Late Effects of Chronic Low Dose Rate Total Body Irradiation on the Heart Proteome of ApoE−/− Mice Resemble Premature Cardiac Ageing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Irradiation
2.2. Protein Extraction and Quantification
2.3. Protein-Protein Interaction and Signalling Network
2.4. Immunoblot Analysis
2.5. SMAD and Phospho-SMAD Assay
2.6. Total SIRT Activity Assay
2.7. AMPK Assay
2.8. mTOR Assay
2.9. GSH/GSSG Ratio Detection Assay
2.10. GLRX Activity Assay
2.11. GPX Activity Assay (ab102530)
2.12. Lipid Peroxidation Assay
2.13. Pathway-Focused Gene Expression Profiling with qRT-PCR
2.14. Histology
2.15. Statistical Analysis
2.16. Data Availability
3. Results
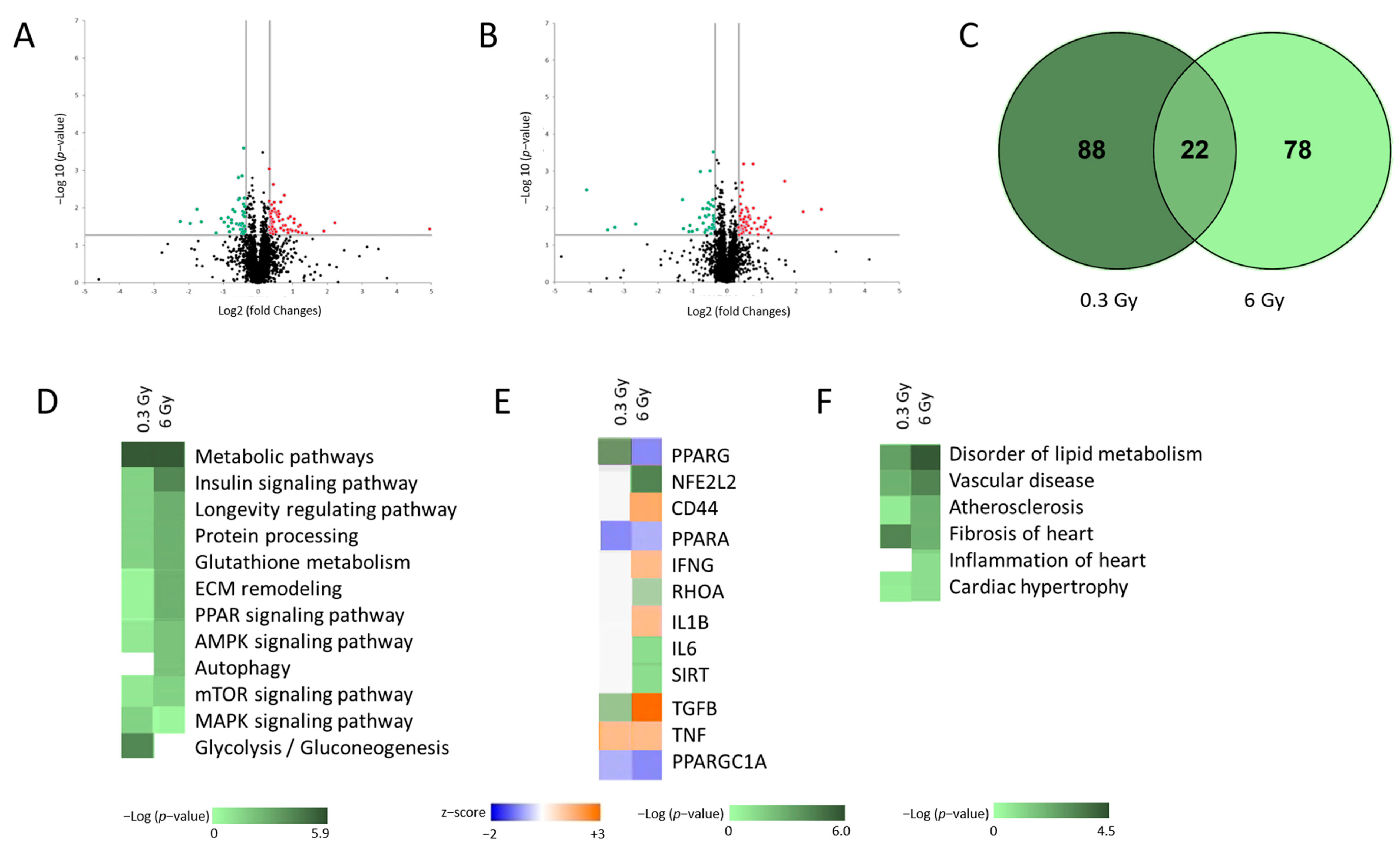
3.1. Low Dose Chronic Irradiation Altered the Cardiac Proteome of ApoE −/− Mice
3.2. Chronic Irradiation Suppresses the Activity of PPARα Signalling Pathway
3.3. Cardiac Oxidative Stress Response Is Affected in Irradiated Hearts
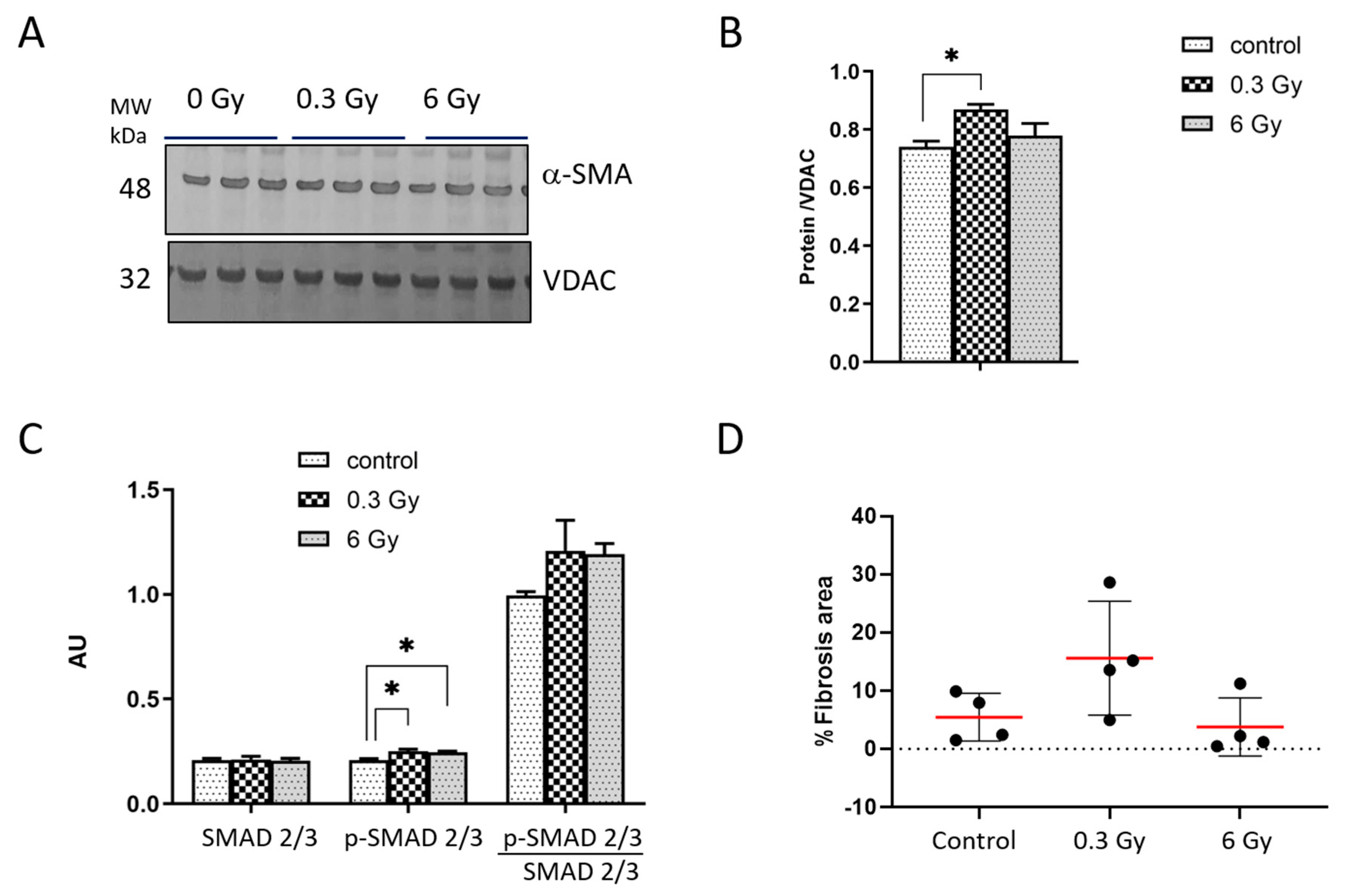
3.4. Chronic Irradiation Affects the Cardiac Fibrogenic Pathway
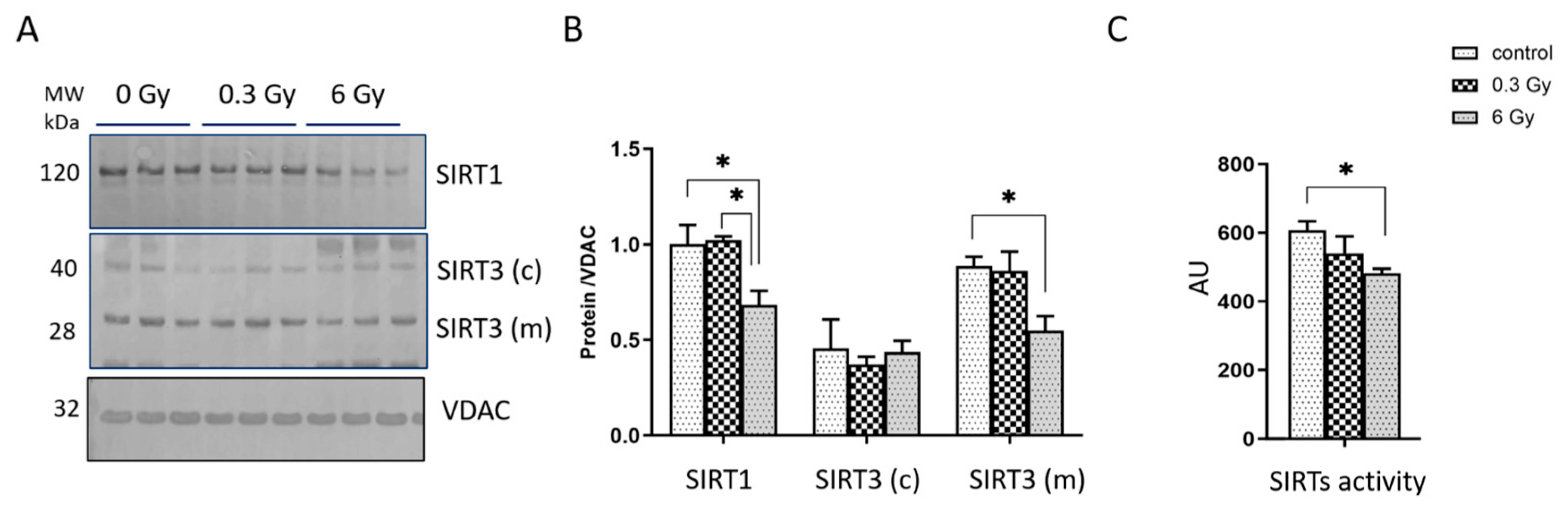
3.5. Expression and Activity of Sirtuins Were Affected in Chronically Irradiated Heart
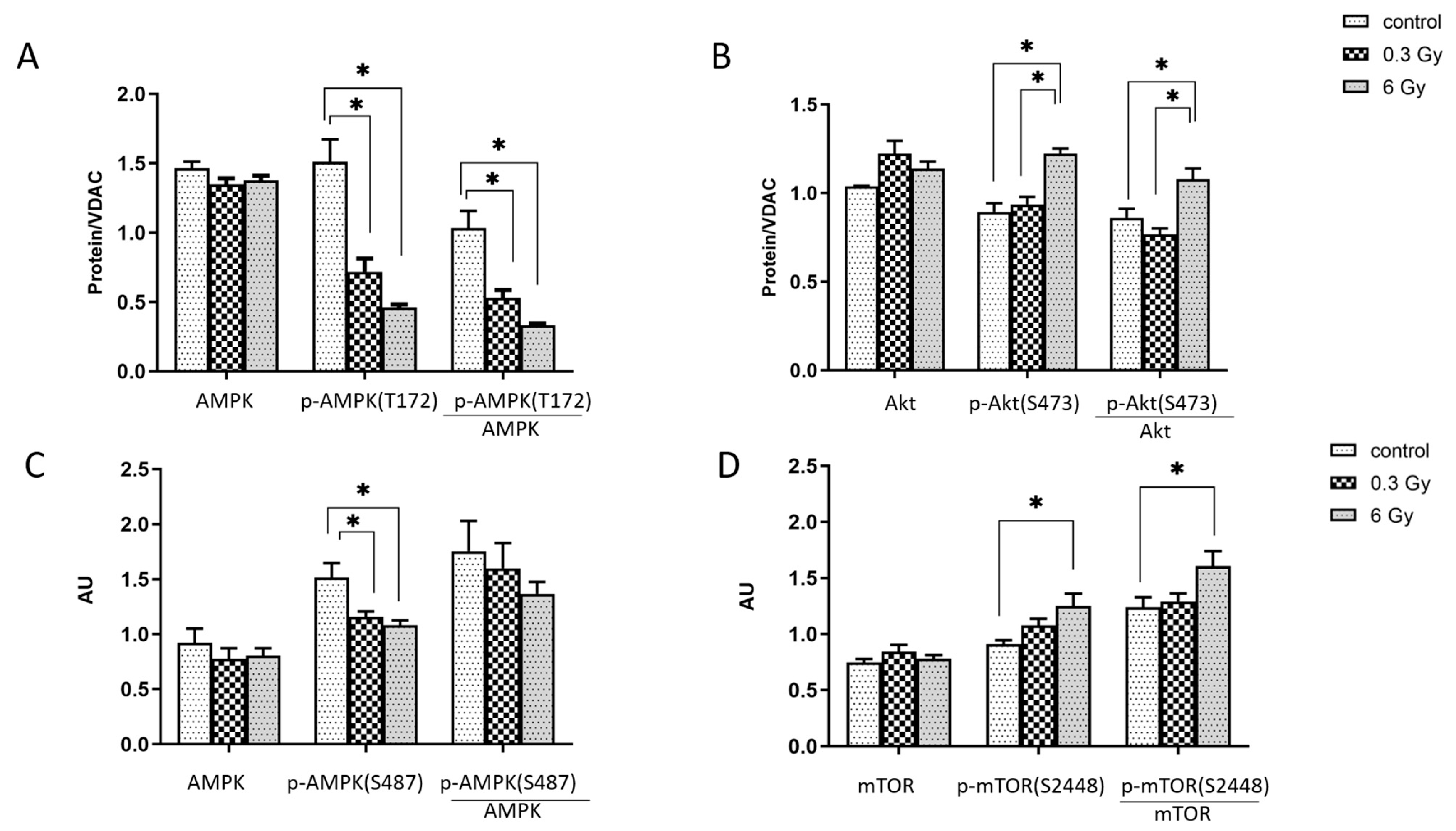
3.6. mTOR and AMPK Signalling Pathways were Affected by Chronic Irradiation
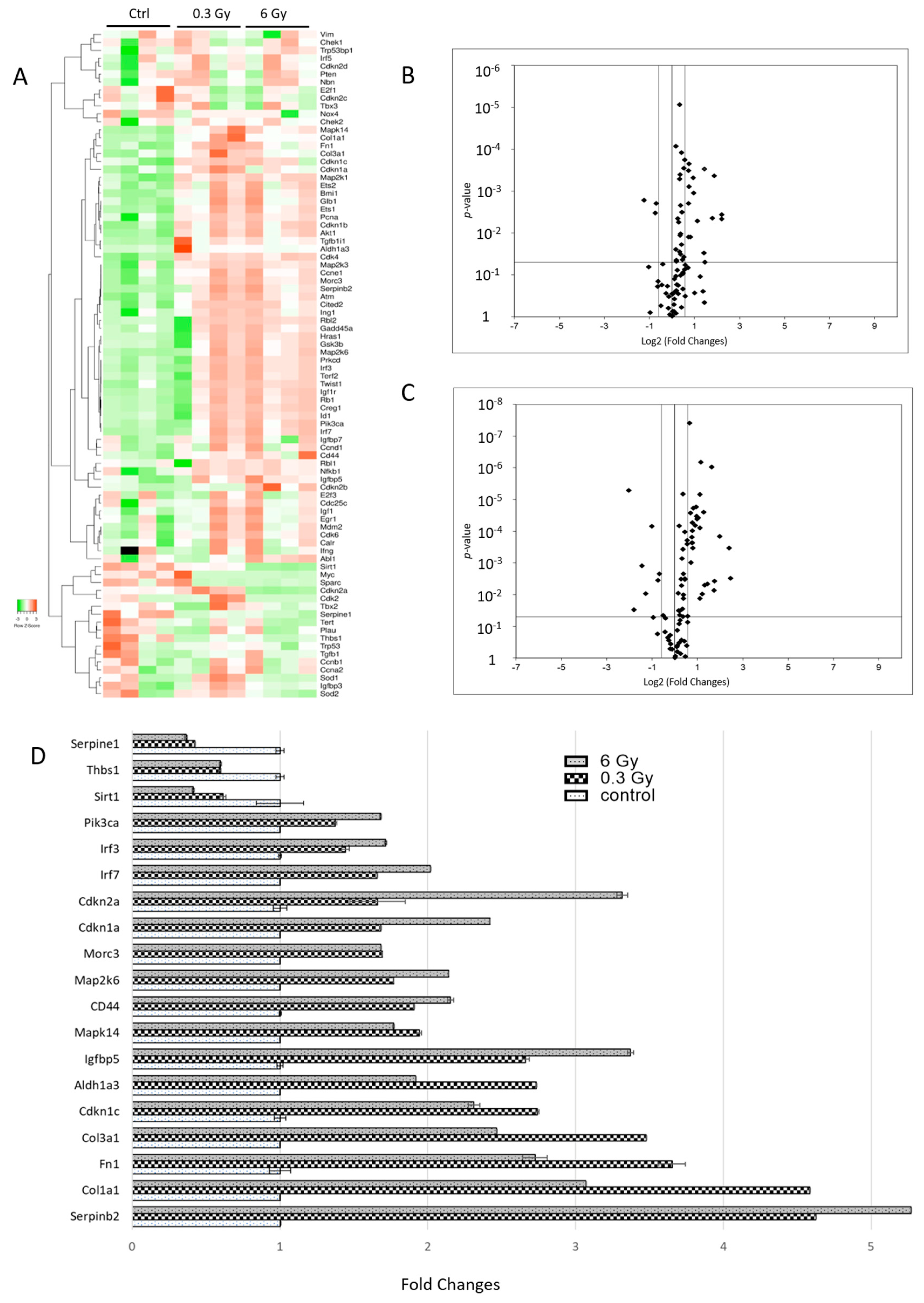
3.7. Senescence-Associated Genes Were Altered in Chronically Irradiated Hearts
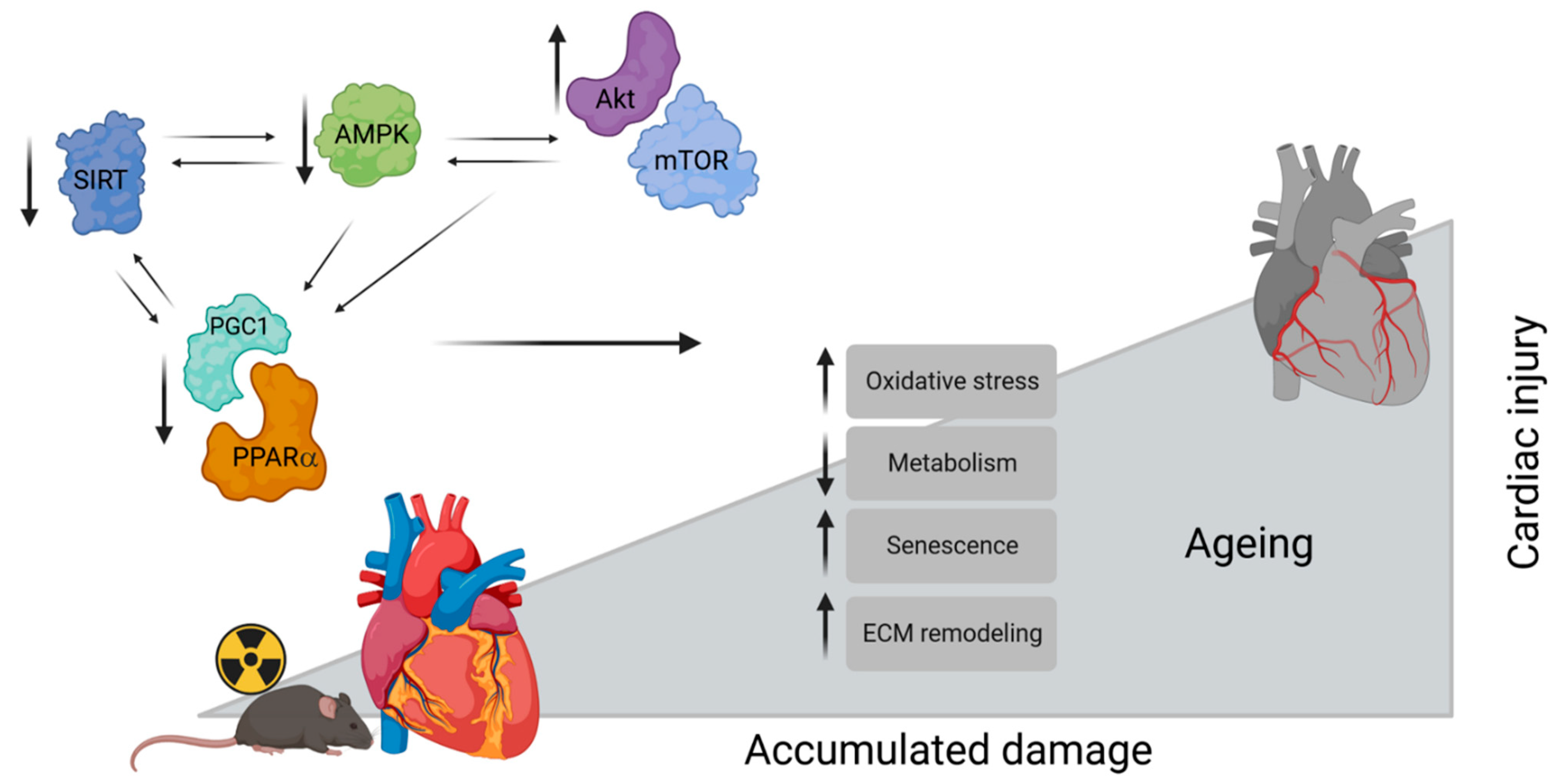
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tapio, S. Pathology and biology of radiation-induced cardiac disease. J. Radiat. Res. 2016, 57, 439–448. [Google Scholar] [CrossRef]
- Little, M.P.; Azizova, T.V.; Bazyka, D.; Bouffler, S.D.; Cardis, E.; Chekin, S.; Chumak, V.V.; Cucinotta, F.A.; de Vathaire, F.; Hall, P.; et al. Systematic review and meta-analysis of circulatory disease from exposure to low-level ionizing radiation and estimates of potential population mortality risks. Environ. Health Perspect. 2012, 120, 1503–1511. [Google Scholar] [CrossRef]
- Tapio, S.; Little, M.P.; Kaiser, J.C.; Impens, N.; Hamada, N.; Georgakilas, A.G.; Simar, D.; Salomaa, S. Ionizing radiation-induced circulatory and metabolic diseases. Env. Int. 2021, 146, 106235. [Google Scholar] [CrossRef]
- Little, M.P.; Azizova, T.V.; Hamada, N. Low- and moderate-dose non-cancer effects of ionizing radiation in directly exposed individuals, especially circulatory and ocular diseases: A review of the epidemiology. Int. J. Radiat. Biol. 2021, 97, 782–803. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, M.; Auvinen, A.; Cardis, E.; Hall, J.; Jourdain, J.R.; Laurier, D.; Little, M.P.; Peters, A.; Raj, K.; Russell, N.S.; et al. Low-dose ionising radiation and cardiovascular diseases--Strategies for molecular epidemiological studies in Europe. Mutat. Res. Rev. Mutat. Res. 2015, 764, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Azizova, T.V.; Grigoryeva, E.S.; Hamada, N. Dose rate effect on mortality from ischemic heart disease in the cohort of Russian Mayak Production Association workers. Sci. Rep. 2023, 13, 1926. [Google Scholar] [CrossRef] [PubMed]
- Little, M.P.; Azizova, T.V.; Richardson, D.B.; Tapio, S.; Bernier, M.O.; Kreuzer, M.; Cucinotta, F.A.; Bazyka, D.; Chumak, V.; Ivanov, V.K.; et al. Ionising radiation and cardiovascular disease: Systematic review and meta-analysis. BMJ 2023, 380, e072924. [Google Scholar] [CrossRef]
- Auvinen, A. Increased cardiovascular disease risk after exposure to low dose radiation. BMJ 2023, 380, e074589. [Google Scholar] [CrossRef]
- Mancuso, M.; Pasquali, E.; Braga-Tanaka, I., 3rd; Tanaka, S.; Pannicelli, A.; Giardullo, P.; Pazzaglia, S.; Tapio, S.; Atkinson, M.J.; Saran, A. Acceleration of atherogenesis in ApoE-/- mice exposed to acute or low-dose-rate ionizing radiation. Oncotarget 2015, 6, 31263–31271. [Google Scholar] [CrossRef]
- Barjaktarovic, Z.; Merl-Pham, J.; Braga-Tanaka, I.; Tanaka, S.; Hauck, S.M.; Saran, A.; Mancuso, M.; Atkinson, M.J.; Tapio, S.; Azimzadeh, O. Hyperacetylation of Cardiac Mitochondrial Proteins Is Associated with Metabolic Impairment and Sirtuin Downregulation after Chronic Total Body Irradiation of ApoE−/− Mice. Int. J. Mol. Sci. 2019, 20, 5239. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Azizova, T.; Merl-Pham, J.; Blutke, A.; Moseeva, M.; Zubkova, O.; Anastasov, N.; Feuchtinger, A.; Hauck, S.M.; Atkinson, M.J.; et al. Chronic Occupational Exposure to Ionizing Radiation Induces Alterations in the Structure and Metabolism of the Heart: A Proteomic Analysis of Human Formalin-Fixed Paraffin-Embedded (FFPE) Cardiac Tissue. Int. J. Mol. Sci. 2020, 21, 6832. [Google Scholar] [CrossRef] [PubMed]
- Mitchel, R.E.; Hasu, M.; Bugden, M.; Wyatt, H.; Little, M.P.; Gola, A.; Hildebrandt, G.; Priest, N.D.; Whitman, S.C. Low-dose radiation exposure and atherosclerosis in ApoE−/− mice. Radiat. Res. 2011, 175, 665–676. [Google Scholar] [CrossRef]
- Ebrahimian, T.G.; Beugnies, L.; Surette, J.; Priest, N.; Gueguen, Y.; Gloaguen, C.; Benderitter, M.; Jourdain, J.R.; Tack, K. Chronic Exposure to External Low-Dose Gamma Radiation Induces an Increase in Anti-inflammatory and Anti-oxidative Parameters Resulting in Atherosclerotic Plaque Size Reduction in ApoE−/− Mice. Radiat. Res. 2018, 189, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Rey, N.; Ebrahimian, T.; Gloaguen, C.; Kereselidze, D.; Magneron, V.; Bontemps, C.A.; Demarquay, C.; Olsson, G.; Haghdoost, S.; Lehoux, S.; et al. Exposure to Low to Moderate Doses of Ionizing Radiation Induces A Reduction of Pro-Inflammatory Ly6chigh Monocytes and a U-Curved Response of T Cells in APOE−/− Mice. Dose Response 2021, 19, 16237. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Tanaka, I.B., 3rd; Sasagawa, S.; Ichinohe, K.; Takabatake, T.; Matsushita, S.; Matsumoto, T.; Otsu, H.; Sato, F. No lengthening of life span in mice continuously exposed to gamma rays at very low dose rates. Radiat. Res. 2003, 160, 376–379. [Google Scholar] [CrossRef]
- Tanaka, I.B., 3rd; Tanaka, S.; Ichinohe, K.; Matsushita, S.; Matsumoto, T.; Otsu, H.; Oghiso, Y.; Sato, F. Cause of death and neoplasia in mice continuously exposed to very low dose rates of gamma rays. Radiat. Res. 2007, 167, 417–437. [Google Scholar] [CrossRef]
- Dalke, C.; Neff, F.; Bains, S.K.; Bright, S.; Lord, D.; Reitmeir, P.; Rößler, U.; Samaga, D.; Unger, K.; Braselmann, H.; et al. Lifetime study in mice after acute low-dose ionizing radiation: A multifactorial study with special focus on cataract risk. Radiat. Environ. Biophys. 2018, 57, 99–113. [Google Scholar] [CrossRef]
- Nakamura, S.; Tanaka, I.B., 3rd; Tanaka, S.; Nakaya, K.; Sakata, N.; Oghiso, Y. Adiposity in female B6C3F1 mice continuously irradiated with low-dose-rate gamma rays. Radiat. Res. 2010, 173, 333–341. [Google Scholar] [CrossRef]
- Dong, J.; Yang, S.; Zhuang, Q.; Sun, J.; Wei, P.; Zhao, X.; Chen, Y.; Chen, X.; Li, M.; Wei, L.; et al. The Associations of Lipid Profiles with Cardiovascular Diseases and Death in a 10-Year Prospective Cohort Study. Front. Cardiovasc. Med. 2021, 8, 745539. [Google Scholar] [CrossRef]
- Shiragai, A. Estimation of the absorbed dose to mice in prolonged irradiation by low-dose rate γ-rays from 137Cs sources. Radioisotopes 1997, 46, 904–911. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Sievert, W.; Sarioglu, H.; Merl-Pham, J.; Yentrapalli, R.; Bakshi, M.V.; Janik, D.; Ueffing, M.; Atkinson, M.J.; Multhoff, G.; et al. Integrative proteomics and targeted transcriptomics analyses in cardiac endothelial cells unravel mechanisms of long-term radiation-induced vascular dysfunction. J. Proteome Res. 2015, 14, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Hauck, S.M.; Dietter, J.; Kramer, R.L.; Hofmaier, F.; Zipplies, J.K.; Amann, B.; Feuchtinger, A.; Deeg, C.A.; Ueffing, M. Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol. Cell. Proteom. 2010, 9, 2292–2305. [Google Scholar] [CrossRef] [PubMed]
- Merl, J.; Ueffing, M.; Hauck, S.M.; von Toerne, C. Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Muller cells. Proteomics 2012, 12, 1902–1911. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Hawley, S.A.; Ross, F.A.; Gowans, G.J.; Tibarewal, P.; Leslie, N.R.; Hardie, D.G. Phosphorylation by Akt within the ST loop of AMPK-α1 down-regulates its activation in tumour cells. Biochem. J. 2014, 459, 275–287. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Azizova, T.; Merl-Pham, J.; Subramanian, V.; Bakshi, M.V.; Moseeva, M.; Zubkova, O.; Hauck, S.M.; Anastasov, N.; Atkinson, M.J.; et al. A dose-dependent perturbation in cardiac energy metabolism is linked to radiation-induced ischemic heart disease in Mayak nuclear workers. Oncotarget 2017, 8, 9067–9078. [Google Scholar] [CrossRef]
- Papiez, A.; Azimzadeh, O.; Azizova, T.; Moseeva, M.; Anastasov, N.; Smida, J.; Tapio, S.; Polanska, J. Integrative multiomics study for validation of mechanisms in radiation-induced ischemic heart disease in Mayak workers. PLoS ONE 2018, 13, e0209626. [Google Scholar] [CrossRef]
- Grant, J.E.; Bradshaw, A.D.; Schwacke, J.H.; Baicu, C.F.; Zile, M.R.; Schey, K.L. Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. J. Proteome Res. 2009, 8, 4252–4263. [Google Scholar] [CrossRef]
- Chiao, Y.A.; Rabinovitch, P.S. The Aging Heart. Cold Spring Harb. Perspect. Med. 2015, 5, a025148. [Google Scholar] [CrossRef] [PubMed]
- Steenman, M.; Lande, G. Cardiac aging and heart disease in humans. Biophys. Rev. 2017, 9, 131–137. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, N.; Servillo, L.; Balestrieri, M.L. SIRT1 and SIRT6 Signaling Pathways in Cardiovascular Disease Protection. Antioxid. Redox Signal. 2018, 28, 711–732. [Google Scholar] [CrossRef] [PubMed]
- Tocchi, A.; Quarles, E.K.; Basisty, N.; Gitari, L.; Rabinovitch, P.S. Mitochondrial dysfunction in cardiac aging. Biochim. Biophys. Acta. 2015, 1847, 1424–1433. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, H.; Wang, B.; Zhang, Y.; Zheng, X.; Shao, B.; Zhuge, Q.; Jin, K. Key Signaling Pathways in Aging and Potential Interventions for Healthy Aging. Cells 2021, 10, 660. [Google Scholar] [CrossRef]
- Packer, M. Longevity genes, cardiac ageing, and the pathogenesis of cardiomyopathy: Implications for understanding the effects of current and future treatments for heart failure. Eur. Heart J. 2020, 41, 3856–3861. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hastings, M.H.; Rhee, J.; Trager, L.E.; Roh, J.D.; Rosenzweig, A. Targeting Age-Related Pathways in Heart Failure. Circ. Res. 2020, 126, 533–551. [Google Scholar] [CrossRef]
- Dai, D.F.; Chen, T.; Johnson, S.C.; Szeto, H.; Rabinovitch, P.S. Cardiac aging: From molecular mechanisms to significance in human health and disease. Antioxid. Redox Signal. 2012, 16, 1492–1526. [Google Scholar] [CrossRef]
- Sadria, M.; Layton, A.T. Interactions among mTORC, AMPK and SIRT: A computational model for cell energy balance and metabolism. Cell Commun. Signal. 2021, 19, 57. [Google Scholar] [CrossRef]
- Liberale, L.; Kraler, S.; Camici, G.G.; Lüscher, T.F. Ageing and longevity genes in cardiovascular diseases. Basic. Clin. Pharm. Toxicol. 2020, 127, 120–131. [Google Scholar] [CrossRef]
- Pillai, V.B.; Sundaresan, N.R.; Gupta, M.P. Regulation of Akt signaling by sirtuins: Its implication in cardiac hypertrophy and aging. Circ. Res. 2014, 114, 368–378. [Google Scholar] [CrossRef]
- Yentrapalli, R.; Azimzadeh, O.; Barjaktarovic, Z.; Sarioglu, H.; Wojcik, A.; Harms-Ringdahl, M.; Atkinson, M.J.; Haghdoost, S.; Tapio, S. Quantitative proteomic analysis reveals induction of premature senescence in human umbilical vein endothelial cells exposed to chronic low-dose rate gamma-radiation. Proteomics 2013, 13, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, O.; Atkinson, M.J.; Tapio, S. Quantitative Proteomic Analysis Using Formalin-Fixed, Paraffin-Embedded (FFPE) Human Cardiac Tissue. Methods Mol. Biol. 2021, 2261, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Hei, T.K. Aging and age-related health effects of ionizing radiation. Radiat. Med. Prot. 2020, 1, 15–23. [Google Scholar] [CrossRef]
- Wu, S.; Zou, M.H. AMPK, Mitochondrial Function, and Cardiovascular Disease. Int. J. Mol. Sci. 2020, 21, 4987. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS signaling under metabolic stress: Cross-talk between AMPK and AKT pathway. Mol. Cancer 2017, 16, 79. [Google Scholar] [CrossRef]
- Hua, Y.; Zhang, Y.; Ceylan-Isik, A.F.; Wold, L.E.; Nunn, J.M.; Ren, J. Chronic Akt activation accentuates aging-induced cardiac hypertrophy and myocardial contractile dysfunction: Role of autophagy. Basic. Res. Cardiol. 2011, 106, 1173–1191. [Google Scholar] [CrossRef] [PubMed]
- Kovacic, S.; Soltys, C.L.; Barr, A.J.; Shiojima, I.; Walsh, K.; Dyck, J.R. Akt activity negatively regulates phosphorylation of AMP-activated protein kinase in the heart. J. Biol. Chem. 2003, 278, 39422–39427. [Google Scholar] [CrossRef]
- Levine, Y.C.; Li, G.K.; Michel, T. Agonist-modulated regulation of AMP-activated protein kinase (AMPK) in endothelial cells. Evidence for an AMPK -> Rac1 -> Akt -> endothelial nitric-oxide synthase pathway. J. Biol. Chem. 2007, 282, 20351–20364. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kandadi, M.R.; Ren, J. Double knockout of Akt2 and AMPK predisposes cardiac aging without affecting lifespan: Role of autophagy and mitophagy. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1865–1875. [Google Scholar] [CrossRef]
- Aoyagi, T.; Matsui, T. Phosphoinositide-3 kinase signaling in cardiac hypertrophy and heart failure. Curr. Pharm. Des. 2011, 17, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khanbabapour Sasi, A.; Hussen, B.M.; Shoorei, H.; Siddiq, A.; Taheri, M.; Ayatollahi, S.A. Interplay between PI3K/AKT pathway and heart disorders. Mol. Biol. Rep. 2022, 49, 9767–9781. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S. Autophagy and cardiac aging. Cell. Death Differ. 2019, 26, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Papadopoli, D.; Boulay, K.; Kazak, L.; Pollak, M.; Mallette, F.; Topisirovic, I.; Hulea, L. mTOR as a central regulator of lifespan and aging. F1000Res 2019, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.; Raj, K. Premature aging induced by radiation exhibits pro-atherosclerotic effects mediated by epigenetic activation of CD44 expression. Aging Cell. 2014, 13, 900–910. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Song, L.; Yu, L.; Zhang, L.; Zhang, Y.; Xing, Y.; Yin, Y.; Ma, H. SIRT3 deficiency exacerbates p53/Parkinmediated mitophagy inhibition and promotes mitochondrial dysfunction: Implication for aged hearts. Int. J. Mol. Med. 2018, 41, 3517–3526. [Google Scholar] [CrossRef]
- Lin, S.; Wang, Y.; Zhang, X.; Kong, Q.; Li, C.; Li, Y.; Ding, Z.; Liu, L. HSP27 Alleviates Cardiac Aging in Mice via a Mechanism Involving Antioxidation and Mitophagy Activation. Oxid. Med. Cell. Longev. 2016, 2016, 2586706. [Google Scholar] [CrossRef]
- Owens, W.A.; Walaszczyk, A.; Spyridopoulos, I.; Dookun, E.; Richardson, G.D. Senescence and senolytics in cardiovascular disease: Promise and potential pitfalls. Mech. Ageing Dev. 2021, 198, 111540. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Sievert, W.; Sarioglu, H.; Yentrapalli, R.; Barjaktarovic, Z.; Sriharshan, A.; Ueffing, M.; Janik, D.; Aichler, M.; Atkinson, M.J.; et al. PPAR Alpha: A Novel Radiation Target in Locally Exposed Mus musculus Heart Revealed by Quantitative Proteomics. J. Proteome Res. 2013, 12, 2700–2714. [Google Scholar] [CrossRef]
- Subramanian, V.; Borchard, S.; Azimzadeh, O.; Sievert, W.; Merl-Pham, J.; Mancuso, M.; Pasquali, E.; Multhoff, G.; Popper, B.; Zischka, H.; et al. PPARalpha Is Necessary for Radiation-Induced Activation of Noncanonical TGFbeta Signaling in the Heart. J. Proteome Res. 2018, 17, 1677–1689. [Google Scholar] [CrossRef]
- Subramanian, V.; Seemann, I.; Merl-Pham, J.; Hauck, S.M.; Stewart, F.A.; Atkinson, M.J.; Tapio, S.; Azimzadeh, O. The Role of TGF Beta and PPAR Alpha Signalling Pathways in Radiation Response of Locally Exposed Heart: Integrated Global Transcriptomics and Proteomics Analysis. J. Proteome Res. 2016, 16, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Kates, A.M.; Herrero, P.; Dence, C.; Soto, P.; Srinivasan, M.; Delano, D.G.; Ehsani, A.; Gropler, R.J. Impact of aging on substrate metabolism by the human heart. J. Am. Coll. Cardiol. 2003, 41, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Pol, C.J.; Lieu, M.; Drosatos, K. PPARs: Protectors or Opponents of Myocardial Function? PPAR Res. 2015, 2015, 835985. [Google Scholar] [CrossRef]
- Meng, R.; Pei, Z.; Zhang, A.; Zhou, Y.; Cai, X.; Chen, B.; Liu, G.; Mai, W.; Wei, J.; Dong, Y. AMPK activation enhances PPARα activity to inhibit cardiac hypertrophy via ERK1/2 MAPK signaling pathway. Arch. Biochem. Biophys. 2011, 511, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sciarretta, S.; Volpe, M.; Sadoshima, J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ. Res. 2014, 114, 549–564. [Google Scholar] [CrossRef]
- Khan, S.A.; Sathyanarayan, A.; Mashek, M.T.; Ong, K.T.; Wollaston-Hayden, E.E.; Mashek, D.G. ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1alpha/PPAR-alpha signaling. Diabetes 2015, 64, 418–426. [Google Scholar] [CrossRef]
- Kong, X.; Wang, R.; Xue, Y.; Liu, X.; Zhang, H.; Chen, Y.; Fang, F.; Chang, Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE 2010, 5, e11707. [Google Scholar] [CrossRef]
- Jeninga, E.H.; Schoonjans, K.; Auwerx, J. Reversible acetylation of PGC-1: Connecting energy sensors and effectors to guarantee metabolic flexibility. Oncogene 2010, 29, 4617–4624. [Google Scholar] [CrossRef]
- Barjaktarovic, Z.; Schmaltz, D.; Shyla, A.; Azimzadeh, O.; Schulz, S.; Haagen, J.; Dorr, W.; Sarioglu, H.; Schafer, A.; Atkinson, M.J.; et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS ONE 2011, 6, e27811. [Google Scholar] [CrossRef]
- Barjaktarovic, Z.; Shyla, A.; Azimzadeh, O.; Schulz, S.; Haagen, J.; Dorr, W.; Sarioglu, H.; Atkinson, M.J.; Zischka, H.; Tapio, S. Ionising radiation induces persistent alterations in the cardiac mitochondrial function of C57BL/6 mice 40 weeks after local heart exposure. Radiother. Oncol. 2013, 106, 404–410. [Google Scholar] [CrossRef]
- Tai, Y.; Inoue, H.; Sakurai, T.; Yamada, H.; Morito, M.; Ide, F.; Mishima, K.; Saito, I. Protective effect of lecithinized SOD on reactive oxygen species-induced xerostomia. Radiat. Res. 2009, 172, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Dukan, S.; Farewell, A.; Ballesteros, M.; Taddei, F.; Radman, M.; Nyström, T. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 2000, 97, 5746–5749. [Google Scholar] [CrossRef] [PubMed]
- Izzo, C.; Vitillo, P.; Di Pietro, P.; Visco, V.; Strianese, A.; Virtuoso, N.; Ciccarelli, M.; Galasso, G.; Carrizzo, A.; Vecchione, C. The Role of Oxidative Stress in Cardiovascular Aging and Cardiovascular Diseases. Life 2021, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Pagan, L.U.; Gomes, M.J.; Gatto, M.; Mota, G.A.F.; Okoshi, K.; Okoshi, M.P. The Role of Oxidative Stress in the Aging Heart. Antioxidants 2022, 11, 336. [Google Scholar] [CrossRef]
- Menezes, K.M.; Wang, H.; Hada, M.; Saganti, P.B. Radiation Matters of the Heart: A Mini Review. Front. Cardiovasc. Med. 2018, 5, 83. [Google Scholar] [CrossRef]
- Rybkina, V.L.; Azizova, T.V.; Scherthan, H.; Meineke, V.; Doerr, H.; Adamova, G.V.; Teplyakova, O.V.; Osovets, S.V.; Bannikova, M.V.; Zurochka, A.V. Expression of blood serum proteins and lymphocyte differentiation clusters after chronic occupational exposure to ionizing radiation. Radiat. Environ. Biophys. 2014, 53, 659–670. [Google Scholar] [CrossRef]
- Biernacka, A.; Frangogiannis, N.G. Aging and Cardiac Fibrosis. Aging Dis. 2011, 2, 158–173. [Google Scholar]
- Lu, L.; Guo, J.; Hua, Y.; Huang, K.; Magaye, R.; Cornell, J.; Kelly, D.J.; Reid, C.; Liew, D.; Zhou, Y.; et al. Cardiac fibrosis in the ageing heart: Contributors and mechanisms. Clin. Exp. Pharm. Physiol. 2017, 44 (Suppl. 1), 55–63. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Subramanian, V.; Sievert, W.; Merl-Pham, J.; Oleksenko, K.; Rosemann, M.; Multhoff, G.; Atkinson, M.J.; Tapio, S. Activation of PPARα by Fenofibrate Attenuates the Effect of Local Heart High Dose Irradiation on the Mouse Cardiac Proteome. Biomedicines 2021, 9, 1845. [Google Scholar] [CrossRef]
- Azimzadeh, O.; Moertl, S.; Ramadan, R.; Baselet, B.; Laiakis, E.C.; Sebastian, S.; Beaton, D.; Hartikainen, J.M.; Kaiser, J.C.; Beheshti, A.; et al. Application of radiation omics in the development of adverse outcome pathway networks: An example of radiation-induced cardiovascular disease. Int. J. Radiat. Biol. 2022, 98, 1722–1751. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azimzadeh, O.; Merl-Pham, J.; Subramanian, V.; Oleksenko, K.; Krumm, F.; Mancuso, M.; Pasquali, E.; Tanaka, I.B., III; Tanaka, S.; Atkinson, M.J.; et al. Late Effects of Chronic Low Dose Rate Total Body Irradiation on the Heart Proteome of ApoE−/− Mice Resemble Premature Cardiac Ageing. Cancers 2023, 15, 3417. https://doi.org/10.3390/cancers15133417
Azimzadeh O, Merl-Pham J, Subramanian V, Oleksenko K, Krumm F, Mancuso M, Pasquali E, Tanaka IB III, Tanaka S, Atkinson MJ, et al. Late Effects of Chronic Low Dose Rate Total Body Irradiation on the Heart Proteome of ApoE−/− Mice Resemble Premature Cardiac Ageing. Cancers. 2023; 15(13):3417. https://doi.org/10.3390/cancers15133417
Chicago/Turabian StyleAzimzadeh, Omid, Juliane Merl-Pham, Vikram Subramanian, Kateryna Oleksenko, Franziska Krumm, Mariateresa Mancuso, Emanuela Pasquali, Ignacia B. Tanaka, III, Satoshi Tanaka, Michael J. Atkinson, and et al. 2023. "Late Effects of Chronic Low Dose Rate Total Body Irradiation on the Heart Proteome of ApoE−/− Mice Resemble Premature Cardiac Ageing" Cancers 15, no. 13: 3417. https://doi.org/10.3390/cancers15133417
APA StyleAzimzadeh, O., Merl-Pham, J., Subramanian, V., Oleksenko, K., Krumm, F., Mancuso, M., Pasquali, E., Tanaka, I. B., III, Tanaka, S., Atkinson, M. J., Tapio, S., & Moertl, S. (2023). Late Effects of Chronic Low Dose Rate Total Body Irradiation on the Heart Proteome of ApoE−/− Mice Resemble Premature Cardiac Ageing. Cancers, 15(13), 3417. https://doi.org/10.3390/cancers15133417






