RUNX Family as a Promising Biomarker and a Therapeutic Target in Bone Cancers: A Review on Its Molecular Mechanism(s) behind Tumorigenesis
Abstract
Simple Summary
Abstract
1. Introduction
2. The RUNX Family
3. RUNX2’s Physiological and Pathological Functions
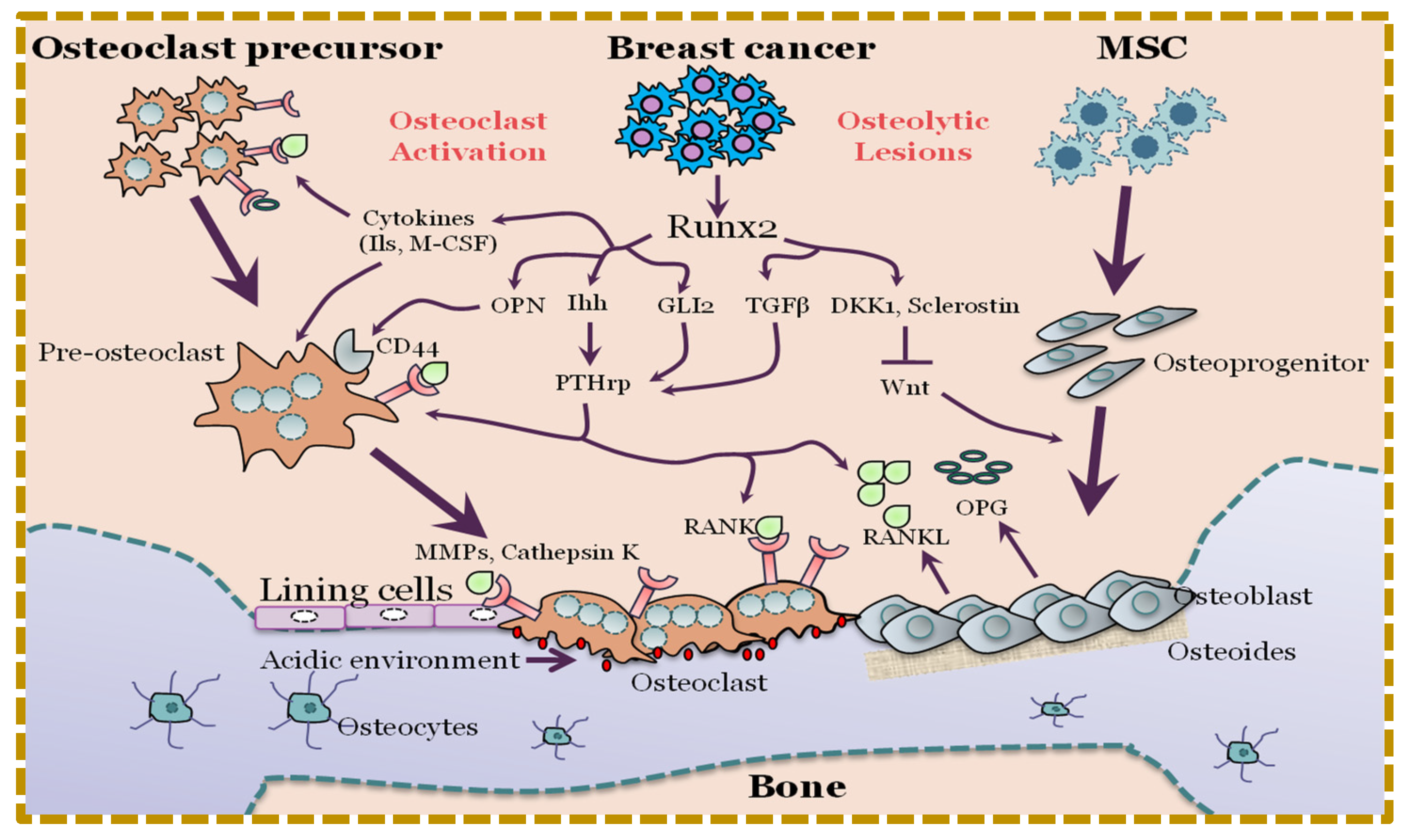
4. Deregulation of RUNX2 Signaling in Osteogenesis May Lead to Oncogenesis
5. RUNX2 May Be an Important Player in Osteosarcoma
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Mutsaers, A.J.; Walkley, C.R. Cells of origin in osteosarcoma: Mesenchymal stem cells or osteoblast committed cells? Bone 2014, 62, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.; Occean, B.-V.; Pacquement, H.; Bompas, E.; Bouvier, C.; Brisse, H.J.; Castex, M.-P.; Cheurfa, N.; Corradini, N.; Delaye, J.; et al. Results of methotrexate-etoposide-ifosfamide based regimen (M-EI) in osteosarcoma patients included in the French OS2006/sarcome-09 study. Eur. J. Cancer 2018, 88, 57–66. [Google Scholar] [CrossRef]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex but Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef]
- Li, Y.; Ge, C.; Franceschi, R.T. Role of Runx2 in prostate development and stem cell function. Prostate 2021, 81, 231–241. [Google Scholar] [CrossRef]
- Westendorf, J.J. Transcriptional co-repressors of Runx2. J. Cell. Biochem. 2006, 98, 54–64. [Google Scholar] [CrossRef]
- Zhang, Y.; Duan, X. A Novel 90-kbp Deletion of RUNX2 Associated with Cleidocranial Dysplasia. Genes 2022, 13, 1128. [Google Scholar] [CrossRef]
- Martin, J.W.; Zielenska, M.; Stein, G.S.; van Wijnen, A.J.; Squire, J.A. The Role of RUNX2 in Osteosarcoma Oncogenesis. Sarcoma 2011, 2011, 282745. [Google Scholar] [CrossRef]
- Miyoshi, H.; Shimizu, K.; Kozu, T.; Maseki, N.; Kaneko, Y.; Ohki, M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 1991, 88, 10431–10434. [Google Scholar] [CrossRef]
- Schlegelberger, B.; Heller, P.G. RUNX1 deficiency (familial platelet disorder with predisposition to myeloid leukemia, FPDMM). Semin. Hematol. 2017, 54, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Bae, S.-C.; Chuang, L.S.H. The RUNX family: Developmental regulators in cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, D.; Brandi, J.; Manfredi, M.; Serena, M.; Carbonare, L.D.; Deiana, M.; Cheri, S.; Parolini, F.; Gandini, A.; Marchetto, G.; et al. Runx2 stimulates neoangiogenesis through the Runt domain in melanoma. Sci. Rep. 2019, 9, 8052. [Google Scholar] [CrossRef] [PubMed]
- Vitale, E.; Sauta, E.; Gugnoni, M.; Torricelli, F.; Manicardi, V.; Ciarrocchi, A. A multimodal integrative approach to model transcriptional addiction of thyroid cancer on RUNX2. Cancer Commun. 2022, 42, 892–896. [Google Scholar] [CrossRef]
- Guan, Y.; Zhang, Y.; Hao, L.; Nie, Z. CircRNA_102272 Promotes Cisplatin-Resistance in Hepatocellular Carcinoma by Decreasing MiR-326 Targeting of RUNX2. Cancer Manag. Res. 2020, 12, 12527–12534. [Google Scholar] [CrossRef] [PubMed]
- Matthijssens, F.; Sharma, N.D.; Nysus, M.; Nickl, C.K.; Kang, H.; Perez, D.R.; Lintermans, B.; Van Loocke, W.; Roels, J.; Peirs, S.; et al. RUNX2 regulates leukemic cell metabolism and chemotaxis in high-risk T cell acute lymphoblastic leukemia. J. Clin. Investig. 2021, 131, e141566. [Google Scholar] [CrossRef]
- Ji, Q.; Cai, G.; Liu, X.; Zhang, Y.; Wang, Y.; Zhou, L.; Sui, H.; Li, Q. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019, 10, 378. [Google Scholar] [CrossRef]
- Zhang, F.; Su, T.; Xiao, M. RUNX3-regulated circRNA METTL3 inhibits colorectal cancer proliferation and metastasis via miR-107/PER3 axis. Cell Death Dis. 2022, 13, 550. [Google Scholar] [CrossRef]
- Liu, H.; Xue, Q.; Cai, H.; Jiang, X.; Cao, G.; Chen, T.; Chen, Y.; Wang, D. RUNX3-mediated circDYRK1A inhibits glutamine metabolism in gastric cancer by up-regulating microRNA-889-3p-dependent FBXO4. J. Transl. Med. 2022, 20, 120. [Google Scholar] [CrossRef]
- Wang, L.; Tang, W.; Yang, S.; He, P.; Wang, J.; Gaedcke, J.; Ströbel, P.; Azizian, A.; Ried, T.; Gaida, M.M.; et al. NO•/RUNX3/kynurenine metabolic signaling enhances disease aggressiveness in pancreatic cancer. Int. J. Cancer 2020, 146, 3160–3169. [Google Scholar] [CrossRef]
- Mevel, R.; Draper, J.E.; Lie-A-Ling, M.; Kouskoff, V.; Lacaud, G. RUNX transcription factors: Orchestrators of development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef]
- Nagata, K.; Hojo, H.; Chang, S.H.; Okada, H.; Yano, F.; Chijimatsu, R.; Omata, Y.; Mori, D.; Makii, Y.; Kawata, M.; et al. Runx2 and Runx3 differentially regulate articular chondrocytes during surgically induced osteoarthritis development. Nat. Commun. 2022, 13, 6187. [Google Scholar] [CrossRef]
- Villanueva, F.; Araya, H.; Briceño, P.; Varela, N.; Stevenson, A.; Jerez, S.; Tempio, F.; Chnaiderman, J.; Perez, C.; Villarroel, M.; et al. The cancer-related transcription factor RUNX2 modulates expression and secretion of the matricellular protein osteopontin in osteosarcoma cells to promote adhesion to endothelial pulmonary cells and lung metastasis. J. Cell. Physiol. 2019, 234, 13659–13679. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; AlJohani, H.; Majumdar, S.; Chellaiah, M.A. Characterization of CD44 intracellular domain interaction with RUNX2 in PC3 human prostate cancer cells. Cell Commun. Signal. 2019, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Teng, X.; Ma, T.; Yang, T.; Zhang, J.; Huo, M.; Liu, W.; Yang, Y.; Yuan, B.; Yu, H.; et al. RUNX2 recruits the NuRD(MTA1)/CRL4B complex to promote breast cancer progression and bone metastasis. Cell Death Differ. 2022, 29, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.W.; Tan, Z.; To, M.K.T.; Chan, D. Regulation and Role of Transcription Factors in Osteogenesis. Int. J. Mol. Sci. 2021, 22, 5445. [Google Scholar] [CrossRef] [PubMed]
- Coffman, J.A. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 2003, 27, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Feng, J.; Guo, T.; Loh, Y.-H.E.; Yuan, Y.; Ho, T.-V.; Cho, C.K.; Li, J.; Jing, J.; Janeckova, E.; et al. Runx2-Twist1 interaction coordinates cranial neural crest guidance of soft palate myogenesis. eLife 2021, 10, e62387. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.S.; Liu, Y.L.; Tang, X.T.; Zhang, X.S.; Zhou, B.; Zou, W.; Zhou, B.O. Tracing the skeletal progenitor transition during postnatal bone formation. Cell Stem Cell 2021, 28, 2122–2136.e3. [Google Scholar] [CrossRef]
- Liu, T.M.; Lee, E.H. Transcriptional Regulatory Cascades in Runx2-Dependent Bone Development. Tissue Eng. Part B Rev. 2013, 19, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Jiang, Q.; Nagano, K.; Moriishi, T.; Miyazaki, T.; Komori, H.; Ito, K.; Von Der Mark, K.; Sakane, C.; Kaneko, H.; et al. Runx2 is essential for the transdifferentiation of chondrocytes into osteoblasts. PLoS Genet. 2020, 16, e1009169. [Google Scholar] [CrossRef] [PubMed]
- Owens, T.W.; Rogers, R.L.; Best, S.A.; Ledger, A.; Mooney, A.-M.; Ferguson, A.; Shore, P.; Swarbrick, A.; Ormandy, C.J.; Simpson, P.T.; et al. Runx2 Is a Novel Regulator of Mammary Epithelial Cell Fate in Development and Breast Cancer. Cancer Res. 2014, 74, 5277–5286. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, N.; Riggio, A.I.; Mason, S.; McDonald, L.; King, A.; Higgins, T.; Rosewell, I.; Neil, J.C.; Smalley, M.J.; Sansom, O.J.; et al. Runx2 contributes to the regenerative potential of the mammary epithelium. Sci. Rep. 2015, 5, 15658. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.; Ferrari, N.; Terry, A.; Bell, M.; Mohammed, Z.M.; Orange, C.; Jenkins, A.; Muller, W.J.; Gusterson, B.A.; Neil, J.C.; et al. RUNX2 correlates with subtype-specific breast cancer in a human tissue microarray, and ectopic expression of Runx2 perturbs differentiation in the mouse mammary gland. Dis. Model. Mech. 2014, 7, 525–534. [Google Scholar] [CrossRef]
- De Winter, T.J.J.; Nusse, R. Running Against the Wnt: How Wnt/β-Catenin Suppresses Adipogenesis. Front. Cell Dev. Biol. 2021, 9, 627429. [Google Scholar] [CrossRef]
- Kawane, T.; Qin, X.; Jiang, Q.; Miyazaki, T.; Komori, H.; Yoshida, C.A.; Matsuura-Kawata, V.K.D.S.; Sakane, C.; Matsuo, Y.; Nagai, K.; et al. Runx2 is required for the proliferation of osteoblast progenitors and induces proliferation by regulating Fgfr2 and Fgfr3. Sci. Rep. 2018, 8, 13551. [Google Scholar] [CrossRef]
- Moorer, M.C.; Riddle, R.C. Regulation of Osteoblast Metabolism by Wnt Signaling. Endocrinol. Metab. 2018, 33, 318–330. [Google Scholar] [CrossRef]
- Felber, K.; Elks, P.M.; Lecca, M.; Roehl, H.H. Expression of osterix Is Regulated by FGF and Wnt/β-Catenin Signalling during Osteoblast Differentiation. PLoS ONE 2015, 10, e0144982. [Google Scholar] [CrossRef]
- Kim, J.-H.; Kim, M.; Hong, S.; Kim, E.-Y.; Lee, H.; Jung, H.-S.; Sohn, Y. Albiflorin Promotes Osteoblast Differentiation and Healing of Rat Femoral Fractures Through Enhancing BMP-2/Smad and Wnt/β-Catenin Signaling. Front. Pharmacol. 2021, 12, 690113. [Google Scholar] [CrossRef]
- Ogasawara, T.; Kawaguchi, H.; Jinno, S.; Hoshi, K.; Itaka, K.; Takato, T.; Nakamura, K.; Okayama, H. Bone Morphogenetic Protein 2-Induced Osteoblast Differentiation Requires Smad-Mediated Down-Regulation of Cdk6. Mol. Cell. Biol. 2004, 24, 6560–6568. [Google Scholar] [CrossRef]
- Jacobsen, C.M.; Barber, L.A.; Ayturk, U.M.; Roberts, H.J.; Deal, L.E.; Schwartz, M.A.; Weis, M.; Eyre, D.; Zurakowski, D.; Robling, A.G.; et al. Targeting the LRP5 Pathway Improves Bone Properties in a Mouse Model of Osteogenesis Imperfecta. J. Bone Miner. Res. 2014, 29, 2297–2306. [Google Scholar] [CrossRef]
- Rangaswami, H.; Schwappacher, R.; Tran, T.; Chan, G.C.; Zhuang, S.; Boss, G.R.; Pilz, R.B. Protein Kinase G and Focal Adhesion Kinase Converge on Src/Akt/β-Catenin Signaling Module in Osteoblast Mechanotransduction. J. Biol. Chem. 2012, 287, 21509–21519. [Google Scholar] [CrossRef]
- Almeida, M.; Han, L.; Bellido, T.; Manolagas, S.C.; Kousteni, S. Wnt Proteins Prevent Apoptosis of Both Uncommitted Osteoblast Progenitors and Differentiated Osteoblasts by β-Catenin-dependent and -independent Signaling Cascades Involving Src/ERK and Phosphatidylinositol 3-Kinase/AKT. J. Biol. Chem. 2005, 280, 41342–41351. [Google Scholar] [CrossRef]
- Ali, S.A.; Zaidi, S.K.; Dacwag, C.S.; Salma, N.; Young, D.W.; Shakoori, A.R.; Montecino, M.A.; Lian, J.B.; van Wijnen, A.J.; Imbalzano, A.N.; et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc. Natl. Acad. Sci. USA 2008, 105, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.K.; Young, D.W.; Montecino, M.A.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Mitotic bookmarking of genes: A novel dimension to epigenetic control. Nat. Rev. Genet. 2010, 11, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.K.; Young, D.W.; Montecino, M.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Bookmarking the Genome: Maintenance of Epigenetic Information. J. Biol. Chem. 2011, 286, 18355–18361. [Google Scholar] [CrossRef]
- Li, Z.; Hong, Z.; Peng, Z.; Zhao, Y.; Shao, R. Acetylshikonin from Zicao ameliorates renal dysfunction and fibrosis in diabetic mice by inhibiting TGF-β1/Smad pathway. Hum. Cell 2018, 31, 199–209. [Google Scholar] [CrossRef]
- Westendorf, J.J.; Zaidi, S.K.; Cascino, J.E.; Kahler, R.; Van Wijnen, A.J.; Lian, J.B.; Yoshida, M.; Stein, G.S.; Li, X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21CIP1/WAF1 promoter. Mol. Cell. Biol. 2002, 22, 7982–7992. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.-P.; Chan, W.-P.; Hsieh, M.-S.; Chen, R.-M. Runx2-mediated bcl-2 gene expression contributes to nitric oxide protection against hydrogen peroxide-induced osteoblast apoptosis. J. Cell. Biochem. 2009, 108, 1084–1093. [Google Scholar] [CrossRef]
- Zaragoza, C.; López-Rivera, E.; García-Rama, C.; Saura, M.; Martínez-Ruíz, A.; Lizarbe, T.R.; Martín-De-Lara, F.; Lamas, S. Cbfa-1 mediates nitric oxide regulation of MMP-13 in osteoblasts. J. Cell Sci. 2006, 119, 1896–1902. [Google Scholar] [CrossRef]
- Ortega, N.; Behonick, D.J.; Werb, Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004, 14, 86–93. [Google Scholar] [CrossRef]
- Fukuyama, R.; Fujita, T.; Azuma, Y.; Hirano, A.; Nakamuta, H.; Koida, M.; Komori, T. Statins inhibit osteoblast migration by inhibiting Rac-Akt signaling. Biochem. Biophys. Res. Commun. 2004, 315, 636–642. [Google Scholar] [CrossRef]
- Hughes, F.J.; Aubin, J.E.; Heersche, J.N. Differential chemotactic responses of different populations of fetal rat calvaria cells to platelet-derived growth factor and transforming growth factor β. Bone Miner. 1992, 19, 63–74. [Google Scholar] [CrossRef]
- Panagakos, F. Insulin-like growth factors-I and -II stimulate chemotaxis of osteoblasts isolated from fetal rat calvaria. Biochimie 1993, 75, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Azuma, Y.; Fukuyama, R.; Hattori, Y.; Yoshida, C.; Koida, M.; Ogita, K.; Komori, T. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J. Cell Biol. 2004, 166, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Johnson, S.A.; Sims, N.; Trivett, M.K.; Slavin, J.L.; Rubin, B.P.; Waring, P.; McArthur, G.A.; Walkley, C.; Holloway, A.J.; et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J. Cell Biol. 2004, 167, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Berman, S.D.; Calo, E.; Landman, A.S.; Danielian, P.S.; Miller, E.S.; West, J.C.; Fonhoue, B.D.; Caron, A.; Bronson, R.; Bouxsein, M.L.; et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc. Natl. Acad. Sci. USA 2008, 105, 11851–11856. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Thomas, D.M.; Gutierrez, G.; Carty, S.A.; Yanagawa, S.-I.; Hinds, P.W. HES1 Cooperates With pRb to Activate RUNX2-Dependent Transcription. J. Bone Miner. Res. 2006, 21, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.M.; Carty, S.A.; Piscopo, D.M.; Lee, J.S.; Wang, W.F.; Forrester, W.C.; Hinds, P.W. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 2001, 8, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.; Villagra, A.; Paredes, R.; Cruzat, F.; Gutierrez, S.; Javed, A.; Arriagada, G.; Olate, J.; Imschenetzky, M.; Van Wijnen, A.J.; et al. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 2003, 23, 3339–3351. [Google Scholar] [CrossRef]
- Schroeder, T.M.; Kahler, R.A.; Li, X.; Westendorf, J.J. Histone Deacetylase 3 Interacts with Runx2 to Repress the Osteocalcin Promoter and Regulate Osteoblast Differentiation. J. Biol. Chem. 2004, 279, 41998–42007. [Google Scholar] [CrossRef]
- Pelletier, N.; Champagne, N.; Stifani, S.; Yang, X.-J. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene 2002, 21, 2729–2740. [Google Scholar] [CrossRef]
- Zaidi, S.K.; Sullivan, A.J.; Medina, R.; Ito, Y.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004, 23, 790–799. [Google Scholar] [CrossRef]
- Vega, R.B.; Matsuda, K.; Oh, J.; Barbosa, A.C.; Yang, X.; Meadows, E.; McAnally, J.; Pomajzl, C.; Shelton, J.M.; Richardson, J.A.; et al. Histone Deacetylase 4 Controls Chondrocyte Hypertrophy during Skeletogenesis. Cell 2004, 119, 555–566. [Google Scholar] [CrossRef]
- Andela, V.B.; Siddiqui, F.; Groman, A.; Rosier, R.N. An immunohistochemical analysis to evaluate an inverse correlation between Runx2/Cbfa1 and NFκB in human osteosarcoma. J. Clin. Pathol. 2005, 58, 328–330. [Google Scholar] [CrossRef]
- Sadikovic, B.; Thorner, P.; Chilton-MacNeill, S.; Martin, J.W.; Cervigne, N.K.; Squire, J.; Zielenska, M. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer 2010, 10, 202. [Google Scholar] [CrossRef]
- Maruyama, Z.; Yoshida, C.A.; Furuichi, T.; Amizuka, N.; Ito, M.; Fukuyama, R.; Miyazaki, T.; Kitaura, H.; Nakamura, K.; Fujita, T.; et al. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev. Dyn. 2007, 236, 1876–1890. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Song, W.-X.; Luo, J.; Haydon, R.C.; He, T.-C. Osteosarcoma Development and Stem Cell Differentiation. Clin. Orthop. Relat. Res. 2008, 466, 2114–2130. [Google Scholar] [CrossRef]
- Won, K.Y.; Park, H.-R.; Park, Y.-K. Prognostic Implication of Immunohistochemical Runx2 Expression in Osteosarcoma. Tumori J. 2009, 95, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Kurek, K.C.; Del Mare, S.; Salah, Z.; Abdeen, S.; Sadiq, H.; Lee, S.-H.; Gaudio, E.; Zanesi, N.; Jones, K.B.; DeYoung, B.; et al. Frequent Attenuation of the WWOX Tumor Suppressor in Osteosarcoma Is Associated with Increased Tumorigenicity and Aberrant RUNX2 Expression. Cancer Res. 2010, 70, 5577–5586. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.A.S.; Varela, N.; Gaete, M.; Villegas, K.; Osorio, M.; Tapia, J.C.; Antonelli, M.; Mancilla, E.E.; Pereira, B.P.; Nathan, S.S.; et al. Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfβ in osteosarcoma cells. J. Cell. Physiol. 2009, 221, 560–571. [Google Scholar] [CrossRef]
- Berman, S.D.; Yuan, T.L.; Miller, E.S.; Lee, E.Y.; Caron, A.; Lees, J.A. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol. Cancer Res. 2008, 6, 1440–1451. [Google Scholar] [CrossRef]
- Goodrich, D.W. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 2006, 25, 5233–5243. [Google Scholar] [CrossRef]
- Van Harn, T.; Foijer, F.; van Vugt, M.; Banerjee, R.; Yang, F.; Oostra, A.; Joenje, H.; Riele, H.T. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010, 24, 1377–1388. [Google Scholar] [CrossRef]
- Zambetti, G.P.; Horwitz, E.M.; Schipani, E. Skeletons in the p53 tumor suppressor closet: Genetic evidence that p53 blocks bone differentiation and development. J. Cell Biol. 2006, 172, 795–797. [Google Scholar] [CrossRef]
- Walkley, C.R.; Qudsi, R.; Sankaran, V.G.; Perry, J.A.; Gostissa, M.; Roth, S.I.; Rodda, S.J.; Snay, E.; Dunning, P.; Fahey, F.H.; et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008, 22, 1662–1676. [Google Scholar] [CrossRef] [PubMed]
- Lengner, C.J.; Steinman, H.A.; Gagnon, J.; Smith, T.W.; Henderson, J.E.; Kream, B.E.; Stein, G.S.; Lian, J.B.; Jones, S.N. Osteoblast differentiation and skeletal development are regulated by Mdm2–p53 signaling. J. Cell Biol. 2006, 172, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.; Koff, A. Cell-cycle inhibitors: Three families united by a common cause. Gene 2000, 247, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Engin, F.; Yao, Z.; Yang, T.; Zhou, G.; Bertin, T.; Jiang, M.M.; Chen, Y.; Wang, L.; Zheng, H.; Sutton, R.E.; et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 2008, 14, 299–305. [Google Scholar] [CrossRef]
- Shen, R.; Wang, X.; Drissi, H.; Liu, F.; O’Keefe, R.J.; Chen, D. Cyclin D1-Cdk4 Induce Runx2 Ubiquitination and Degradation. J. Biol. Chem. 2006, 281, 16347–16353. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, Y.; Zweidler-McKay, P.A.; Hughes, D.P.M. Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin. Cancer Res. 2008, 14, 2962–2969. [Google Scholar] [CrossRef]
- Zaidi, S.K.; Pande, S.; Pratap, J.; Gaur, T.; Grigoriu, S.; Ali, S.A.; Stein, J.L.; Lian, J.B.; Van Wijnen, A.J.; Stein, G.S. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc. Natl. Acad. Sci. USA 2007, 104, 19861–19866. [Google Scholar] [CrossRef]
- Todosenko, N.; Khlusov, I.; Yurova, K.; Khaziakhmatova, O.; Litvinova, L. Signal Pathways and microRNAs in Osteosarcoma Growth and the Dual Role of Mesenchymal Stem Cells in Oncogenesis. Int. J. Mol. Sci. 2023, 24, 8993. [Google Scholar] [CrossRef]
- Tsai, H.-C.; Cheng, S.-P.; Han, C.-K.; Huang, Y.-L.; Wang, S.-W.; Lee, J.-J.; Lai, C.-T.; Fong, Y.-C.; Tang, C.-H. Resistin enhances angiogenesis in osteosarcoma via the MAPK signaling pathway. Aging 2019, 11, 9767–9777. [Google Scholar] [CrossRef] [PubMed]
- Verrecchia, F.; Rédini, F. Transforming Growth Factor-β Signaling Plays a Pivotal Role in the Interplay Between Osteosarcoma Cells and Their Microenvironment. Front. Oncol. 2018, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, X.-H.; Yan, Y.-G.; Wang, C.; Wang, W.-J. PI3K/Akt signaling in osteosarcoma. Clin. Chim. Acta 2015, 444, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Soki, F.N.; Park, S.I.; McCauley, L.K. The multifaceted actions of PTHrP in skeletal metastasis. Future Oncol. 2012, 8, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Dapunt, U.; Giese, T.; Stegmaier, S.; Moghaddam, A.; Hänsch, G.M. The osteoblast as an inflammatory cell: Production of cytokines in response to bacteria and components of bacterial biofilms. BMC Musculoskelet. Disord. 2016, 17, 243. [Google Scholar] [CrossRef]
- Bendre, M.S.; Margulies, A.G.; Walser, B.; Akel, N.S.; Bhattacharrya, S.; Skinner, R.A.; Swain, F.; Ramani, V.; Mohammad, K.S.; Wessner, L.L.; et al. Tumor-derived interleukin-8 stimulates osteolysis independent of the receptor activator of nuclear factor-κB ligand pathway. Cancer Res. 2005, 65, 11001–11009. [Google Scholar] [CrossRef]
- Branstetter, D.; Rohrbach, K.; Huang, L.-Y.; Soriano, R.; Tometsko, M.; Blake, M.; Jacob, A.P.; Dougall, W.C. RANK and RANK ligand expression in primary human osteosarcoma. J. Bone Oncol. 2015, 4, 59–68. [Google Scholar] [CrossRef]
- Lim, M.; Zhong, C.; Yang, S.; Bell, A.M.; Cohen, M.B.; Roy-Burman, P. Runx2 regulates survivin expression in prostate cancer cells. Lab. Investig. 2010, 90, 222–233. [Google Scholar] [CrossRef]
- Van der Deen, M.; Akech, J.; Wang, T.; FitzGerald, T.J.; Altieri, D.C.; Languino, L.R.; Lian, J.B.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFβ in prostate cancer cells. J. Cell. Biochem. 2010, 109, 828–837. [Google Scholar] [CrossRef]
- Ashe, H.; Krakowiak, P.; Hasterok, S.; Sleppy, R.; Roller, D.G.; Gioeli, D. Role of the runt-related transcription factor (RUNX) family in prostate cancer. FEBS J. 2021, 288, 6112–6126. [Google Scholar] [CrossRef]
- Wysokinski, D.; Blasiak, J.; Pawlowska, E. Role of RUNX2 in Breast Carcinogenesis. Int. J. Mol. Sci. 2015, 16, 20969–20993. [Google Scholar] [CrossRef]
- Pratap, J.; Wixted, J.J.; Gaur, T.; Zaidi, S.K.; Dobson, J.; Gokul, K.D.; Hussain, S.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Runx2 Transcriptional Activation of Indian Hedgehog and a Downstream Bone Metastatic Pathway in Breast Cancer Cells. Cancer Res. 2008, 68, 7795–7802. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC Activation Is a Hallmark of Cancer Initiation and Maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef] [PubMed]
- Righi, A.; Morandi, L.; Leonardi, E.; Farnedi, A.; Marucci, G.; Sisto, A.; Frank, G.; Faustini-Fustini, M.; Zoli, M.; Mazzatenta, D.; et al. Galectin-3 expression in pituitary adenomas as a marker of aggressive behavior. Hum. Pathol. 2013, 44, 2400–2409. [Google Scholar] [CrossRef]
- Ahmed, H.; AlSadek, D.M.M. Galectin-3 as a Potential Target to Prevent Cancer Metastasis. Clin. Med. Insights Oncol. 2015, 9, 113–121. [Google Scholar] [CrossRef]
- Silva, B.C.; Bilezikian, J.P. Parathyroid hormone: Anabolic and catabolic actions on the skeleton. Curr. Opin. Pharmacol. 2015, 22, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; de Andrés, M.C.; Hashimoto, K.; Itoi, E.; Otero, M.; Goldring, M.B.; Oreffo, R.O.C. DNA methylation of the RUNX2 P1 promoter mediates MMP13 transcription in chondrocytes. Sci. Rep. 2017, 7, 7771. [Google Scholar] [CrossRef] [PubMed]
- D’Alonzo, R.C.; Selvamurugan, N.; Karsenty, G.; Partridge, N.C. Physical Interaction of the Activator Protein-1 Factors c-Fos and c-Jun with Cbfa1 for Collagenase-3 Promoter Activation. J. Biol. Chem. 2002, 277, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Papachristou, D.J.; Papachristou, G.J.; Papaefthimiou, O.A.; Agnantis, N.J.; Basdra, E.K.; Papavassiliou, A.G. The MAPK-AP-1/-Runx2 signalling axes are implicated in chondrosarcoma pathobiology either independently or via up-regulation of VEGF. Histopathology 2005, 47, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Vishal, M.; Swetha, R.; Thejaswini, G.; Arumugam, B.; Selvamurugan, N. Role of Runx2 in breast cancer-mediated bone metastasis. Int. J. Biol. Macromol. 2017, 99, 608–614. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vimalraj, S.; Sekaran, S. RUNX Family as a Promising Biomarker and a Therapeutic Target in Bone Cancers: A Review on Its Molecular Mechanism(s) behind Tumorigenesis. Cancers 2023, 15, 3247. https://doi.org/10.3390/cancers15123247
Vimalraj S, Sekaran S. RUNX Family as a Promising Biomarker and a Therapeutic Target in Bone Cancers: A Review on Its Molecular Mechanism(s) behind Tumorigenesis. Cancers. 2023; 15(12):3247. https://doi.org/10.3390/cancers15123247
Chicago/Turabian StyleVimalraj, Selvaraj, and Saravanan Sekaran. 2023. "RUNX Family as a Promising Biomarker and a Therapeutic Target in Bone Cancers: A Review on Its Molecular Mechanism(s) behind Tumorigenesis" Cancers 15, no. 12: 3247. https://doi.org/10.3390/cancers15123247
APA StyleVimalraj, S., & Sekaran, S. (2023). RUNX Family as a Promising Biomarker and a Therapeutic Target in Bone Cancers: A Review on Its Molecular Mechanism(s) behind Tumorigenesis. Cancers, 15(12), 3247. https://doi.org/10.3390/cancers15123247




