Enhancement of Tumor Cell Immunogenicity and Antitumor Properties Derived from Platinum-Conjugated Iron Nanoparticles
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Chemistry
2.2.1. Preparation and Characterization of Platinum Derivate
2.2.2. Synthesis of IONPs
2.2.3. Incubation of IONPs with [PtCl(GUDCA)en]
2.2.4. Physical Characterization
2.2.5. Release of the Platinum Complex Bound to the NPs
2.3. Biological Assays
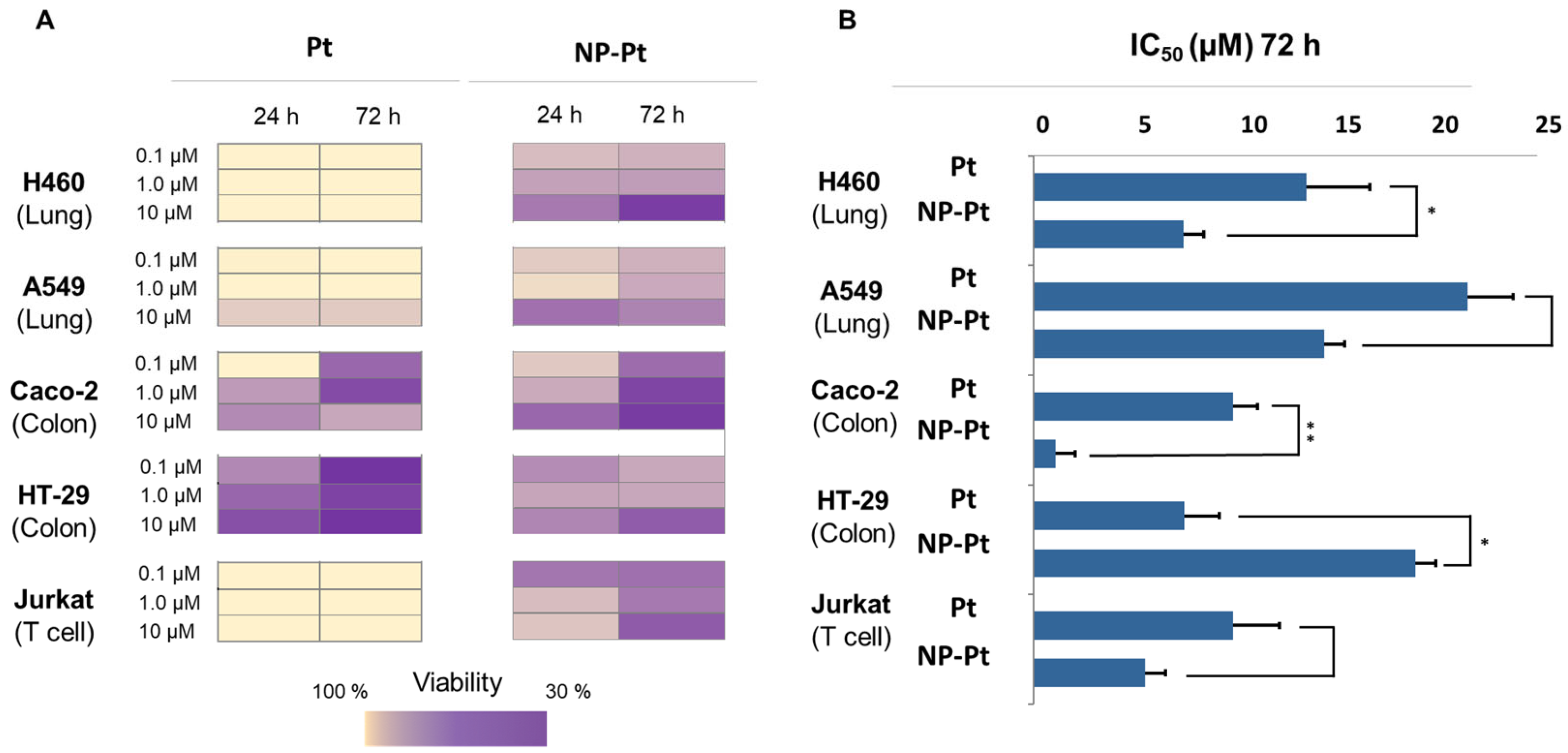
2.3.1. Cell Viability
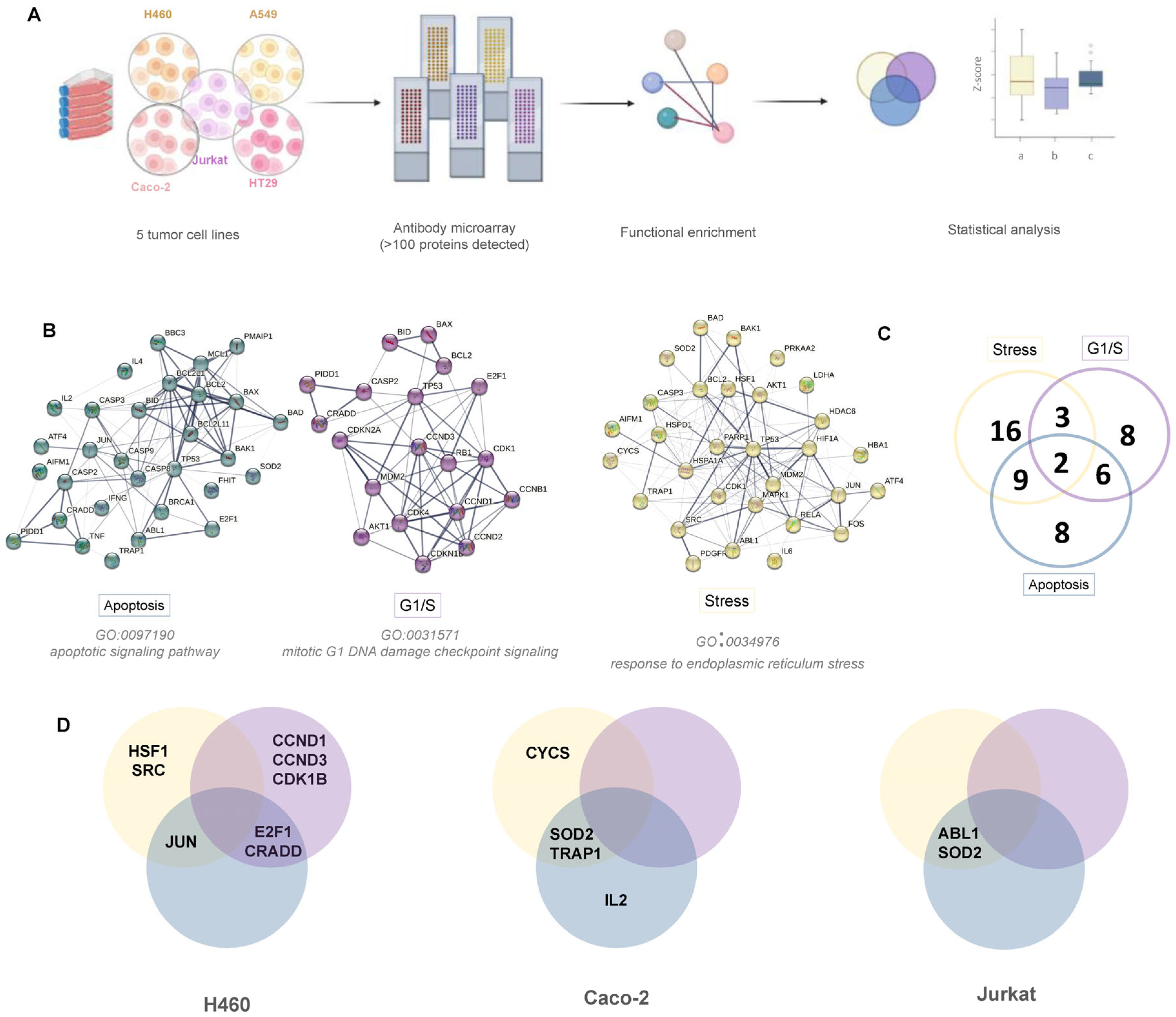
2.3.2. Protein Microarray Assays
- Cell lysis, protein extraction, and biotin labelling
- Protein arrays processing, image acquisition, and data processing
3. Results and Discussion
3.1. Chemistry
Preparation and Characterization of the Platinum Compound with Glycoursodeoxycholic Acid, [PtCl(GUDCA)en]
3.2. Biological Assays
3.2.1. Tumor Cell Cytotoxicity by Viability Assays
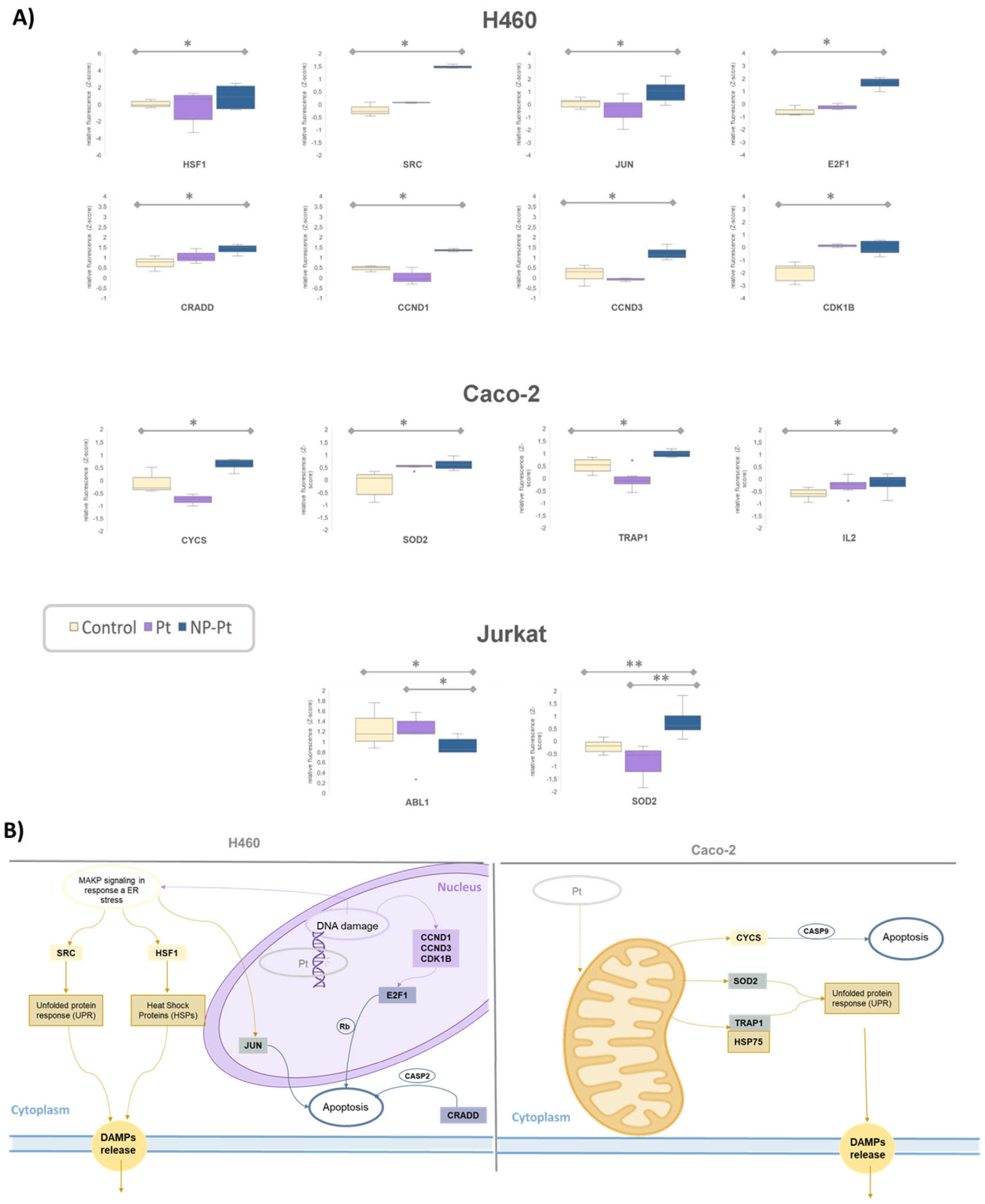
3.2.2. Analysis of Signaling Pathways by Antibody Microarrays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Yang, B.; Shi, J. Developing New Cancer Nanomedicines by Repurposing Old Drugs. Angew. Chem. Int. Ed. 2020, 59, 21829–21838. [Google Scholar] [CrossRef] [PubMed]
- Rottenberg, S.; Disler, C.; Perego, P. The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer 2021, 21, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Pourmadadi, M.; Eshaghi, M.M.; Rahmani, E.; Ajalli, N.; Bakhshi, S.; Mirkhaef, H.; Lasemi, M.V.; Rahdar, A.; Behzadmehr, R.; Diez-Pascual, A.M. Cisplatin-loaded nanoformulations for cancer therapy: A comprehensive review. J. Drug Deliv. Sci. Technol. 2022, 77, 103928. [Google Scholar] [CrossRef]
- Pavan, S.R.; Prabhu, A. Advanced cisplatin nanoformulations as targeted drug delivery platforms for lung carcinoma treatment: A review. J. Mater. Sci. 2022, 57, 16192–16227. [Google Scholar] [CrossRef]
- Han, Y.; Wen, P.; Li, J.; Kataoka, K. Targeted nanomedicine in cisplatin-based cancer therapeutics. J. Control. Release 2022, 345, 709–720. [Google Scholar] [CrossRef]
- Acebes-Fernandez, V.; Landeira-Vinuela, A.; Juanes-Velasco, P.; Hernandez, A.-P.; Otazo-Perez, A.; Manzano-Roman, R.; Gongora, R.; Fuentes, M. Nanomedicine and Onco-Immunotherapy: From the Bench to Bedside to Biomarkers. Nanomaterials 2020, 10, 1274. [Google Scholar] [CrossRef] [PubMed]
- Su, S.H.; Chen, Y.T.; Zhang, P.F.; Ma, R.J.; Zhang, W.; Liu, J.N.; Li, T.; Niu, H.J.; Cao, Y.; Hu, B.; et al. The role of Platinum(IV)-based antitumor drugs and the anticancer immune response in medicinal inorganic chemistry. A systematic review from 2017 to 2022. Eur. J. Med. Chem. 2022, 243, 114680. [Google Scholar] [CrossRef]
- Hernandez, A.P.; Juanes-Velasco, P.; Landeira-Vinuela, A.; Bareke, H.; Montalvillo, E.; Gongora, R.; Fuentes, M. Restoring the Immunity in the Tumor Microenvironment: Insights into Immunogenic Cell Death in Onco-Therapies. Cancers 2021, 13, 281. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Krysko, D.V.; Garg, A.D.; Kaczmarek, A.; Krysko, O.; Agostinis, P.; Vandenabeele, P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer 2012, 12, 860–875. [Google Scholar] [CrossRef]
- Fucikova, J.; Moserova, I.; Urbanova, L.; Bezu, L.; Kepp, O.; Cremer, I.; Salek, C.; Strnad, P.; Kroemer, G.; Galluzzi, L.; et al. Prognostic and predictive value of DAMPs and DAMP-associated processes in cancer. Front. Immunol. 2015, 6, 402. [Google Scholar] [CrossRef]
- Fucikova, J.; Kepp, O.; Kasikova, L.; Petroni, G.; Yamazaki, T.; Liu, P.; Zhao, L.; Spisek, R.; Kroemer, G.; Galluzzi, L. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. 2020, 11, 1013. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Huang, S. Magnetic nanoparticles in cancer diagnosis, drug delivery and treatment. Mol. Clin. Oncol. 2017, 7, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.M.A.; Shaker, S.A.; Pinol, R.; Millan, A.; Hanafy, M.Y.; Helmy, M.H.; Kamel, M.A.; Mahmoud, S.A. Effect of superparamagnetic iron oxide nanoparticles on glucose homeostasis on type 2 diabetes experimental model. Life Sci. 2020, 245, 11. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Vallejo, V.; Puigivila, M.; Plaza-Garcia, S.; Szczupak, B.; Pinol, R.; Murillo, J.L.; Sorribas, V.; Lou, G.; Veintemillas, S.; Ramos-Cabrer, P.; et al. PEG-copolymer-coated iron oxide nanoparticles that avoid the reticuloendothelial system and act as kidney MRI contrast agents. Nanoscale 2018, 10, 14153–14164. [Google Scholar] [CrossRef]
- Pinol, R.; Brites, C.D.S.; Bustamante, R.; Martinez, A.; Silva, N.J.O.; Murillo, J.L.; Cases, R.; Carrey, J.; Estepa, C.; Sosa, C.; et al. Joining Time-Resolved Thermometry and Magnetic-Induced Heating in a Single Nanoparticle Unveils Intriguing Thermal Properties. ACS Nano 2015, 9, 3134–3142. [Google Scholar] [CrossRef]
- Soetaert, F.; Korangath, P.; Serantes, D.; Fiering, S.; Ivkov, R. Cancer therapy with iron oxide nanoparticles: Agents of thermal and immune therapies. Adv. Drug Deliv. Rev. 2020, 163, 65–83. [Google Scholar] [CrossRef]
- Wang, M.; Li, Y.; Liu, K.L.; Hoover, A.R.; Li, M.; Towner, R.A.; Mukherjee, P.; Zhou, F.F.; Qu, J.L.; Chen, W.R. Synergistic interventional photothermal therapy and immunotherapy using an iron oxide nanoplatform for the treatment of pancreatic cancer. Acta Biomater. 2022, 138, 453–462. [Google Scholar] [CrossRef]
- Ali, L.M.A.; Gutierrez, M.; Cornudella, R.; Moreno, J.A.; Pinol, R.; Gabilondo, L.; Millan, A.; Palacio, F. Hemostasis Disorders Caused by Polymer Coated Iron Oxide Nanoparticles. J. Biomed. Nanotechnol. 2013, 9, 1272–1285. [Google Scholar] [CrossRef]
- Sanchez-Paradinas, S.; Perez-Andres, M.; Almendral-Parra, M.J.; Rodriguez-Fernandez, E.; Milian, A.; Palacio, F.; Orfao, A.; Criado, J.J.; Fuentes, M. Enhanced cytotoxic activity of bile acid cisplatin derivatives by conjugation with gold nanoparticles. J. Inorg. Biochem. 2014, 131, 8–11. [Google Scholar] [CrossRef]
- Diez, P.; Gonzalez-Munoz, M.; Gonzalez-Gonzalez, M.; Degano, R.M.; Jara-Acevedo, R.; Sanchez-Paradinas, S.; Pinol, R.; Murillo, J.L.; Lou, G.; Palacio, F.; et al. Functional insights into the cellular response triggered by a bile-acid platinum compound conjugated to biocompatible ferric nanoparticles using quantitative proteomic approaches. Nanoscale 2017, 9, 9960–9972. [Google Scholar] [CrossRef]
- Hernandez, A.-P.; Micaelo, A.; Pinol, R.; Garcia-Vaquero, M.L.; Aramayona, J.J.; Criado, J.J.; Rodriguez, E.; Sanchez-Gallego, J.I.; Landeira-Vinuela, A.; Juanes-Velasco, P.; et al. Comprehensive and systematic characterization of multi-functionalized cisplatin nano-conjugate: From the chemistry and proteomic biocompatibility to the animal model. J. Nanobiotechnol. 2022, 20, 341. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, A.P.; Diez, P.; García, P.A.; Miguel del Corral, J.M.; Pérez-Andrés, M.; Diez, D.; San Feliciano, A.; Fuentes, M.; Castro, M.Á. New Hybrids Derived from Podophyllic Aldehyde and Diterpenylhydroquinones with Selectivity toward Osteosarcoma Cells. ACS Med. Chem. Lett. 2018, 9, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Hommel, G.; Blettner, M. Linear Regression Analysis Part 14 of a Series on Evaluation of Scientific Publications. Dtsch. Arztebl. Int. 2010, 107, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Juanes-Velasco, P.; Landeira-Vinuela, A.; Garcia-Vaquero, M.L.; Lecrevisse, Q.; Herrero, R.; Ferruelo, A.; Gongora, R.; Corrales, F.; Rivas, J.D.L.; Lorente, J.A.; et al. SARS-CoV-2 Infection Triggers Auto-Immune Response in ARDS. Front. Immunol. 2022, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Landeira-Vinuela, A.; Diez, P.; Juanes-Velasco, P.; Lecrevisse, Q.; Orfao, A.; de las Rivas, J.; Fuentes, M. Deepening into Intracellular Signaling Landscape through Integrative Spatial Proteomics and Transcriptomics in a Lymphoma Model. Biomolecules 2021, 11, 1716. [Google Scholar] [CrossRef] [PubMed]
- González-González, M.; Bartolome, R.; Jara-Acevedo, R.; Casado-Vela, J.; Dasilva, N.; Matarraz, S.; García, J.; Alcazar, J.A.; Sayagues, J.M.; Orfao, A. Evaluation of homo-and hetero-functionally activated glass surfaces for optimized antibody arrays. Anal. Biochem. 2014, 450, 37–45. [Google Scholar] [CrossRef]
- Sierra-Sanchez, A.; Garrido-Martin, D.; Lourido, L.; Gonzalez-Gonzalez, M.; Diez, P.; Ruiz-Romero, C.; Sjober, R.; Droste, C.; De Las Rivas, J.; Nilsson, P.; et al. Screening and Validation of Novel Biomarkers in Osteoarticular Pathologies by Comprehensive Combination of Protein Array Technologies. J. Proteome Res. 2017, 16, 1890–1899. [Google Scholar] [CrossRef]
- Juanes-Velasco, P.; Landeira-Vinuela, A.; Hernandez, A.-P.; Fuentes, M. Systematic and Rational Design of Protein Arrays in Noncontact Printers: Pipeline and Critical Aspects. Methods Mol. Biol. 2021, 2344, 9–29. [Google Scholar] [CrossRef]
- Diez, P.; Dasilva, N.; Gonzalez-Gonzalez, M.; Matarraz, S.; Casado-Vela, J.; Orfao, A.; Fuentes, M. Data Analysis Strategies for Protein Microarrays. Microarrays 2012, 1, 64–83. [Google Scholar] [CrossRef] [PubMed]
- Criado, J.J.; Fernandez, E.R.; Manzano, J.L.; Alonso, A.; Barrena, S.; Medarde, M.; Pelaez, R.; Tabernero, M.D.; Orfao, A. Intrinsically fluorescent cytotoxic cisplatin analogues as DNA marker molecules. Bioconjugate Chem. 2005, 16, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Rezatabar, S.; Karimian, A.; Rameshknia, V.; Parsian, H.; Majidinia, M.; Kopi, T.A.; Bishayee, A.; Sadeghinia, A.; Yousefi, M.; Monirialamdari, M.; et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J. Cell. Physiol. 2019, 234, 14951–14965. [Google Scholar] [CrossRef]
- Kepp, O.; Senovilla, L.; Vitale, I.; Vacchelli, E.; Adjemian, S.; Agostinis, P.; Apetoh, L.; Aranda, F.; Barnaba, V.; Bloy, N.; et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014, 3, e955691. [Google Scholar] [CrossRef] [PubMed]
- Darling, N.J.; Cook, S.J. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta-Mol. Cell Res. 2014, 1843, 2150–2163. [Google Scholar] [CrossRef]
- Shi, T.; Soest, D.M.K.V.; Polderman, P.E.; Burgering, B.M.T.; Dansen, T.B. DNA damage and oxidant stress activate p53 through differential upstream signaling pathways. Free Radic. Biol. Med. 2021, 172, 298–311. [Google Scholar] [CrossRef]
- He, C.X.; Hart, P.C.; Germain, D.; Bonini, M.G. SOD2 and the Mitochondrial UPR: Partners Regulating Cellular Phenotypic Transitions. Trends Biochem. Sci. 2016, 41, 568–577. [Google Scholar] [CrossRef]
- Kroemer, G.; Galassi, C.; Zitvogel, L.; Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol. 2022, 23, 487–500. [Google Scholar] [CrossRef]
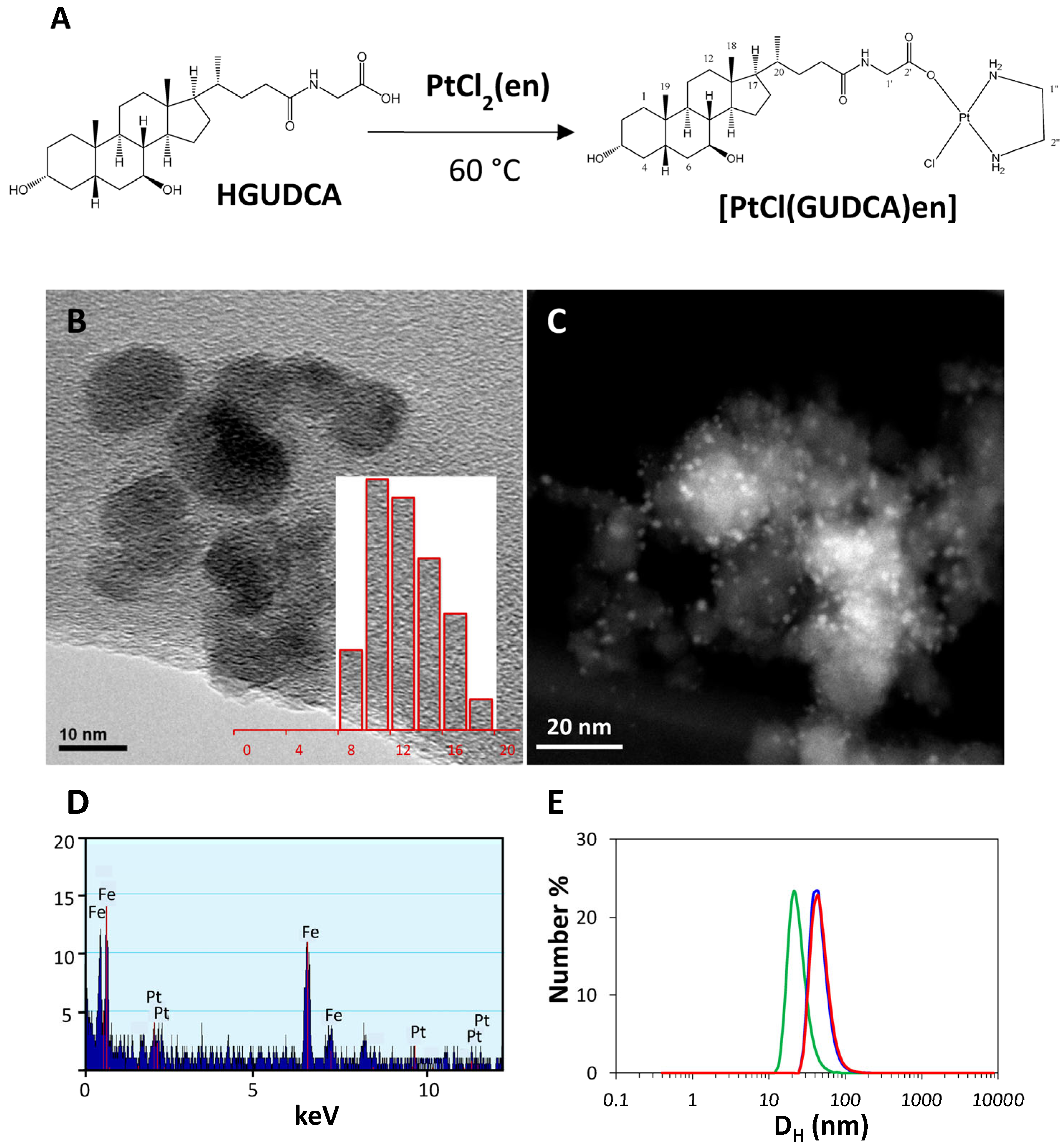
| Sample | DH (nm) | SD (nm) | PDI | z (mV) |
|---|---|---|---|---|
| IONP | 24.2 | 7.5 | 0.18 | +40 |
| IONP@BCP | 46.5 | 14.2 | 0.14 | −30 |
| IONP@BCP@[PtCl(GUDCA)en] | 47.9 | 14.8 | 0.22 | −9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández, Á.-P.; Iglesias-Anciones, L.; Vaquero-González, J.J.; Piñol, R.; Criado, J.J.; Rodriguez, E.; Juanes-Velasco, P.; García-Vaquero, M.L.; Arias-Hidalgo, C.; Orfao, A.; et al. Enhancement of Tumor Cell Immunogenicity and Antitumor Properties Derived from Platinum-Conjugated Iron Nanoparticles. Cancers 2023, 15, 3204. https://doi.org/10.3390/cancers15123204
Hernández Á-P, Iglesias-Anciones L, Vaquero-González JJ, Piñol R, Criado JJ, Rodriguez E, Juanes-Velasco P, García-Vaquero ML, Arias-Hidalgo C, Orfao A, et al. Enhancement of Tumor Cell Immunogenicity and Antitumor Properties Derived from Platinum-Conjugated Iron Nanoparticles. Cancers. 2023; 15(12):3204. https://doi.org/10.3390/cancers15123204
Chicago/Turabian StyleHernández, Ángela-Patricia, Laura Iglesias-Anciones, José Javier Vaquero-González, Rafael Piñol, Julio J. Criado, Emilio Rodriguez, Pablo Juanes-Velasco, Marina L. García-Vaquero, Carlota Arias-Hidalgo, Alberto Orfao, and et al. 2023. "Enhancement of Tumor Cell Immunogenicity and Antitumor Properties Derived from Platinum-Conjugated Iron Nanoparticles" Cancers 15, no. 12: 3204. https://doi.org/10.3390/cancers15123204
APA StyleHernández, Á.-P., Iglesias-Anciones, L., Vaquero-González, J. J., Piñol, R., Criado, J. J., Rodriguez, E., Juanes-Velasco, P., García-Vaquero, M. L., Arias-Hidalgo, C., Orfao, A., Millán, Á., & Fuentes, M. (2023). Enhancement of Tumor Cell Immunogenicity and Antitumor Properties Derived from Platinum-Conjugated Iron Nanoparticles. Cancers, 15(12), 3204. https://doi.org/10.3390/cancers15123204








