Spatial Distribution of Immune Cells in Primary and Recurrent Glioblastoma: A Small Case Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Tissue Microarray (TMA)
2.3. Immunohistochemistry and Its Quantification
2.4. Antibodies Labelling with Digital Space Profiling GeoMXTM
2.5. Nanostring Genomic Arrays
3. Results
3.1. Patient Characteristics
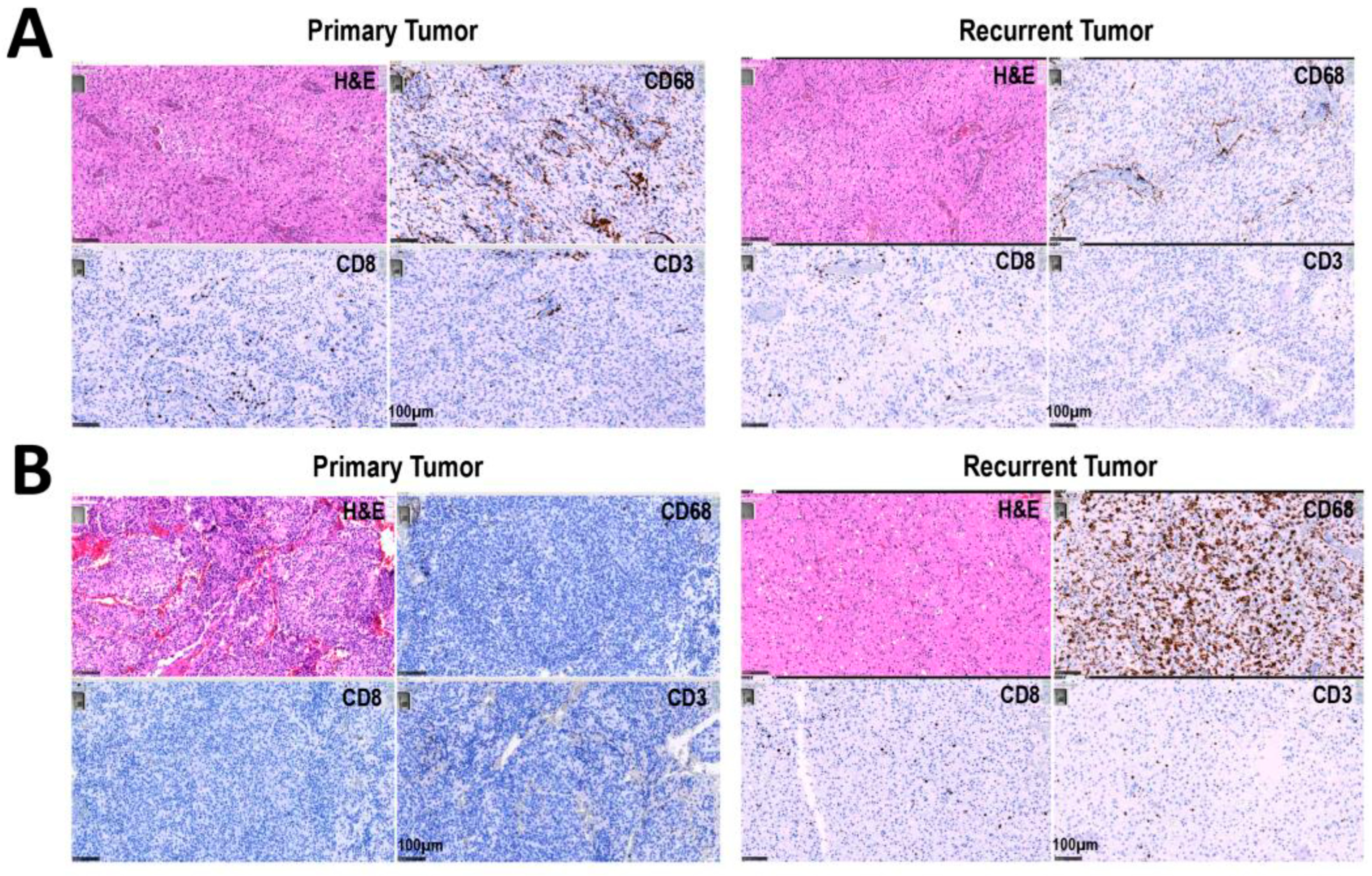
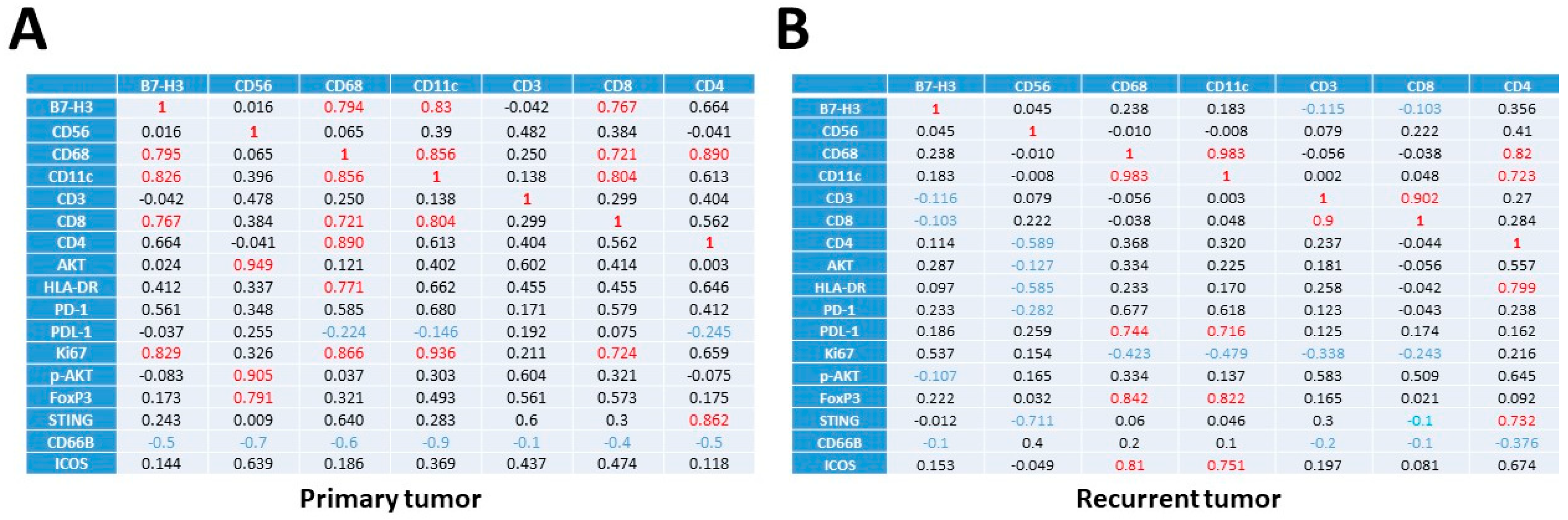
3.2. Increased Percentage of CD8+ and CD68+ Cells in Recurrent versus Primary Tumors
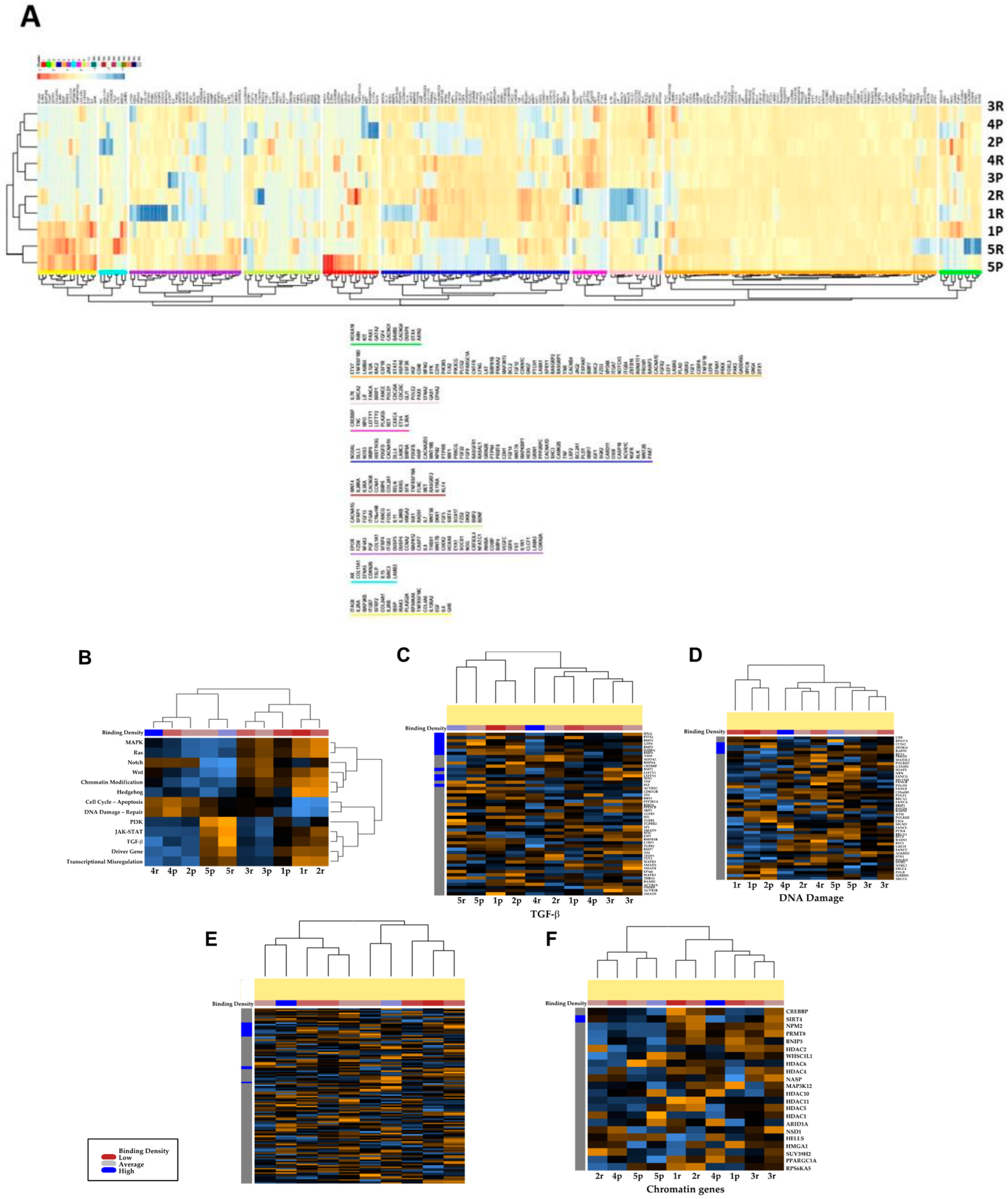
3.3. Transcriptional Profiles of Recurrent vs. Primary Glioblastoma
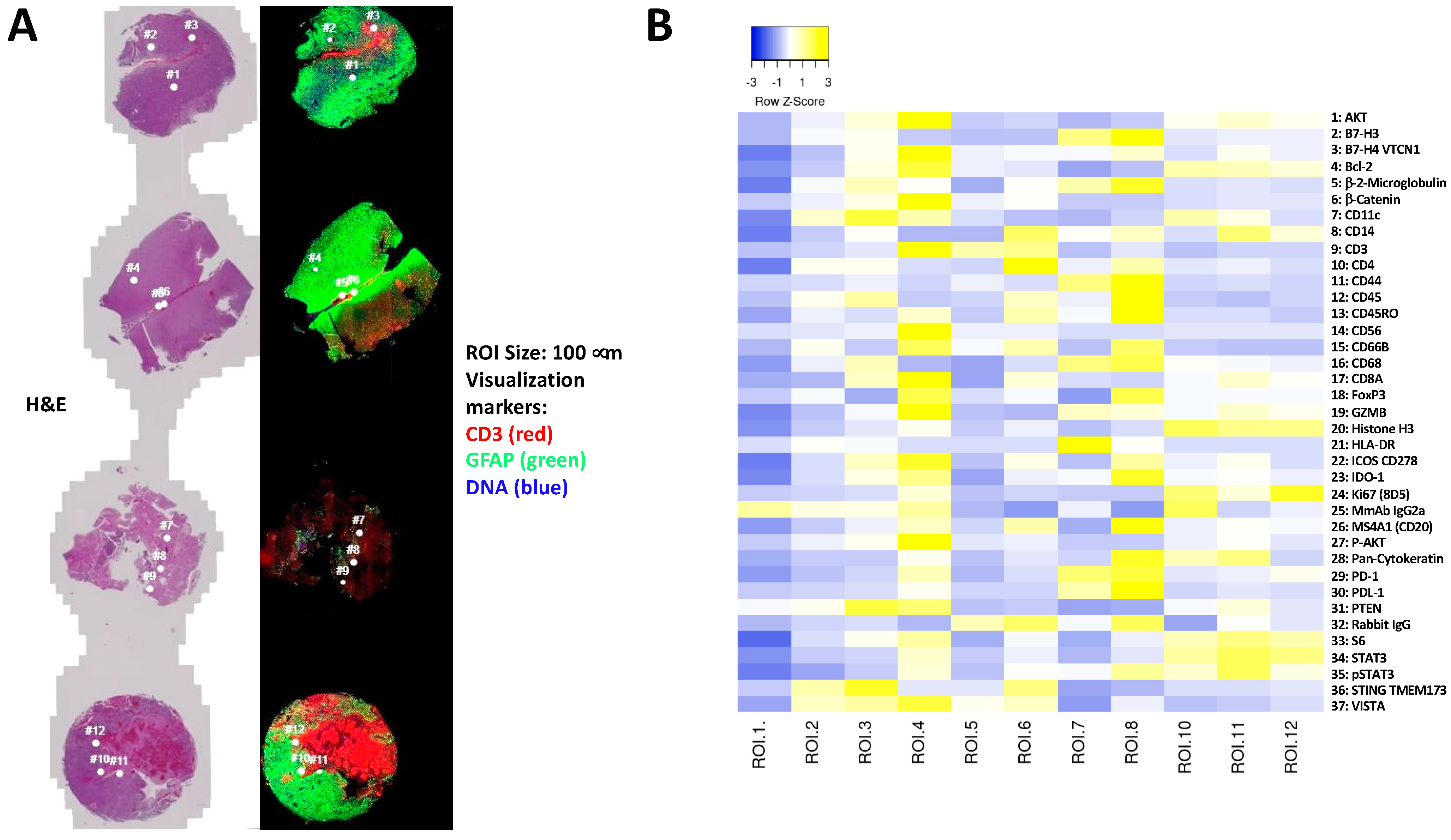
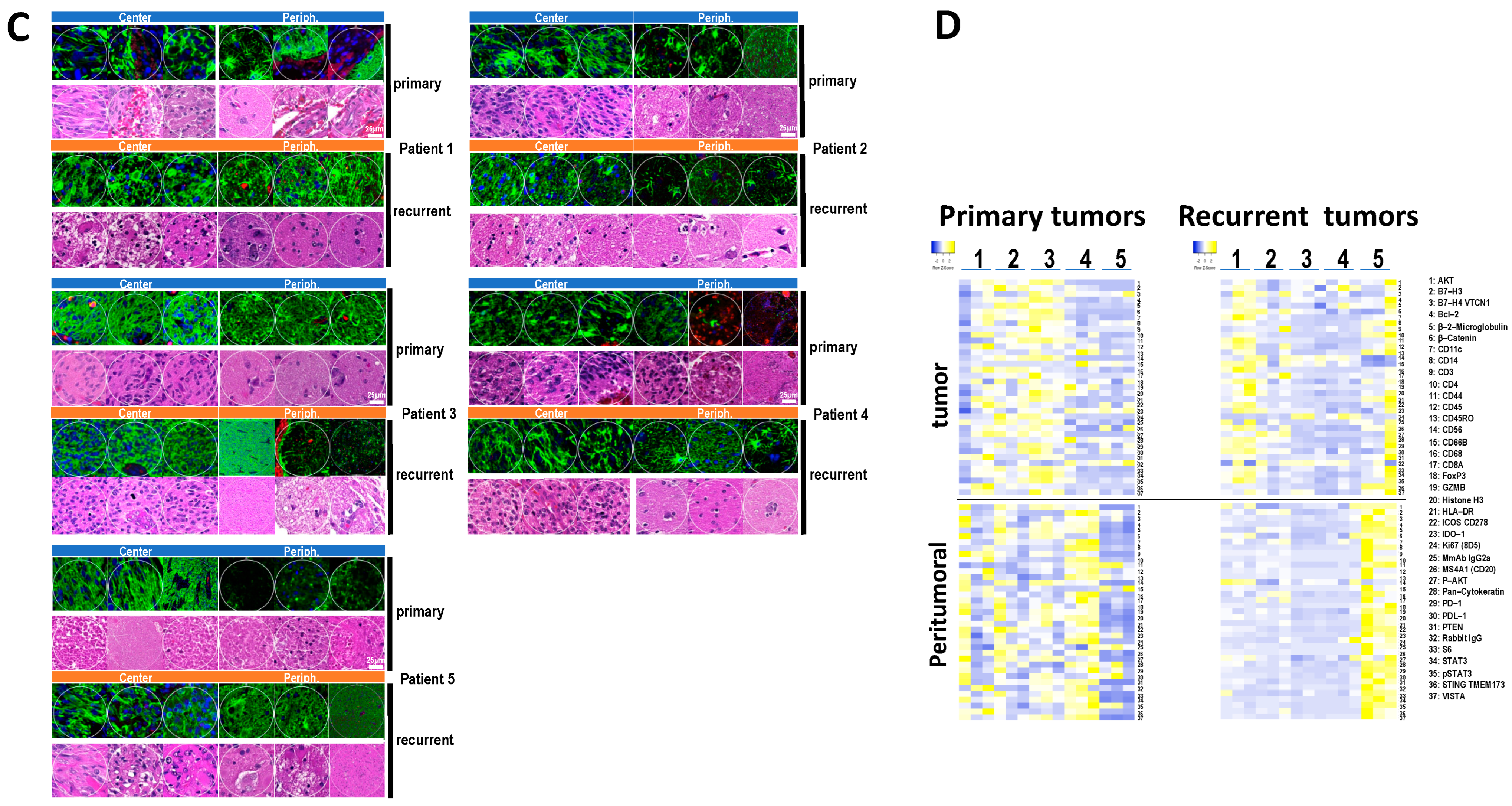
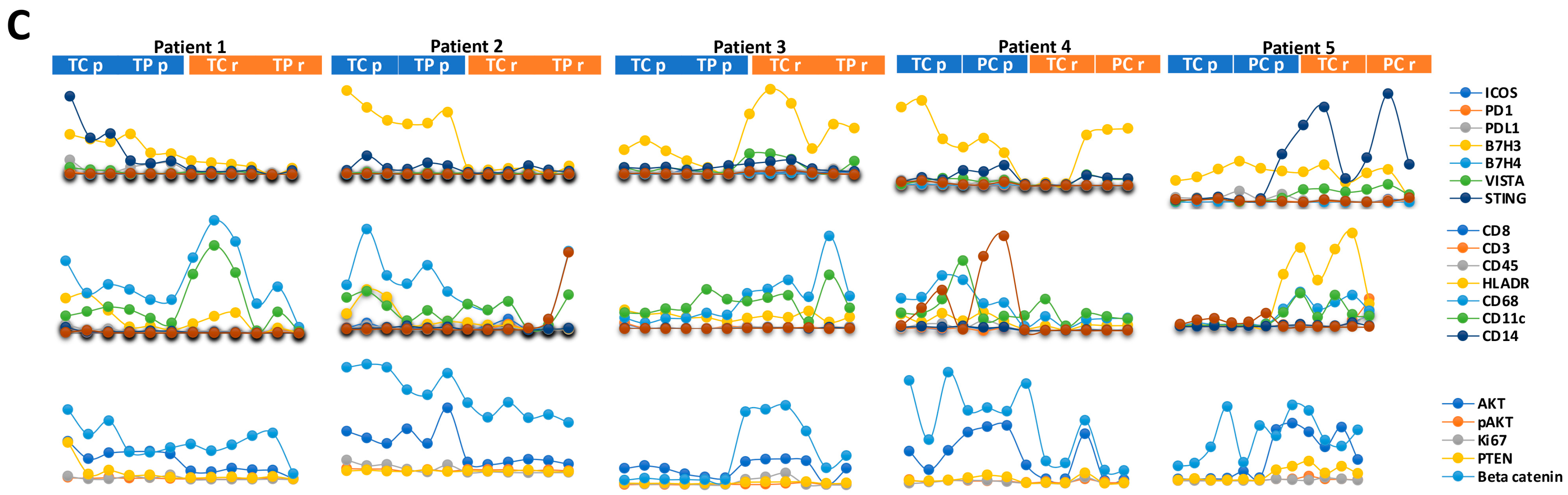
3.4. Immune and Cancer Spatial Distribution in Intra- and Peri-Tumoral Regions in Paired Primary and Recurrent GBM
- Homogenous cellular populations;
- Distinct tumoral or normal features;
- Intra- to peri-tumoral or microenvironmental localizations;
- Vascularized regions.
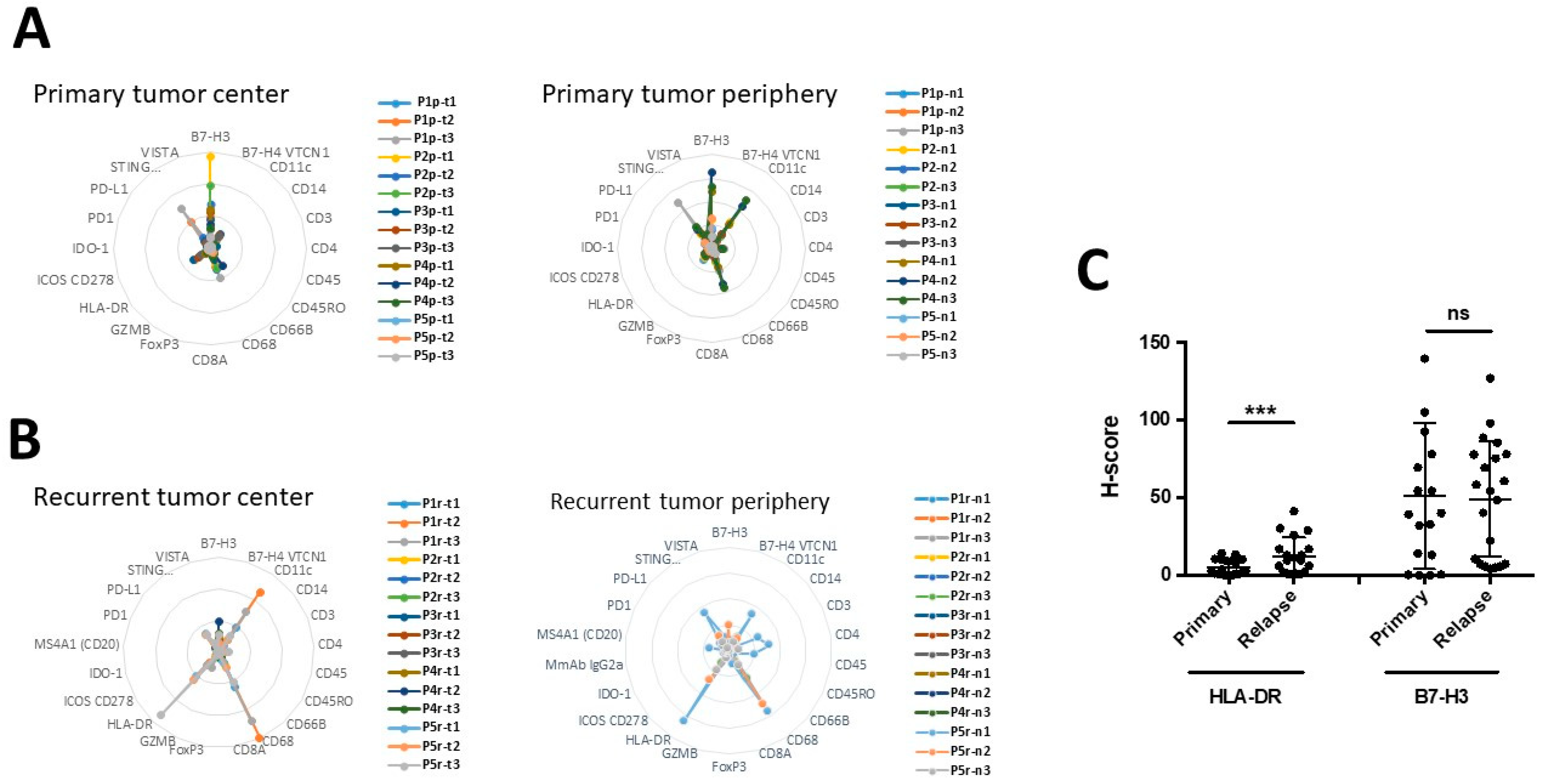
3.5. The Expression of B7-H3 and HLA-DR in Primary versus Recurrent Paired Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bikfalvi, A.; da Costa, C.A.; Avril, T.; Barnier, J.-V.; Bauchet, L.; Brisson, L.; Cartron, P.F.; Castel, H.; Chevet, E.; Chneiweiss, H.; et al. Challenges in glioblastoma research: Focus on the tumor microenvironment. Trends Cancer 2023, 9, 9–27. [Google Scholar] [CrossRef]
- Schritz, A.; Aouali, N.; Fischer, A.; Dessenne, C.; Adams, R.; Berchem, G.; Huiart, L.; Schmitz, S. Systematic review and network meta-analysis of the efficacy of existing treatments for patients with recurrent glioblastoma. Neurooncol. Adv. 2021, 3, vdab052. [Google Scholar] [CrossRef]
- Yang, K.; Ellenbogen, Y.; Martyniuk, A.; Sourour, M.; Takroni, R.; Somji, M.; Gardiner, E.; Hui, K.; Odedra, D.; Larrazabal, R.; et al. Reoperation in adult patients with recurrent glioblastoma: A matched cohort analysis. Neurooncol. Adv. 2022, 4, vdac115. [Google Scholar] [CrossRef]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef]
- Wang, X.; Lu, J.; Guo, G.; Yu, J. Immunotherapy for recurrent glioblastoma: Practical insights and challenging prospects. Cell Death Dis. 2021, 12, 299. [Google Scholar] [CrossRef]
- Engelhardt, B.; Vajkoczy, P.; Weller, R.O. The movers and shapers in immune privilege of the CNS. Nat. Immunol. 2017, 18, 123–131. [Google Scholar] [CrossRef]
- Louveau, A.; Herz, J.; Alme, M.N.; Salvador, A.F.; Dong, M.Q.; Viar, K.E.; Herod, S.G.; Knopp, J.; Setliff, J.C.; Lupi, A.L.; et al. CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat. Neurosci. 2018, 21, 1380–1391. [Google Scholar] [CrossRef]
- Chen, Z.; Hambardzumyan, D. Immune Microenvironment in Glioblastoma Subtypes. Front. Immunol. 2018, 9, 1004. [Google Scholar] [CrossRef]
- Chongsathidkiet, P.; Jackson, C.; Koyama, S.; Loebel, F.; Cui, X.; Farber, S.H.; Woroniecka, K.; Elsamadicy, A.A.; Dechant, C.A.; Kemeny, H.R.; et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat. Med. 2018, 24, 1459–1468. [Google Scholar] [CrossRef]
- Sampson, J.H.; Gunn, M.D.; Fecci, P.E.; Ashley, D.M. Brain immunology and immunotherapy in brain tumours. Nat. Rev. Cancer 2020, 20, 12–25. [Google Scholar] [CrossRef]
- Jackson, C.M.; Choi, J.; Lim, M. Mechanisms of immunotherapy resistance: Lessons from glioblastoma. Nat. Immunol. 2019, 20, 1100–1109. [Google Scholar] [CrossRef]
- Wang, E.J.; Chen, J.-S.; Jain, S.; Morshed, R.A.; Haddad, A.F.; Gill, S.; Beniwal, A.S.; Aghi, M.K. Immunotherapy Resistance in Glioblastoma. Front. Genet. 2021, 12, 750675. [Google Scholar] [CrossRef]
- Buonfiglioli, A.; Hambardzumyan, D. Macrophages and microglia: The cerberus of glioblastoma. Acta Neuropathol. Commun. 2021, 9, 54. [Google Scholar] [CrossRef]
- Sevenich, L. Brain-Resident Microglia and Blood-Borne Macrophages Orchestrate Central Nervous System Inflammation in Neurodegenerative Disorders and Brain Cancer. Front. Immunol. 2018, 9, 697. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Martinez-Lage, M.; Lynch, T.M.; Bi, Y.; Cocito, C.; Way, G.P.; Pal, S.; Haller, J.; Yan, R.E.; Ziober, A.; Nguyen, A.; et al. Immune landscapes associated with different glioblastoma molecular subtypes. Acta Neuropathol. Commun. 2019, 7, 203. [Google Scholar] [CrossRef]
- Hernandez, S.; Lazcano, R.; Serrano, A.; Powell, S.; Kostousov, L.; Mehta, J.; Khan, K.; Lu, W.; Solis, L.M. Challenges and Opportunities for Immunoprofiling Using a Spatial High-Plex Technology: The NanoString GeoMx® Digital Spatial Profiler. Front. Oncol. 2022, 12, 890410. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Marzese, D.M.; Wang, X.; Yang, Z.; Li, C.; Zhang, H.; Zhang, J.; Chen, C.C.; Kelly, D.F.; et al. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab. Investig. 2019, 99, 1117–1129. [Google Scholar] [CrossRef]
- Kokubu, Y.; Tabu, K.; Muramatsu, N.; Wang, W.; Murota, Y.; Nobuhisa, I.; Jinushi, M.; Taga, T. Induction of protumoral CD11chighmacrophages by glioma cancer stem cells through GM-CSF. Genes Cells 2016, 21, 241–251. [Google Scholar] [CrossRef]
- Berger, G.; Knelson, E.H.; Jimenez-Macias, J.L.; Nowicki, M.O.; Han, S.; Panagioti, E.; Lizotte, P.H.; Adu-Berchie, K.; Stafford, A.; Dimitrakakis, N.; et al. STING activation promotes robust immune response and NK cell–mediated tumor regression in glioblastoma models. Proc. Natl. Acad. Sci. USA 2022, 119, e2111003119. [Google Scholar] [CrossRef]
- Ghouzlani, A.; Lakhdar, A.; Rafii, S.; Karkouri, M.; Badou, A. The immune checkpoint VISTA exhibits high expression levels in human gliomas and associates with a poor prognosis. Sci. Rep. 2021, 11, 21504. [Google Scholar] [CrossRef]
- Swann, J.B.; Smyth, M.J. Immune surveillance of tumors. J. Clin. Investig. 2007, 117, 1137–1146. [Google Scholar] [CrossRef]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Pitt, J.M.; Vétizou, M.; Daillère, R.; Roberti, M.P.; Yamazaki, T.; Routy, B.; Lepage, P.; Boneca, I.G.; Chamaillard, M.; Kroemer, G.; et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016, 44, 1255–1269. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Bausart, M.; Préat, V.; Malfanti, A. Immunotherapy for glioblastoma: The promise of combination strategies. J. Exp. Clin. Cancer Res. 2022, 41, 35. [Google Scholar] [CrossRef]
- Xuan, W.; Lesniak, M.S.; James, C.D.; Heimberger, A.B.; Chen, P. Context-Dependent Glioblastoma–Macrophage/Microglia Symbiosis and Associated Mechanisms. Trends Immunol. 2021, 42, 280–292. [Google Scholar] [CrossRef]
- Walker, D.G.; Lue, L.-F. Immune phenotypes of microglia in human neurodegenerative disease: Challenges to detecting microglial polarization in human brains. Alzheimers Res. Ther. 2015, 7, 56. [Google Scholar] [CrossRef]
- Diao, J.; Xia, T.; Zhao, H.; Liu, J.; Li, B.; Zhang, Z. Overexpression of HLA-DR is associated with prognosis of glioma patients. Int. J. Clin. Exp. Pathol. 2015, 8, 5485–5490. [Google Scholar]
- Wang, Z.; Wang, Z.; Zhang, C.; Liu, X.; Li, G.; Liu, S.; Sun, L.; Liang, J.; Hu, H.; Liu, Y.; et al. Genetic and clinical characterization of B7-H3 (CD276) expression and epigenetic regulation in diffuse brain glioma. Cancer Sci. 2018, 109, 2697–2705. [Google Scholar] [CrossRef]
- Nehama, D.; Di Ianni, N.; Musio, S.; Du, H.; Patané, M.; Pollo, B.; Finocchiaro, G.; Park, J.J.; Dunn, D.E.; Edwards, D.S.; et al. B7-H3-redirected chimeric antigen receptor T cells target glioblastoma and neurospheres. eBioMedicine 2019, 47, 33–43. [Google Scholar] [CrossRef]

| Patients | Sex | Age | Period between Surgeries | Survival after 2nd Surgery | Overall Survival | Localization | Karnofsky (Diagnosis) | Karnofsky (Recurrent) | IDH | ATRX | P53 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary | Recurrent | Primary | Recurrent | Primary | Recurrent | |||||||||
| 1 | M | 55 | 10.6 | 11 | 21.5 | left temporal | 90% | 80% | wt | wt | + | + | + | − |
| 2 | F | 56 | 8.6 | 8 | 16.6 | left frontal | 70% | 50% | wt | wt | + | + | + | − |
| 3 | M | 54 | 12.4 | 13 | 25.4 | left frontal | 90% | 60% | wt | wt | + | partial loss | + | + |
| 4 | M | 48 | 18.4 | 12 | 30.4 | left frontal | 90% | 70% | wt | wt | + | + | + | − |
| 5 | M | 42 | 15.7 | 16.5 | 26.7 | right frontal | 90% | 60% | wt | wt | + | + | + | + |
| 6 | M | 62 | 9 | 15.5 | 24.5 | left temporal | 70% | 80% | wt | wt | + | + | − | + |
| 7 | F | 51 | 26.8 | 25 | 51.8 | right frontal | 80% | 80% | wt | wt | + | + | − | − |
| 8 | M | 52 | 13.8 | 5 | 18.8 | right frontal | 50% | 70% | wt | wt | + | + | + | − |
| 9 | M | 43 | 16.2 | 9 | 25.2 | left frontal | 70% | 80% | wt | wt | + | + | + | − |
| 10 | F | 64 | 17.2 | 6 | 23.2 | left temporal | 70% | 60% | wt | wt | nd | + | − | + |
| 11 | M | 72 | 6.8 | 6.5 | 13.3 | right frontal | 80% | 70% | wt | wt | + | partial loss | + | + |
| 12 | F | 48 | 13.3 | 12 | 25.3 | right parietal | 50% | 90% | wt | wt | + | + | + | + |
| 13 | M | 72 | 12.6 | 19.2 | 31.8 | right frontal | 90% | 90% | wt | wt | + | + | + | + |
| 14 | F | 56 | 14.2 | 6.5 | 20.7 | right parieto–occipital | 80% | 70% | wt | wt | + | + | + | − |
| 15 | M | 49 | 12.7 | 11 | 23.7 | left parietal | 90% | 80% | wt | wt | + | + | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loussouarn, D.; Oliver, L.; Salaud, C.; Samarut, E.; Bourgade, R.; Béroud, C.; Morenton, E.; Heymann, D.; Vallette, F.M. Spatial Distribution of Immune Cells in Primary and Recurrent Glioblastoma: A Small Case Study. Cancers 2023, 15, 3256. https://doi.org/10.3390/cancers15123256
Loussouarn D, Oliver L, Salaud C, Samarut E, Bourgade R, Béroud C, Morenton E, Heymann D, Vallette FM. Spatial Distribution of Immune Cells in Primary and Recurrent Glioblastoma: A Small Case Study. Cancers. 2023; 15(12):3256. https://doi.org/10.3390/cancers15123256
Chicago/Turabian StyleLoussouarn, Delphine, Lisa Oliver, Celine Salaud, Edouard Samarut, Raphaël Bourgade, Christophe Béroud, Emilie Morenton, Dominique Heymann, and Francois M. Vallette. 2023. "Spatial Distribution of Immune Cells in Primary and Recurrent Glioblastoma: A Small Case Study" Cancers 15, no. 12: 3256. https://doi.org/10.3390/cancers15123256
APA StyleLoussouarn, D., Oliver, L., Salaud, C., Samarut, E., Bourgade, R., Béroud, C., Morenton, E., Heymann, D., & Vallette, F. M. (2023). Spatial Distribution of Immune Cells in Primary and Recurrent Glioblastoma: A Small Case Study. Cancers, 15(12), 3256. https://doi.org/10.3390/cancers15123256






