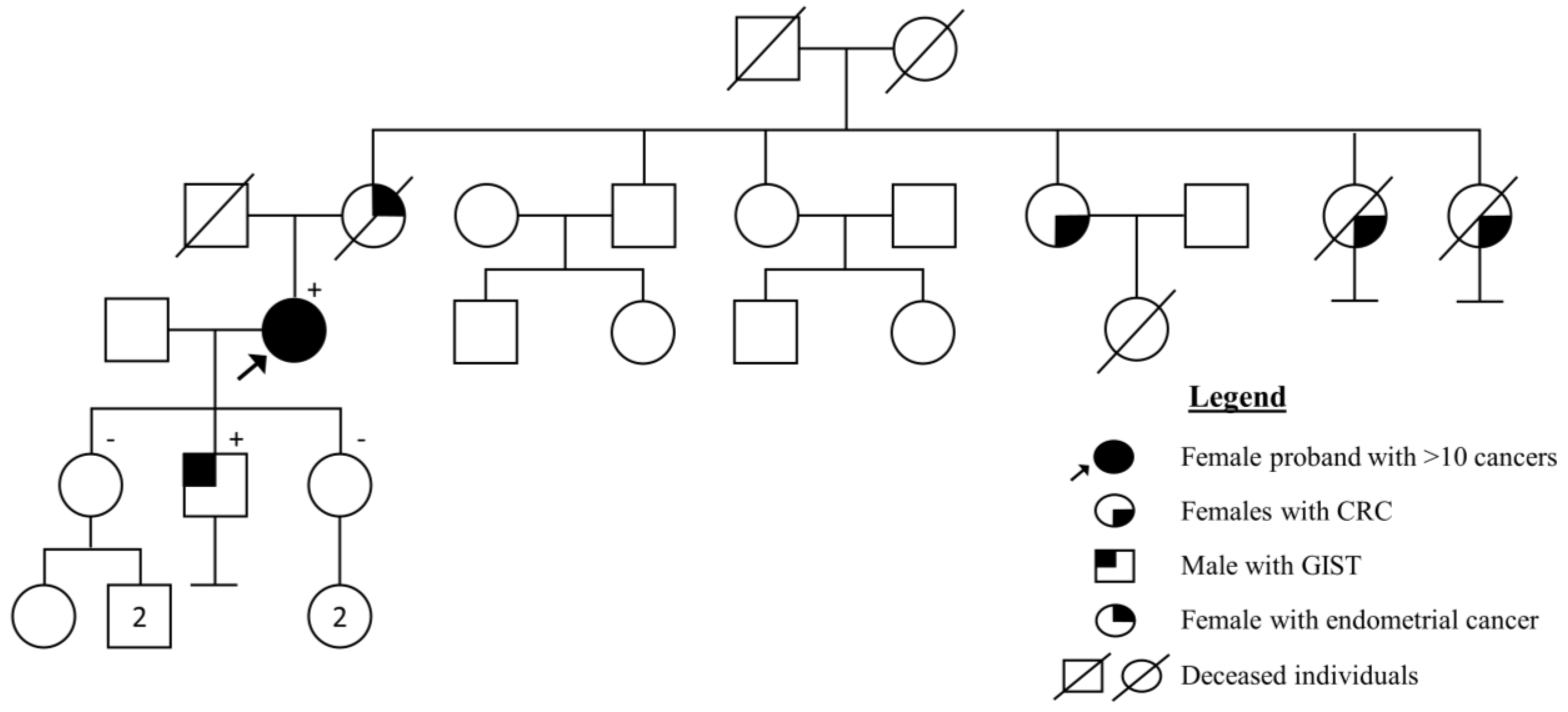
4.5. Case Report: Highest Number of Cancers in Health History
4.6. MMR IHC Pilot Study
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.C.; Bodmer, W.F. Genomic landscape of colorectal carcinogenesis. J. Cancer Res. Clin. Oncol. 2022, 148, 533–545. [Google Scholar] [CrossRef]
- Abu-Ghazaleh, N.; Kaushik, V.; Gorelik, A.; Jenkins, M.; Macrae, F. Worldwide prevalence of Lynch syndrome in patients with colorectal cancer: Systematic review and meta-analysis. Genet. Med. 2022, 24, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Snyder, C.L.; Shaw, T.G.; Heinen, C.D.; Hitchins, M.P. Milestones of Lynch syndrome: 1895-2015. Nat. Rev. Cancer 2015, 15, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.H.; Syngal, S.; Yurgelun, M.B. Recent advances in Lynch syndrome. Fam. Cancer 2019, 18, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.H.; Horiguchi, M.; Uno, H.; Ukaegbu, C.; Syngal, S.; Yurgelun, M.B. Familial Burden and Other Clinical Factors Associated with Various Types of Cancer in Individuals With Lynch Syndrome. Gastroenterology 2021, 161, 143–150 e144. [Google Scholar] [CrossRef]
- Ponti, G.; Castellsague, E.; Ruini, C.; Percesepe, A.; Tomasi, A. Mismatch repair genes founder mutations and cancer susceptibility in Lynch syndrome. Clin. Genet. 2015, 87, 507–516. [Google Scholar] [CrossRef]
- Peltomaki, P.; Olkinuora, A.; Nieminen, T.T. Updates in the field of hereditary nonpolyposis colorectal cancer. Expert. Rev. Gastroenterol. Hepatol. 2020, 14, 707–720. [Google Scholar] [CrossRef]
- Kunkel, T.A. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 91–101. [Google Scholar] [CrossRef]
- Pecina-Slaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Roht, L.; Pajusalu, S.; Samarina, U.; Kahre, T.; Ounap, K. The prevalence of MMR gene mutations in Estonian general population. Eur. J. Hum. Genet. 2020, 28, 530. [Google Scholar]
- Jurgens, H.; Roht, L.; Leitsalu, L.; Noukas, M.; Palover, M.; Nikopensius, T.; Reigo, A.; Kals, M.; Kallak, K.; Kutner, R.; et al. Precise, Genotype-First Breast Cancer Prevention: Experience with Transferring Monogenic Findings From a Population Biobank to the Clinical Setting. Front. Genet. 2022, 13, 881100. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shiel, J.A.; Thomas, N.S.; Abeysinghe, S.; Krawczak, M.; Cooper, D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003, 21, 577–581. [Google Scholar] [CrossRef]
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002, 30, e57. [Google Scholar] [CrossRef]
- Fowler, A.; Mahamdallie, S.; Ruark, E.; Seal, S.; Ramsay, E.; Clarke, M.; Uddin, I.; Wylie, H.; Strydom, A.; Lunter, G.; et al. Accurate clinical detection of exon copy number variants in a targeted NGS panel using DECoN. Wellcome Open Res. 2016, 1, 20. [Google Scholar] [CrossRef]
- Roht, L.; Tooming, M.; Rekker, K.; Roomere, H.; Toome, K.; Murumets, U.; Samarina, U.; Ounap, K.; Kahre, T. The prevalence of germline pathogenic variants in Estonian colorectal cancer patients: Results from routine clinical setting 2016-2021. Front. Genet. 2022, 13, 1020543. [Google Scholar] [CrossRef]
- Leitsalu, L.; Palover, M.; Sikka, T.T.; Reigo, A.; Kals, M.; Parn, K.; Nikopensius, T.; Esko, T.; Metspalu, A.; Padrik, P.; et al. Genotype-first approach to the detection of hereditary breast and ovarian cancer risk, and effects of risk disclosure to biobank participants. Eur. J. Hum. Genet. 2021, 29, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Begaud, B.; Martin, K.; Abouelfath, A.; Tubert-Bitter, P.; Moore, N.; Moride, Y. An easy to use method to approximate Poisson confidence limits. Eur. J. Epidemiol. 2005, 20, 213–216. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- McCarthy, A.J.; Capo-Chichi, J.M.; Spence, T.; Grenier, S.; Stockley, T.; Kamel-Reid, S.; Serra, S.; Sabatini, P.; Chetty, R. Heterogenous loss of mismatch repair (MMR) protein expression: A challenge for immunohistochemical interpretation and microsatellite instability (MSI) evaluation. J. Pathol. Clin. Res. 2019, 5, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Soplepmann, J.; Laidre, P. Teenage colorectal polyposis and cancer may be caused by constitutional mismatch repair deficiency (CMMRD). Acta Oncol. 2016, 55, 1503–1505. [Google Scholar] [CrossRef] [PubMed]
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531. [Google Scholar] [CrossRef]
- Win, A.K.; Jenkins, M.A.; Dowty, J.G.; Antoniou, A.C.; Lee, A.; Giles, G.G.; Buchanan, D.D.; Clendenning, M.; Rosty, C.; Ahnen, D.J.; et al. Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2017, 26, 404–412. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, Z.; Huang, T.; Tam, B.; Ruan, Y.; Guo, M.; Wu, X.; Li, J.; Zhao, B.; Chian, J.S.; et al. Prevalence and spectrum of DNA mismatch repair gene variation in the general Chinese population. J. Med. Genet. 2022, 59, 652–661. [Google Scholar] [CrossRef]
- Lagerstedt-Robinson, K.; Rohlin, A.; Aravidis, C.; Melin, B.; Nordling, M.; Stenmark-Askmalm, M.; Lindblom, A.; Nilbert, M. Mismatch repair gene mutation spectrum in the Swedish Lynch syndrome population. Oncol. Rep. 2016, 36, 2823–2835. [Google Scholar] [CrossRef]
- Lin-Hurtubise, K.M.; Yheulon, C.G.; Gagliano, R.A., Jr.; Lynch, H.T. Excess of extracolonic non-endometrial multiple primary cancers in MSH2 germline mutation carriers over MLH1. J. Surg. Oncol. 2013, 108, 433–437. [Google Scholar] [CrossRef]
- Moller, P.; Seppala, T.; Dowty, J.G.; Haupt, S.; Dominguez-Valentin, M.; Sunde, L.; Bernstein, I.; Engel, C.; Aretz, S.; Nielsen, M.; et al. Colorectal cancer incidences in Lynch syndrome: A comparison of results from the prospective lynch syndrome database and the international mismatch repair consortium. Hered. Cancer Clin. Pr. 2022, 20, 36. [Google Scholar] [CrossRef]

| Total | Not Enough Detailed Information | Healthy or Benign Changes | One Cancer in Health History | Two Cancers at Different Time or Site in Health History | Three or More Cancers in Health History | Most Frequent Cancer Site | Mean Age of First Cancer | Mean Age of LS Diagnosis | MMR IHC Performed | |
|---|---|---|---|---|---|---|---|---|---|---|
| Diagnostic cohort; index cases | 71 | 3 | 18 | 29 | 15 | 6 | CRC | 44.8 years | 44.6 years | 26/50 (52%) for cancer cases; 10/26 (38.5%) MLH1 and PMS2 negative expression |
| Diagnostic cohort; family members | 48 | None | 40 | 7 | 1 | None | CRC | 43.5 years | 36.8 years | 2/8 (25%) for cancer cases |
| Gene | Variant | No. of Individuals (%) | Exon/Intron Position | Class of Variant Based on ACMG Criteria |
|---|---|---|---|---|
| NM_000249.4(MLH1) | ||||
| MLH1 | c.1976G>C, p.(Arg659Pro) ‡ | 13 (31.7%) | 17 | Pathogenic |
| MLH1 | c.1668-1G>T, p.? ‡ | 4 (9.6%) | Intron 14 | Likely pathogenic |
| MLH1 | c.55A>T, p.(I19F) | 4 (9.6%) | 1 | Pathogenic |
| MLH1 | c.92C>T, p.(Ala31Asp) | 3 (7.32%) | 1 | Likely Pathogenic |
| MLH1 | c.751del, p.(Tyr251Thrfs*3) | 3 (7.32%) | 9 | Pathogenic |
| MLH1 | c.1168delG, p.(Glu390Asnfs*11) † | 2 (4.8%) | 12 | New in this study Likely pathogenic |
| MLH1 | c.1918C>T, p.(Pro640Ser) | 2 (4.8%) | 17 | Likely pathogenic |
| MLH1 | c.2128_2131dupAACT, p.(Ser711*) † | 1 (2.4%) | 19 | New in this study Likely pathogenic |
| MLH1 | c.146T>A, p.(Val49Glu) | 1 (2.4%) | 2 | Pathogenic |
| MLH1 | c.840T>G, p.(Tyr280*) | 1 (2.4%) | 10 | Pathogenic |
| MLH1 | c.1685A>C p.(Gln562Pro) | 1 (2.4%) | 15 | Likely pathogenic |
| NM_000251.3(MSH2) | ||||
| MSH2 | c.1283_1284delAC, p.(His428Profs*14) | 4 (14.8%) | 8 | Likely pathogenic |
| MSH2 | MSH2 exon 9–15 deletion NC_000002.11:g.(?_47690170)_(47708011_?)del † | 4 (14.8%) | Deletion ex 9–15 | New in this study Likely pathogenic |
| MSH2 | c.793-1G>A, p.? ‡ | 3 (11.1%) | Intron 4 | Likely pathogenic |
| MSH2 | MSH2 exon 8 deletion NC_000002.11:g.(?_47669476)_(47710098_?)del | 2 (7.4%) | Deletion ex 8 | Pathogenic |
| MSH2 | c.1164_1165delinsGT, p.(Asn388_Arg389delinsLys*) | 2 (7.4%) | 7 | Pathogenic |
| MSH2 | c.289C>T, p.(Gln97*) | 2 (7.4%) | 2 | Pathogenic |
| MSH2 | c.1661+5G>A, p.? | 1 (3.7%) | 10 | Likely pathogenic |
| MSH2 | MSH2 exon 11–14 deletion NC_000002.12:g.(?_47698104)_(47705658_?)del | 1 (3.7%) | Deletion ex 11–14 | Pathogenic |
| MSH2 | c.942+3A>T, p.? | 1 (3.7%) | Intron 5 | Pathogenic |
| MSH2 | c.942+1G>T, p.? | 1 (3.7%) | Intron 5 | Likely pathogenic |
| MSH2 | c.181C>T, p.(Gln61*) | 1 (3.7%) | 1 | Pathogenic |
| MSH2 | c.2131C>T, p.(Arg711*) ‡ | 1 (3.7%) | 13 | Pathogenic |
| MSH2 | c.1942dupA, p.(Ile648Asn*fs6) † | 1 (3.7%) | 12 | New in this study Likely pathogenic |
| MSH2+EPCAM | MSH2 exon 1–7 and EPCAM exon 9 deletion NC_000002.12:g.(?_47614711)_(47657080_?)del † | 2 (7.4%) | MSH2 ex. 1–7 EPCAM ex. 9 | New in this study Likely pathogenic |
| MSH2+EPCAM | MSH2 exon 1–6 and EPCAM exon 8–9 deletion NC_000002.12:g.(?_47612305)_(47643568_?)del † | 1 (3.7%) | MSH2 ex. 1–6 EPCAM ex. 8–9 | New in this study Likely pathogenic |
| NM_000179.3(MSH6) | ||||
| MSH6 | c.3226C>T, p.(Arg1076Cys) | 14 (43.75%) | 5 | Likely pathogenic |
| MSH6 | c.3514dupA, p.(Arg1172Lysfs*5) | 4 (12.5%) | 6 | Pathogenic |
| MSH6 | c.2419G>T, p.(Glu807*) | 3 (9.4%) | 4 | Pathogenic |
| MSH6 | c.1998dupT, p.(Asp667*) † | 3 (9.4%) | 4 | New in this study Likely pathogenic |
| MSH6 | c.3725G>A, p.(Arg1242His) | 1 (3.1%) | 8 | Pathogenic/Likely pathogenic |
| MSH6 | c.3522dup, p.(Thr1175Tyrfs*2) † | 1 (3.1%) | 6 | New in this study Likely pathogenic |
| MSH6 | c.2308G>T, p.(Gly770Cys) † | 1 (3.1%) | 4 | New in this study Likely pathogenic |
| MSH6 | c.3261del, p.(Phe1088fs) | 1 (3.1%) | 5 | Pathogenic |
| MSH6 | c.3195_3199del, p.(Asn1065Lysfs*5) | 1 (3.1%) | 5 | Pathogenic |
| MSH6 | c.2569_2572del, p.(Asp857Phefs*10) | 1 (3.1%) | 4 | Pathogenic |
| NM_000535.7(PMS2) | ||||
| PMS2 | c.861_864del, p.(Arg287Serfs*19) | 11 (26.2%) | 8 | Pathogenic |
| PMS2 | c.1666del, p.(Glu556Lysfs*39) † | 8 (19%) | 11 | New in this study Likely pathogenic |
| PMS2 | c.703C>T, p.(Gln235*) | 4 (9.5%) | 6 | Pathogenic |
| PMS2 | c.2413C>T, p.(Q805*) | 4 (9.5%) | 14 | Pathogenic |
| PMS2 | c.1939A>T, p.(Lys647*) | 3 (7.14%) | 11 | Pathogenic |
| PMS2 | c.2506del, p.(Glu836Argfs*15) | 2 (4.76%) | 15 | Pathogenic |
| PMS2 | c.2445+1G>T, | 2 (4.76%) | 14 | Pathogenic/Likely pathogenic |
| PMS2 | c.2192_2196del, p.(Leu731Cysfs*3) in mosaic level 10% | 2 (4.76%) | 13 | Pathogenic |
| PMS2 | c.2T>A, (p.Met1?) | 1 (2.4%) | 1 | Pathogenic |
| PMS2 | c.634C>T, p.(Gln212*) | 1 (2.4%) | 6 | Pathogenic |
| PMS2 | c.137G>T, p.(Ser46Ile) | 1 (2.4%) | 2 | Likely pathogenic |
| PMS2 | c.319C>T, p.(Arg107Trp) | 1 (2.4%) | 4 | Likely pathogenic |
| PMS2 | c.1588C>T, p.(Gln530*) | 1 (2.4%) | 11 | Pathogenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roht, L.; Laidre, P.; Tooming, M.; Tõnisson, N.; Nõukas, M.; Nurm, M.; Estonian Biobank Research Team; Roomere, H.; Rekker, K.; Toome, K.; et al. The Prevalence and Molecular Landscape of Lynch Syndrome in the Affected and General Population. Cancers 2023, 15, 3663. https://doi.org/10.3390/cancers15143663
Roht L, Laidre P, Tooming M, Tõnisson N, Nõukas M, Nurm M, Estonian Biobank Research Team, Roomere H, Rekker K, Toome K, et al. The Prevalence and Molecular Landscape of Lynch Syndrome in the Affected and General Population. Cancers. 2023; 15(14):3663. https://doi.org/10.3390/cancers15143663
Chicago/Turabian StyleRoht, Laura, Piret Laidre, Mikk Tooming, Neeme Tõnisson, Margit Nõukas, Miriam Nurm, Estonian Biobank Research Team, Hanno Roomere, Kadri Rekker, Kadri Toome, and et al. 2023. "The Prevalence and Molecular Landscape of Lynch Syndrome in the Affected and General Population" Cancers 15, no. 14: 3663. https://doi.org/10.3390/cancers15143663
APA StyleRoht, L., Laidre, P., Tooming, M., Tõnisson, N., Nõukas, M., Nurm, M., Estonian Biobank Research Team, Roomere, H., Rekker, K., Toome, K., Fjodorova, O., Murumets, Ü., Šamarina, U., Pajusalu, S., Aaspõllu, A., Salumäe, L., Muhu, K., Soplepmann, J., Õunap, K., & Kahre, T. (2023). The Prevalence and Molecular Landscape of Lynch Syndrome in the Affected and General Population. Cancers, 15(14), 3663. https://doi.org/10.3390/cancers15143663






