Analysis of the Peritumoral Tissue Unveils Cellular Changes Associated with a High Risk of Recurrence
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples and Patient Information
2.2. Pathological Assessment
- The healthy tissue located more than 1 cm away from the tumor is referred to as HT.
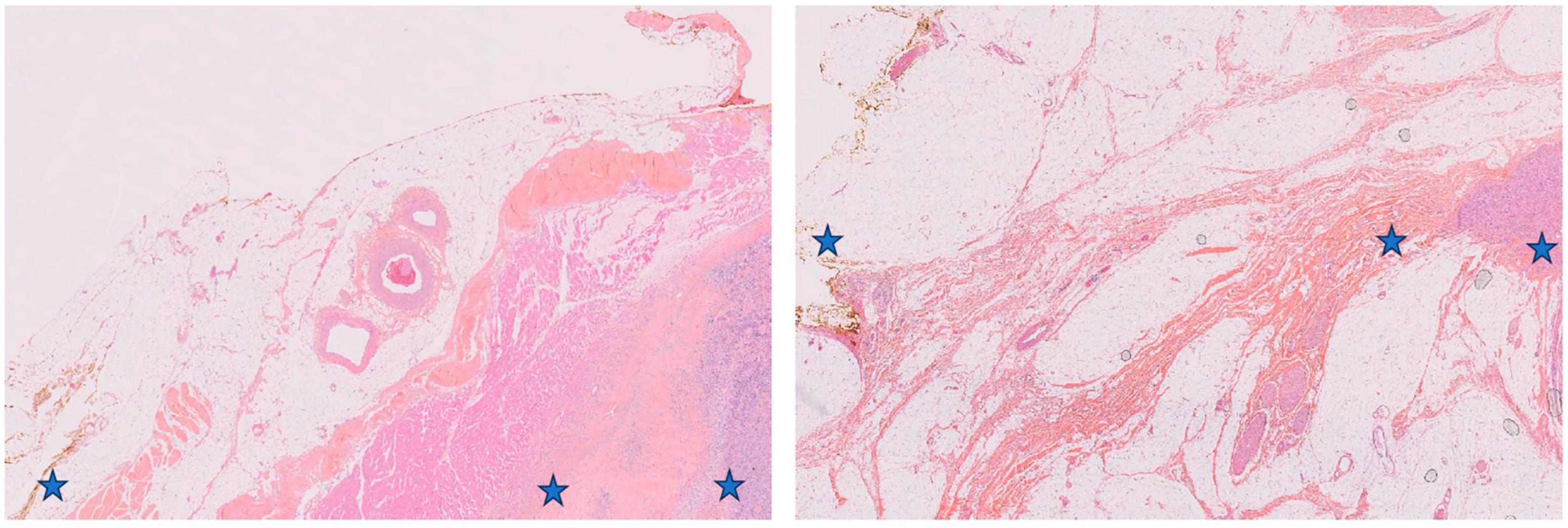
- The tissue that comes into direct contact with the tumor and is free of tumor at the macroscopic level and upon microscopic examination is considered an R1 margin. The term R1 is used to describe this tissue. Additionally, the composition of the capsule was observed.
- The tissue taken from within the tumor mass is referred to as T.
2.3. Nucleic Acid Isolation
2.4. Array CGH
2.5. Gene Expression
2.6. Statistical Analysis
2.7. CIBERSORT
2.8. Immunohistochemistry Stainings
3. Results
3.1. Comparative Transcriptomic Analysis of R1 Areas
3.2. Assessment of GI in Tumor Samples
3.3. Comparative Histological Analysis of R1.h versus R1.t Groups
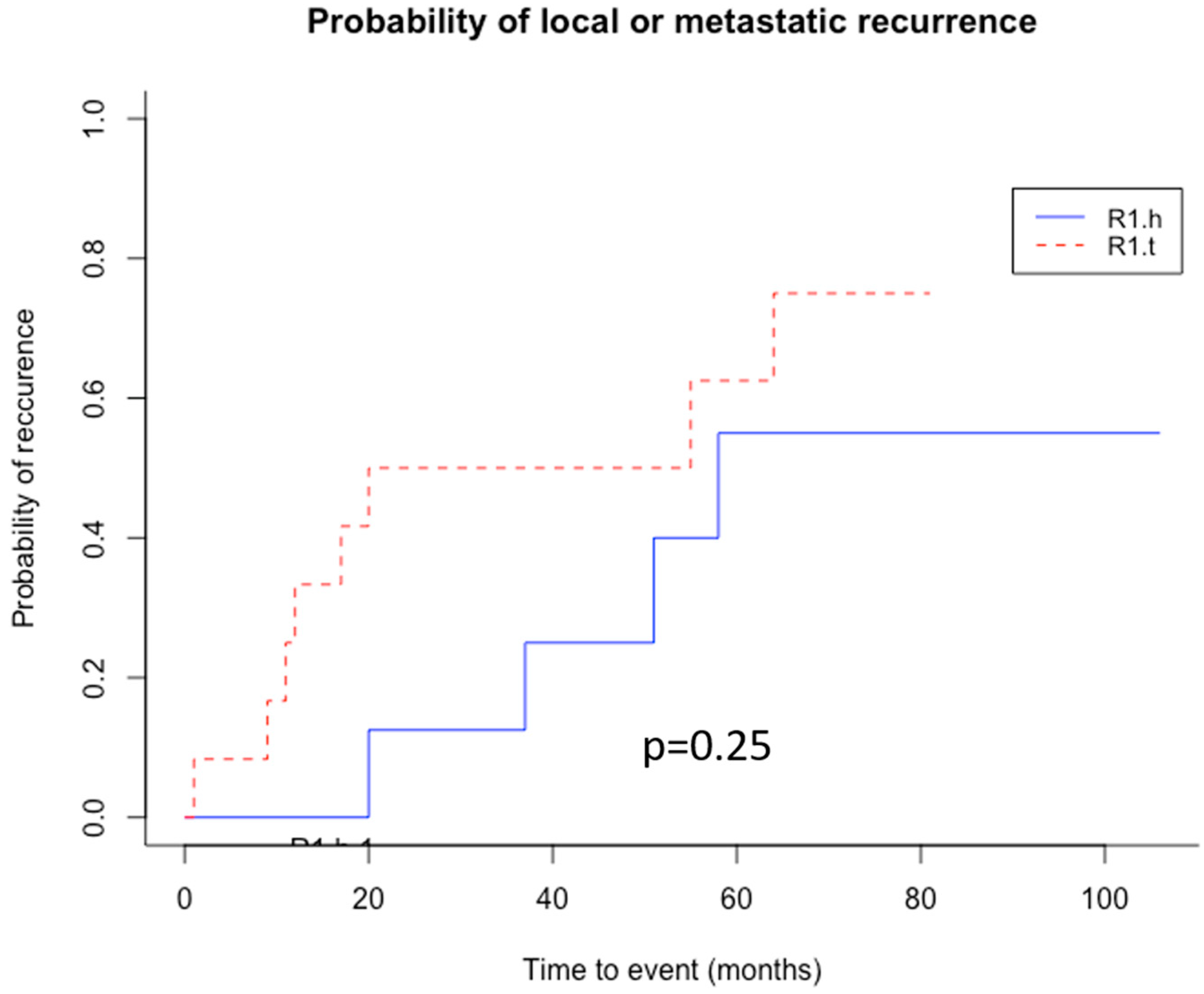
3.4. Clinical Significance of Patient’s Stratification According to Their R1 Tissue Nature
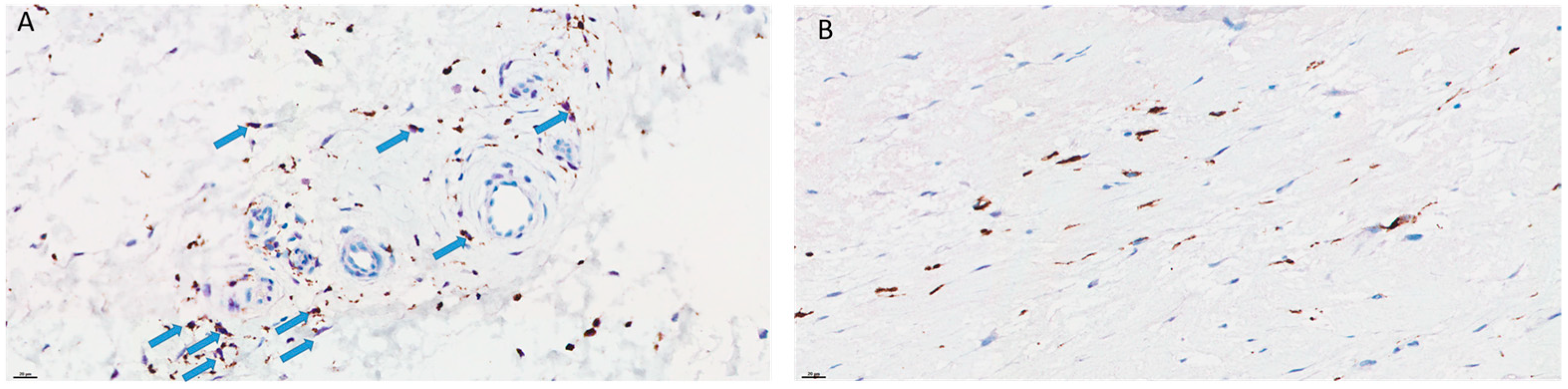
3.5. Comparison of the Immune Reactions between R1.h and R1.t
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| STS | soft tissue sarcoma |
| FNCLCC | fédération nationale des centres de lutte contre le cancer |
| WHO | world health organization |
| HE | hematoxylin and eosin |
| RNA | ribonucleic acid |
| DNA | desoxyribonucleic acid |
| CGH | comparative genomic hybridization |
| ADM-2 | aberration detection method |
| LMS | leiomyosarcoma |
| MFS | myxofibrosarcoma |
| UPS | undifferentiated pleomorphic sarcoma |
| LPS | liposarcoma |
| RMS | rhabdomyosarcoma |
| LFGM | low grade fibromyxoid sarcoma |
| HT | healthy tissue (=TS in Tables/Figures) |
| R1 | healthy tissue at a distance |
| T | tumor (=Tumeur in Tables/Figures) |
| GO | gene ontology |
| ECM | extracellular matrix |
References
- Stiller, C.A.; Trama, A.; Serraino, D.; Rossi, S.; Navarro, C.; Chirlaque, M.D.; Casali, P.G.; RARECARE Working Group. Descriptive Epidemiology of Sarcomas in Europe: Report from the RARECARE Project. Eur. J. Cancer 2013, 49, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Penel, N.; Coindre, J.-M.; Giraud, A.; Terrier, P.; Ranchere-Vince, D.; Collin, F.; Guellec, S.L.E.; Bazille, C.; Lae, M.; de Pinieux, G.; et al. Presentation and Outcome of Frequent and Rare Sarcoma Histologic Subtypes: A Study of 10,262 Patients with Localized Visceral/Soft Tissue Sarcoma Managed in Reference Centers. Cancer 2018, 124, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Stoeckle, E.; Coindre, J.-M.; Kind, M.; Kantor, G.; Bui, B.N. Evaluating Surgery Quality in Soft Tissue Sarcoma. Recent Results Cancer Res. 2009, 179, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Gronchi, A.; Palmerini, E.; Quagliuolo, V.; Martin Broto, J.; Lopez Pousa, A.; Grignani, G.; Brunello, A.; Blay, J.-Y.; Tendero, O.; Diaz Beveridge, R.; et al. Neoadjuvant Chemotherapy in High-Risk Soft Tissue Sarcomas: Final Results of a Randomized Trial From Italian (ISG), Spanish (GEIS), French (FSG), and Polish (PSG) Sarcoma Groups. J. Clin. Oncol. 2020, 38, 2178–2186. [Google Scholar] [CrossRef]
- Chibon, F.; Lagarde, P.; Salas, S.; Pérot, G.; Brouste, V.; Tirode, F.; Lucchesi, C.; de Reynies, A.; Kauffmann, A.; Bui, B.; et al. Validated Prediction of Clinical Outcome in Sarcomas and Multiple Types of Cancer on the Basis of a Gene Expression Signature Related to Genome Complexity. Nat. Med. 2010, 16, 781–787. [Google Scholar] [CrossRef]
- Maretty-Nielsen, K.; Aggerholm-Pedersen, N.; Safwat, A.; Jørgensen, P.H.; Hansen, B.H.; Baerentzen, S.; Pedersen, A.B.; Keller, J. Prognostic Factors for Local Recurrence and Mortality in Adult Soft Tissue Sarcoma of the Extremities and Trunk Wall: A Cohort Study of 922 Consecutive Patients. Acta Orthop. 2014, 85, 323–332. [Google Scholar] [CrossRef]
- Stoeckle, E.; Gardet, H.; Coindre, J.-M.; Kantor, G.; Bonichon, F.; Milbéo, Y.; Thomas, L.; Avril, A.; Bui, B.N. Prospective Evaluation of Quality of Surgery in Soft Tissue Sarcoma. Eur. J. Surg. Oncol. 2006, 32, 1242–1248. [Google Scholar] [CrossRef]
- Bonvalot, S.; Levy, A.; Terrier, P.; Tzanis, D.; Bellefqih, S.; Le Cesne, A.; Le Péchoux, C. Primary Extremity Soft Tissue Sarcomas: Does Local Control Impact Survival? Ann. Surg. Oncol. 2017, 24, 194–201. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours WHO Classification of Tumours, 5th ed.; WHO Classification of Tumours Editorial Board: Geneva, Switzerland, 2020; Volume 3. [Google Scholar]
- Austin, J.L.; Temple, W.J.; Puloski, S.; Schachar, N.S.; Oddone Paolucci, E.; Kurien, E.; Sarkhosh, K.; Mack, L.A. Outcomes of Surgical Treatment Alone in Patients with Superficial Soft Tissue Sarcoma Regardless of Size or Grade. J. Surg. Oncol. 2016, 113, 108–113. [Google Scholar] [CrossRef]
- Bonvalot, S.; Missana, M.-C.; Le Cesne, A.; Missenard, G.; Vanel, D.; Terrier, P.; Le Péchoux, C. Function preserving surgery of limb sarcomas: History and improvements. Cancer Radiother. 2006, 10, 416–424. [Google Scholar] [CrossRef]
- Enneking, W.F.; Spanier, S.S.; Malawer, M.M. The Effect of the Anatomic Setting on the Results of Surgical Procedures for Soft Parts Sarcoma of the Thigh. Cancer 1981, 47, 1005–1022. [Google Scholar] [CrossRef]
- Weskamp, P.; Ufton, D.; Drysch, M.; Wagner, J.M.; Dadras, M.; Lehnhardt, M.; Behr, B.; Wallner, C. Risk Factors for Occurrence and Relapse of Soft Tissue Sarcoma. Cancers 2022, 14, 1273. [Google Scholar] [CrossRef]
- Halama, N.; Zoernig, I.; Berthel, A.; Kahlert, C.; Klupp, F.; Suarez-Carmona, M.; Suetterlin, T.; Brand, K.; Krauss, J.; Lasitschka, F.; et al. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell 2016, 29, 587–601. [Google Scholar] [CrossRef]
- Googe, P.B.; Theocharis, S.; Pergaris, A.; Li, H.; Yan, Y.; McKenna, E.F.; Moschos, S.J. Theragnostic Significance of Tumor-Infiltrating Lymphocytes and Ki67 in BRAFV600-Mutant Metastatic Melanoma (BRIM-3 Trial). Curr. Probl. Cancer 2022, 46, 100862. [Google Scholar] [CrossRef]
- Martínez, R.; Tapia, G.; De Muga, S.; Hernández, A.; Cao, M.G.; Teixidó, C.; Urrea, V.; García, E.; Pedreño-López, S.; Ibarz, L.; et al. Combined Assessment of Peritumoral Th1/Th2 Polarization and Peripheral Immunity as a New Biomarker in the Prediction of BCG Response in Patients with High-Risk NMIBC. Oncoimmunology 2019, 8, 1602460. [Google Scholar] [CrossRef]
- Shen, Z.; Xiao, J.; Wang, J.; Lu, L.; Wan, X.; Cai, X. Peritumoral Ductular Reaction Can Be a Prognostic Factor for Intrahepatic Cholangiocarcinoma. BMC Gastroenterol. 2020, 20, 322. [Google Scholar] [CrossRef]
- White, L.M.; Wunder, J.S.; Bell, R.S.; O’Sullivan, B.; Catton, C.; Ferguson, P.; Blackstein, M.; Kandel, R.A. Histologic Assessment of Peritumoral Edema in Soft Tissue Sarcoma. Int. J. Radiat. Oncol. Biol. Phys. 2005, 61, 1439–1445. [Google Scholar] [CrossRef]
- Fridman, A.; Tainsky, M. Critical Pathways in Cellular Senescence and Immortalization Revealed by Gene Expression Profiling. Oncogene 2008, 27, 5975–5987. [Google Scholar] [CrossRef]
- Braakhuis, B.J.M.; Tabor, M.P.; Kummer, J.A.; Leemans, C.R.; Brakenhoff, R.H. A Genetic Explanation of Slaughter’s Concept of Field Cancerization: Evidence and Clinical Implications. Cancer Res. 2003, 63, 1727–1730. [Google Scholar]
- Leemans, C.R.; Braakhuis, B.J.M.; Brakenhoff, R.H. The Molecular Biology of Head and Neck Cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef]
- Mangiola, A.; Saulnier, N.; De Bonis, P.; Orteschi, D.; Sica, G.; Lama, G.; Pettorini, B.L.; Sabatino, G.; Zollino, M.; Lauriola, L.; et al. Gene Expression Profile of Glioblastoma Peritumoral Tissue: An Ex Vivo Study. PLoS ONE 2013, 8, e57145. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Ke, X.; Xue, C.; Li, S.; Huang, X.; Zhang, B.; Zhou, J. A Nomogram for Predicting Early Recurrence in Patients with High-Grade Gliomas. World Neurosurg. 2022, 164, e619–e628. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, C.; D’Alessio, A.; Sorrentino, S.; Biamonte, F.; Moscato, U.; Mangiola, A.; Sica, G.; Iacopino, F. Immunohistochemical Analysis of DNA Repair- and Drug-Efflux-Associated Molecules in Tumor and Peritumor Areas of Glioblastoma. Int. J. Mol. Sci. 2021, 22, 1620. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.C.; Muñoz-Ballester, C.; Chaunsali, L.; Mills, W.A.; Yang, J.H.; Sontheimer, H.; Robel, S. Potassium and Glutamate Transport Is Impaired in Scar-Forming Tumor-Associated Astrocytes. Neurochem. Int. 2020, 133, 104628. [Google Scholar] [CrossRef]
- Frati, A.; Armocida, D.; Arcidiacono, U.A.; Pesce, A.; D’Andrea, G.; Cofano, F.; Garbossa, D.; Santoro, A. Peritumoral Brain Edema in Relation to Tumor Size Is a Variable That Influences the Risk of Recurrence in Intracranial Meningiomas. Tomography 2022, 8, 1987–1996. [Google Scholar] [CrossRef]
- Guo, X.; Zhu, Y.; Wang, X.; Xu, K.; Hong, Y. Peritumoral Edema Is Associated With Postoperative Hemorrhage and Reoperation Following Vestibular Schwannoma Surgery. Front. Oncol. 2021, 11, 633350. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Lartigue, L.; Neuville, A.; Lagarde, P.; Brulard, C.; Rutkowski, P.; Dei Tos, P.; Wardelmann, E.; Debiec-Rychter, M.; Italiano, A.; Coindre, J.-M.; et al. Genomic Index Predicts Clinical Outcome of Intermediate-Risk Gastrointestinal Stromal Tumours, Providing a New Inclusion Criterion for Imatinib Adjuvant Therapy. Eur. J. Cancer 2015, 51, 75–83. [Google Scholar] [CrossRef]
- Stoeckle, E.; Italiano, A.; Stock, N.; Kind, M.; Kantor, G.; Coindre, J.-M.; Bui, B.N. Surgical margins in soft tissue sarcoma. Bull. Cancer 2008, 95, 1199–1204. [Google Scholar]
- Levy, A.; Bonvalot, S.; Callegaro, D.; Botticella, A.; Le Cesne, A.; Gronchi, A.; Le Péchoux, C. Consistent Margin-Status Reporting in Soft Tissue Sarcoma Is Essential to Establish Risk-Adapted Strategies Integrating Biology and Histotype in Perioperative Treatments. J. Clin. Oncol. 2018, 36, 2357–2358. [Google Scholar] [CrossRef]
- Fujiwara, T.; Sumathi, V.; Parry, M.; Stevenson, J.; Tsuda, Y.; Kaneuchi, Y.; Jeys, L. The Role of Surgical Margin Quality in Myxofibrosarcoma and Undifferentiated Pleomorphic Sarcoma. Eur. J. Surg. Oncol. 2021, 47, 1756–1762. [Google Scholar] [CrossRef]
- Blay, J.-Y.; Honoré, C.; Stoeckle, E.; Meeus, P.; Jafari, M.; Gouin, F.; Anract, P.; Ferron, G.; Rochwerger, A.; Ropars, M.; et al. Surgery in Reference Centers Improves Survival of Sarcoma Patients: A Nationwide Study. Ann. Oncol. 2019, 30, 1143–1153. [Google Scholar] [CrossRef]
- Gronchi, A.; Miah, A.B.; Dei Tos, A.P.; Abecassis, N.; Bajpai, J.; Bauer, S.; Biagini, R.; Bielack, S.; Blay, J.Y.; Bolle, S.; et al. Soft Tissue and Visceral Sarcomas: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up☆. Ann. Oncol. 2021, 32, 1348–1365. [Google Scholar] [CrossRef]
- Mueller, B.M. Different Roles for Plasminogen Activators and Metalloproteinases in Melanoma Metastasis. Curr. Top. Microbiol. Immunol. 1996, 213 (Pt 1), 65–80. [Google Scholar] [CrossRef]
- Luo, W. Nasopharyngeal Carcinoma Ecology Theory: Cancer as Multidimensional Spatiotemporal “Unity of Ecology and Evolution” Pathological Ecosystem. Theranostics 2023, 13, 1607–1631. [Google Scholar] [CrossRef]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer Immunotherapy Comes of Age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef]
- Medrek, C.; Pontén, F.; Jirström, K.; Leandersson, K. The Presence of Tumor Associated Macrophages in Tumor Stroma as a Prognostic Marker for Breast Cancer Patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef]
- Sugimoto, M.; Mitsunaga, S.; Yoshikawa, K.; Kato, Y.; Gotohda, N.; Takahashi, S.; Konishi, M.; Ikeda, M.; Kojima, M.; Ochiai, A.; et al. Prognostic Impact of M2 Macrophages at Neural Invasion in Patients with Invasive Ductal Carcinoma of the Pancreas. Eur. J. Cancer 2014, 50, 1900–1908. [Google Scholar] [CrossRef]
- Jensen, T.O.; Schmidt, H.; Møller, H.J.; Høyer, M.; Maniecki, M.B.; Sjoegren, P.; Christensen, I.J.; Steiniche, T. Macrophage Markers in Serum and Tumor Have Prognostic Impact in American Joint Committee on Cancer Stage I/II Melanoma. J. Clin. Oncol. 2009, 27, 3330–3337. [Google Scholar] [CrossRef]
- Curtius, K.; Wright, N.A.; Graham, T.A. An Evolutionary Perspective on Field Cancerization. Nat. Rev. Cancer 2018, 18, 19–32. [Google Scholar] [CrossRef]
- Polanska, U.M.; Orimo, A. Carcinoma-Associated Fibroblasts: Non-Neoplastic Tumour-Promoting Mesenchymal Cells. J. Cell. Physiol. 2013, 228, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hu, Y.; Liu, T.; Wang, Y. Profiles of Immune Cell Infiltration and Immune-Related Genes in the Tumor Microenvironment of Osteosarcoma Cancer. BMC Cancer 2021, 21, 1345. [Google Scholar] [CrossRef] [PubMed]






| Case ID | Age (yo) | Size (mm) | Location | Tumor Grade | Extension | Type | Margin Status | Evolution | Time to Event (mo) | Follow Up (mo) | Genomic Index | Vital Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 61 | 100 | Limb | 2 | Deep | LMS | R0 | M | 55 | 105 | 61 | DOD |
| 2 | 53 | 80 | Trunk | 3 | Supf | LMS | R0 | M | 12 | 14 | 277 | DOD |
| 3 | 85 | 55 | Limb | 3 | Supf | LMS | R1 | CR | 80 | 49 | Dead | |
| 4 | 62 | 70 | Limb | 3 | Supf | LMS | R0 | M | 1 | 118 | 65 | AWD |
| 5 | 72 | 11 | Limb | 1 | Deep | LMS | R0 | CR | 36 | 109 | NED | |
| 6 | 57 | 80 | Limb | 2 | Deep | MFS | R0 | CR | 97 | 17 | NED | |
| 7 | 46 | 130 | Limb | 3 | Deep | MFS | R0 | M | 37 | 70 | 138 | DOD |
| 8 | 59 | 25 | Limb | 3 | Deep | MFS | R1 | LR | 17 | 33 | 41 | DOD |
| 9 | 66 | 25 | Limb | 3 | Supf | MFS | R0 | M | 20 | 25 | 97 | DOD |
| 10 | 71 | 65 | Trunk | 3 | Supf | UPS | R0 | M | 11 | 18 | 134 | DOD |
| 11 | 65 | 75 | Limb | 3 | Deep | UPS | R0 | LRM | 58 | 61 | 158 | DOD |
| 12 | 54 | 25 | Limb | 3 | Deep | UPS | R0 | CR | 81 | 110 | NED | |
| 13 | 59 | 170 | Trunk | 3 | Deep | UPS | R0 | M | 9 | 10 | 114 | DOD |
| 14 | 75 | 60 | Limb | 3 | Deep | UPS | R0 | CR | 65 | 192 | NED | |
| 15 | 58 | 100 | Limb | 2 | Deep | UPS | R0 | M | 20 | 29 | 16 | DOD |
| 16 | 60 | 110 | Limb | 3 | Deep | UPS | R0 | CR | 106 | 127 | NED | |
| 17 | 76 | 150 | Limb | 3 | Deep | LPS | R1 | M | 64 | 70 | 385 | DOD |
| 18 | 44 | 100 | Limb | 3 | Deep | LGFMS | R0 | CR | 44 | 5 | NED | |
| 19 | 52 | 80 | Limb | 3 | Deep | RMS | R1 | CR | 40 | 9 | Dead | |
| 20 | 75 | 100 | Limb | 2 | Deep | LPS | R1 | LR | 51 | 53 | 49 | DOD |
| abbreviations: | Age in years | Size in mm | Grade FNCLCC | Type of sarcoma (cf abbreviations) | R0 complete microscopic exerese R1 in contact at microscopic level | M metastasis CR complete remission LR local recurrence | Time in months | Follow up in months | DOD dead of disease; Dead of other cause; NED no evolutive disease; AWD alive with disease |
| GO Term | Description | p-Value | FDR q-Value | Enrichment (N, B, n, b) | Corresponding Most Genes Significant |
|---|---|---|---|---|---|
| GO:0030198 | extracellular matrix organization | 4.76 × 10-29 | 5.91 × 10-25 | 4.28 (16,751, 356, 868, 79) | CYP1B1—cytochrome p450, family 1, subfamily b, polypeptide 1 SFRP2—secreted frizzled-related protein 2 ADAMTS18—adam metallopeptidase with thrombospondin type 1 motif, 18 FBN1—fibrillin 1 BGN—biglycan |
| GO:0043062 | extracellular structure organization | 5.82 × 10-29 | 3.61 × 10-25 | 4.27 (16,751, 357, 868, 79) | CYP1B1—cytochrome p450, family 1, subfamily b, polypeptide 1 SFRP2—secreted frizzled-related protein 2 ADAMTS18—adam metallopeptidase with thrombospondin type 1 motif, 18 FBN1—fibrillin 1 BGN—biglycan |
| GO:0002376 | immune system process | 5.44 × 10-19 | 2.25 × 10-15 | 2.01 (16,751, 1586, 868, 165) | CFH—complement factor h LRMP—lymphoid-restricted membrane protein CFB—complement factor b WDFY4—wdfy family member 4 IRF8—interferon regulatory factor 8 |
| GO:0006954 | inflammatory response | 8.22 × 10-15 | 6.81 × 10-12 | 3.12 (16,751, 359, 868, 58) | HFE—hemochromatosis FOLR2—folate receptor 2 (fetal) IL1R1—interleukin 1 receptor, type i CXCR6—chemokine (c-x-c motif) receptor 6 GPR68—g protein-coupled receptor 68 |
| GO:0003012 | muscle system process | 5.5 × 10-14 | 6.82 × 10-10 | 4.99 (16,751, 225, 477, 32) | SNTA1—syntrophin, alpha 1 SLC6A8—solute carrier family 6 (neurotransmitter transporter), member 8 MAP2K6—mitogen-activated protein kinase kinase 6 DTNA—dystrobrevin, alpha HEY2—hairy/enhancer-of-split related with yrpw motif 2 |
| GO:0006936 | muscle contraction | 5.89 × 10-13 | 3.66 × 10-9 | 5.26 (16,751, 187, 477, 28) | SNTA1—syntrophin, alpha 1 MYOM1—myomesin 1 SCN7A—sodium channel, voltage-gated, type vii, alpha subunit SLC6A8—solute carrier family 6 (neurotransmitter transporter), member 8 MAP2K6—mitogen-activated protein kinase kinase 6 |
| GO:0055114 | oxidation-reduction process | 3.98 × 10-8 | 4.95 × 10-5 | 2.17 (16,751, 908, 477, 56) | ACAT1—acetyl-coa acetyltransferase 1 PGM2L1—phosphoglucomutase 2-like 1 NDUFS7—nadh dehydrogenase (ubiquinone) fe-s protein 7, 20kda (nadh-coenzyme q reductase) LYRM7—lyr motif containing 7 COQ9—coenzyme q9 homolog (s. cerevisiae) |
| R1.t | M2 | M0 | Monocytes | Mast Cells Resting | Plasma Cells | B Cells Memory | T Cells CD4 Naive | Neutrophils | T Cells Follicular Helper | M1 | Tregs | NK Cells Resting | T Cells CD4 Memory Activated | Dendritic Cells Resting | Dendritic Cells Activated | T Cells Gamma Delta | B Cells Naive | Eosinophils | Mast Cells Activated | T Cells CD4 Memory Resting | T Cells CD8 | NK Cells Activated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2_R1 | 0.342 | 0 | 0.0141 | 0.17 | 0.0051 | 0.045 | 0 | 0.003 | 0 | 0.01 | 0.03 | 0 | 0 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0.156 | 0.071 | 0.06 |
| 12_R1 | 0.234 | 0.034 | 0.056 | 0.37 | 0 | 0.085 | 0.05 | 0 | 0.015 | 0.01 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.016 | 0.057 | 0.06 |
| 19_HT | 0.016 | 0.083 | 0.3111 | 0.26 | 0.0013 | 0.014 | 0 | 0.001 | 0.005 | 0 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.039 | 0.112 | 0.047 | 0.04 |
| 3_R1 | 0.249 | 0 | 0.113 | 0.18 | 0.0055 | 0 | 0.06 | 0 | 0.031 | 0 | 0 | 0 | 0 | 0.1 | 0.1 | 0 | 0 | 0 | 0 | 0.084 | 0.106 | 0.03 |
| 3_HT | 0.227 | 0 | 0.0503 | 0.33 | 0.0205 | 0.047 | 0 | 0 | 0.025 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.113 | 0.071 | 0.06 |
| 4_HT | 0.222 | 0 | 0.234 | 0.16 | 0.0041 | 0.013 | 0 | 0.064 | 0.008 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.116 | 0.053 | 0.11 |
| 19_R1 | 0.098 | 0 | 0.1881 | 0.03 | 0.0514 | 0.043 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.153 | 0.193 | 0.067 | 0.16 |
| 13_R1 | 0.364 | 0 | 0.1186 | 0 | 0.0015 | 0.007 | 0 | 0.002 | 0.015 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.247 | 0.099 | 0.017 | 0.08 |
| 17_R1 | 0.457 | 0 | 0.064 | 0.02 | 0.0101 | 0.026 | 0 | 0 | 0.005 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.085 | 0.117 | 0.058 | 0.13 |
| 1_R1 | 0.16 | 0.07 | 0.2793 | 0 | 0.0056 | 0.002 | 0 | 0.163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.176 | 0.102 | 0.01 | 0.03 |
| 8_R1 | 0.072 | 0.014 | 0.3202 | 0 | 0.0157 | 0.005 | 0.01 | 0.128 | 0.013 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.245 | 0.015 | 0.007 | 0.13 |
| 9_HT | 0.098 | 0 | 0.2478 | 0 | 0.0101 | 0.021 | 0 | 0.041 | 0 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.09 | 0.161 | 0.114 | 0.18 |
| 5_R1 | 0.366 | 0 | 0.1563 | 0 | 0 | 0.019 | 0 | 0.049 | 0.002 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.152 | 0.106 | 0.028 | 0.11 |
| 4_R1 | 0.285 | 0 | 0.2719 | 0.01 | 0.012 | 0.001 | 0.01 | 0 | 0.001 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.013 | 0.094 | 0.09 | 0.16 |
| 12_HT | 0.424 | 0 | 0.1053 | 0.12 | 0.0317 | 0.039 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.072 | 0.083 | 0.12 |
| 10_R1 | 0.217 | 0.02 | 0.082 | 0.14 | 0.0419 | 0.064 | 0 | 0.006 | 0 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.145 | 0.129 | 0.1 |
| 9_R1 | 0.427 | 0.034 | 0.0464 | 0.09 | 0.0065 | 0.037 | 0 | 0 | 0.003 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.122 | 0.065 | 0.11 |
| R1 h | M2 | M0 | Monocytes | Mast Cells Resting | Plasma Cells | B Cells Memory | T Cells CD4 Naive | Neutrophils | T Cells Follicular Helper | M1 | Tregs | NK Cells Resting | T Cells CD4 Memory Activated | Dendritic Cells Resting | Dendritic Cells Activated | T Cells Gamma Delta | B Cells Naive | Eosinophils | Mast Cells Activated | T Cells CD4 Memory Resting | T Cells CD8 | NK Cells Activated |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16_R1 | 0.283 | 0 | 0.1129 | 0.2 | 0.0807 | 0.039 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.063 | 0.119 | 0.09 |
| 15_HT | 0.319 | 0 | 0.0744 | 0.05 | 0.0533 | 0.008 | 0 | 0 | 0.016 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.131 | 0.09 | 0.078 | 0.16 |
| 15_R1 | 0.33 | 0 | 0.0892 | 0 | 0.0291 | 0.03 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.209 | 0.083 | 0.064 | 0.14 |
| 11_R1 | 0.417 | 0 | 0.0597 | 0.12 | 0.0546 | 0 | 0 | 0 | 0.013 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.033 | 0.078 | 0.046 | 0.15 |
| 8_HT | 0.036 | 0.072 | 0.3012 | 0 | 0.0563 | 0.125 | 0 | 0.068 | 0 | 0 | 0 | 0.07 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.097 | 0.105 | 0.049 | 0.02 |
| 11_HT | 0.11 | 0 | 0.2757 | 0.07 | 0.2012 | 0 | 0 | 0 | 0.076 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0.034 | 0.088 | 0.034 | 0.08 |
| 13_HT | 0.187 | 0 | 0.2923 | 0 | 0.1376 | 0.041 | 0.05 | 0.034 | 0.023 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 | 0 | 0.013 | 0.09 |
| 20_HT | 0.053 | 0 | 0.2066 | 0 | 0.0202 | 0.011 | 0 | 0.099 | 0.04 | 0.04 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.276 | 0.06 | 0.039 | 0.15 |
| 14_HT | 0.153 | 0 | 0.0865 | 0.15 | 0.1158 | 0 | 0 | 0 | 0.068 | 0.04 | 0.05 | 0.15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.163 | 0.021 | 0 |
| 2_HT | 0.167 | 0 | 0.1487 | 0.27 | 0.1484 | 0.046 | 0 | 0 | 0.012 | 0.01 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.103 | 0.08 |
| 17_HT | 0.094 | 0 | 0.2735 | 0.01 | 0.2342 | 0.019 | 0 | 0.042 | 0.028 | 0 | 0.09 | 0.05 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0.055 | 0.053 | 0.04 |
| 5_HT | 0.249 | 0 | 0.1298 | 0.08 | 0.1158 | 0.094 | 0.05 | 0.03 | 0.013 | 0.01 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.037 | 0.066 | 0.12 |
| 14_R1 | 0.202 | 0 | 0.1965 | 0.03 | 0.107 | 0.036 | 0 | 0 | 0.015 | 0 | 0.02 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.091 | 0.061 | 0.075 | 0.16 |
| 6_HT | 0.088 | 0 | 0.169 | 0 | 0.1531 | 0.055 | 0 | 0.027 | 0.033 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.16 | 0.066 | 0.049 | 0.16 |
| 1_HT | 0 | 0.024 | 0.5147 | 0 | 0.027 | 0.053 | 0.02 | 0 | 0.006 | 0 | 0.05 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.087 | 0.045 | 0.024 | 0.13 |
| 7_HT | 0.164 | 0 | 0.3505 | 0.03 | 0.1103 | 0.095 | 0 | 0.009 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.069 | 0.109 | 0.031 | 0.03 |
| 10_HT | 0.197 | 0 | 0.1116 | 0.2 | 0.1466 | 0.075 | 0 | 0 | 0.033 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.068 | 0.067 | 0.1 |
| 18_R1 | 0.198 | 0.006 | 0.2142 | 0.16 | 0.1108 | 0.089 | 0.02 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.092 | 0.08 |
| 18_HT | 0.152 | 0 | 0.1612 | 0 | 0.0866 | 0.059 | 0 | 0 | 0.013 | 0.01 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.178 | 0.101 | 0.087 | 0.14 |
| 6_R1 | 0.479 | 0 | 0.0848 | 0.1 | 0.0106 | 0.058 | 0 | 0 | 0.025 | 0.01 | 0.01 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0.101 | 0.03 | 0.08 |
| 20_R1 | 0.42 | 0 | 0.0456 | 0 | 0.0224 | 0.029 | 0 | 0.005 | 0.028 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.215 | 0.074 | 0.028 | 0.11 |
| 7_R1 | 0.41 | 0 | 0.175 | 0.12 | 0.0188 | 0.058 | 0.06 | 0 | 0 | 0.01 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.114 | 0 | 0.03 |
| 16_HT | 0.354 | 0 | 0.1426 | 0.21 | 0.0301 | 0.021 | 0 | 0 | 0.005 | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.035 | 0.114 | 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michot, A.; Lagarde, P.; Lesluyes, T.; Darbo, E.; Neuville, A.; Baud, J.; Perot, G.; Bonomo, I.; Maire, M.; Michot, M.; et al. Analysis of the Peritumoral Tissue Unveils Cellular Changes Associated with a High Risk of Recurrence. Cancers 2023, 15, 3450. https://doi.org/10.3390/cancers15133450
Michot A, Lagarde P, Lesluyes T, Darbo E, Neuville A, Baud J, Perot G, Bonomo I, Maire M, Michot M, et al. Analysis of the Peritumoral Tissue Unveils Cellular Changes Associated with a High Risk of Recurrence. Cancers. 2023; 15(13):3450. https://doi.org/10.3390/cancers15133450
Chicago/Turabian StyleMichot, Audrey, Pauline Lagarde, Tom Lesluyes, Elodie Darbo, Agnès Neuville, Jessica Baud, Gaëlle Perot, Iris Bonomo, Mathilde Maire, Maxime Michot, and et al. 2023. "Analysis of the Peritumoral Tissue Unveils Cellular Changes Associated with a High Risk of Recurrence" Cancers 15, no. 13: 3450. https://doi.org/10.3390/cancers15133450
APA StyleMichot, A., Lagarde, P., Lesluyes, T., Darbo, E., Neuville, A., Baud, J., Perot, G., Bonomo, I., Maire, M., Michot, M., Coindre, J.-M., Le Loarer, F., & Chibon, F. (2023). Analysis of the Peritumoral Tissue Unveils Cellular Changes Associated with a High Risk of Recurrence. Cancers, 15(13), 3450. https://doi.org/10.3390/cancers15133450







