A Weakly Supervised Deep Learning Model and Human–Machine Fusion for Accurate Grading of Renal Cell Carcinoma from Histopathology Slides
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
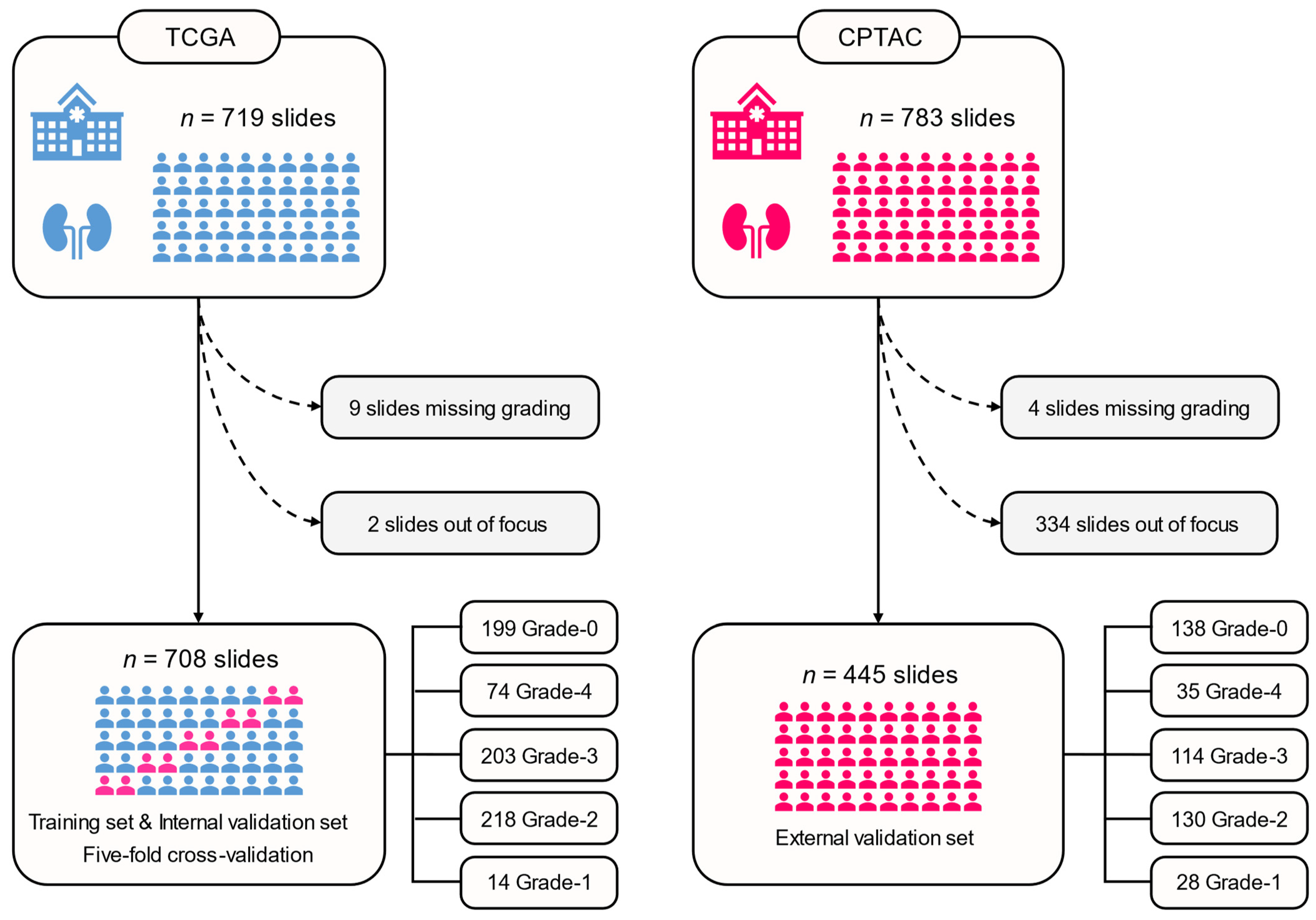
2.1. Patient Cohorts
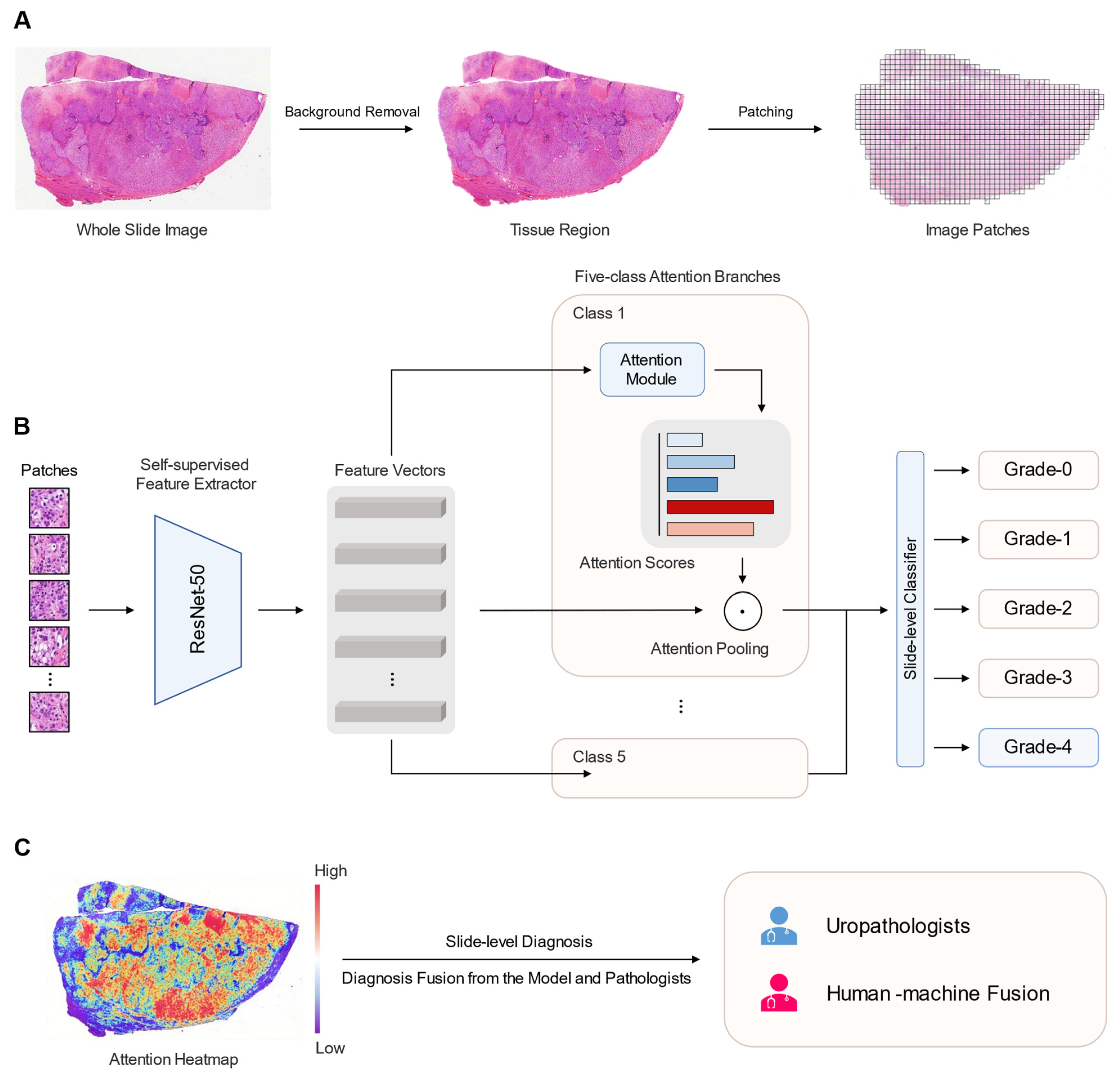
2.2. WSI Preprocessing
2.3. Deep Learning Algorithm
2.4. Human–Machine Fusion
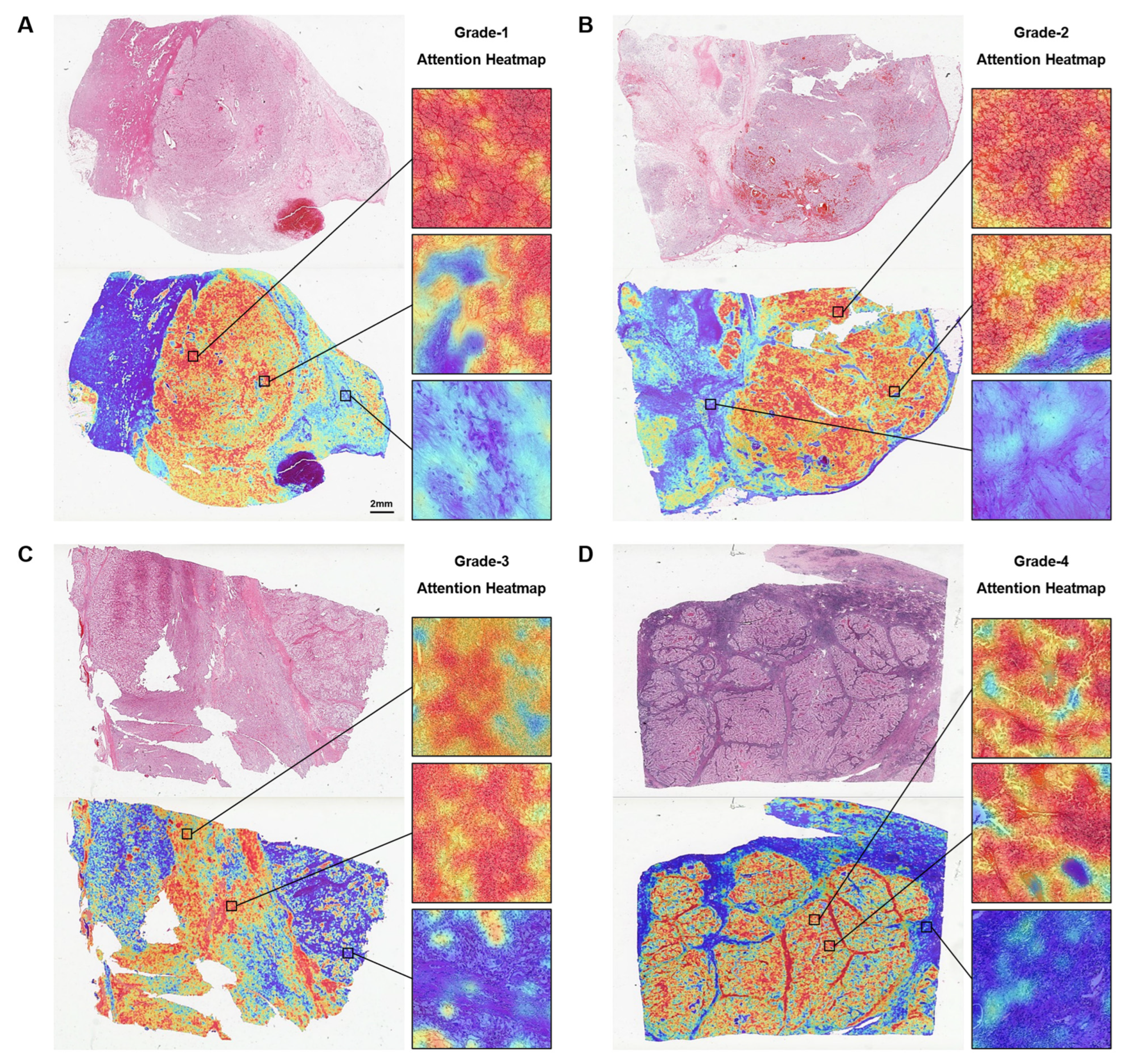
2.5. Interpretability of the Model
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics
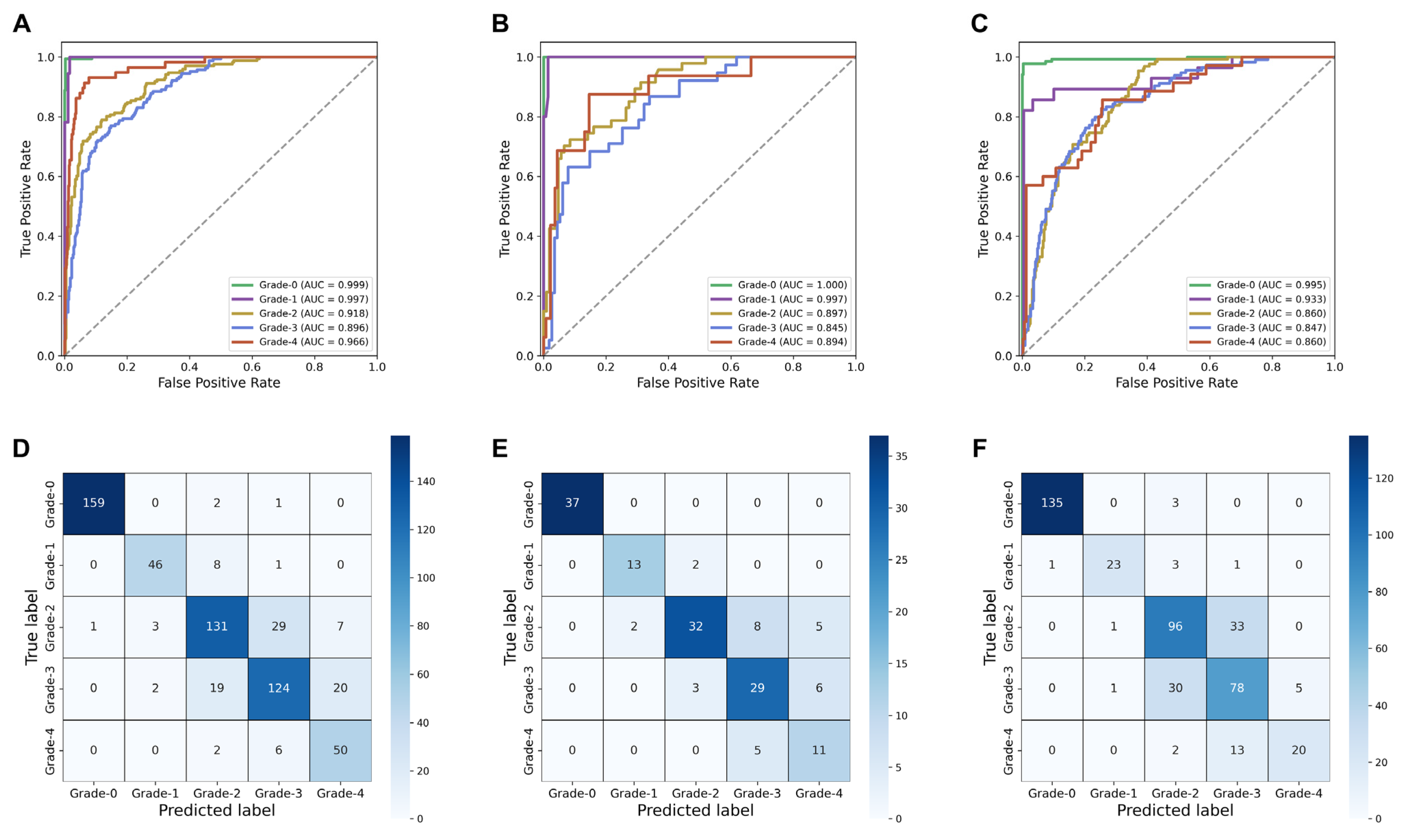
3.2. Diagnostic Performance of the SSL-CLAM Model
3.3. Human–Machine Fusion
3.4. Attention-Based Interpretation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of renal cell carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef]
- Capitanio, U.; Montorsi, F. Renal cancer. Lancet 2016, 387, 894–906. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Linehan, W.M.; Ricketts, C.J. The cancer genome atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Delahunt, B.; Eble, J.N.; Egevad, L.; Samaratunga, H. Grading of renal cell carcinoma. Histopathology 2019, 74, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Janssen, M.; Linxweiler, J.; Terwey, S.; Rugge, S.; Ohlmann, C.; Becker, F.; Thomas, C.; Neisius, A.; Thüroff, J.W.; Siemer, S. Survival outcomes in patients with large (≥7 cm) clear cell renal cell carcinomas treated with nephron-sparing surgery versus radical nephrectomy: Results of a multicenter cohort with long-term follow-up. PLoS ONE 2018, 13, e196427. [Google Scholar] [CrossRef] [PubMed]
- Delahunt, B. Advances and controversies in grading and staging of renal cell carcinoma. Mod. Pathol. 2009, 22, S24–S36. [Google Scholar] [CrossRef] [PubMed]
- Delahunt, B.; Cheville, J.C.; Martignoni, G.; Humphrey, P.A.; Magi-Galluzzi, C.; Mckenney, J.; Egevad, L.; Algaba, F.; Moch, H.; Grignon, D.J. The international society of urological pathology (isup) grading system for renal cell carcinoma and other prognostic parameters. Am. J. Surg. Pathol. 2013, 37, 1490–1504. [Google Scholar] [CrossRef]
- Shuch, B.; Pantuck, A.J.; Pouliot, F.; Finley, D.S.; Said, J.W.; Belldegrun, A.S.; Saigal, C. Quality of pathological reporting for renal cell cancer: Implications for systemic therapy, prognostication and surveillance. BJU Int. 2011, 108, 343–348. [Google Scholar] [CrossRef]
- Bektas, S.; Bahadir, B.; Kandemir, N.O.; Barut, F.; Gul, A.E.; Ozdamar, S.O. Intraobserver and interobserver variability of fuhrman and modified fuhrman grading systems for conventional renal cell carcinoma. Kaohsiung J. Med. Sci. 2009, 25, 596–600. [Google Scholar] [CrossRef]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, M.; Wang, S.; Li, X.; Sun, Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020, 40, 154–166. [Google Scholar] [CrossRef] [PubMed]
- van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Xiong, L.; Yan, D.; Xia, L.; Yuan, J.; Wang, J.; Jiao, P.; et al. Accurate diagnosis and survival prediction of bladder cancer using deep learning on histological slides. Cancers 2022, 14, 5807. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Williamson, D.; Chen, T.Y.; Lipkova, J.; Noor, Z.; Shaban, M.; Shady, M.; Williams, M.; Joo, B.; et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 2022, 40, 865–878. [Google Scholar] [CrossRef]
- Foersch, S.; Eckstein, M.; Wagner, D.C.; Gach, F.; Woerl, A.C.; Geiger, J.; Glasner, C.; Schelbert, S.; Schulz, S.; Porubsky, S.; et al. Deep learning for diagnosis and survival prediction in soft tissue sarcoma. Ann. Oncol. 2021, 32, 1178–1187. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck, K.S.V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Courtiol, P.; Maussion, C.; Moarii, M.; Pronier, E.; Pilcer, S.; Sefta, M.; Manceron, P.; Toldo, S.; Zaslavskiy, M.; Le Stang, N.; et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 2019, 25, 1519–1525. [Google Scholar] [CrossRef]
- Hipp, J.; Flotte, T.; Monaco, J.; Cheng, J.; Madabhushi, A.; Yagi, Y.; Rodriguez-Canales, J.; Emmert-Buck, M.; Dugan, M.C.; Hewitt, S.; et al. Computer aided diagnostic tools aim to empower rather than replace pathologists: Lessons learned from computational chess. J. Pathol. Inform. 2011, 2, 25. [Google Scholar] [CrossRef]
- Marostica, E.; Barber, R.; Denize, T.; Kohane, I.S.; Signoretti, S.; Golden, J.A.; Yu, K.H. Development of a histopathology informatics pipeline for classification and prediction of clinical outcomes in subtypes of renal cell carcinoma. Clin. Cancer Res. 2021, 27, 2868–2878. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, L.N.; Muti, H.S.; Loeffler, C.; Echle, A.; Saldanha, O.L.; Mahmood, F.; Lu, M.Y.; Trautwein, C.; Langer, R.; Dislich, B.; et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med. Image Anal. 2022, 79, 102474. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Y.; Williamson, D.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Schirris, Y.; Gavves, E.; Nederlof, I.; Horlings, H.M.; Teuwen, J. Deepsmile: Contrastive self-supervised pre-training benefits msi and hrd classification directly from h&e whole-slide images in colorectal and breast cancer. Med. Image Anal. 2022, 79, 102464. [Google Scholar]
- Wang, X.; Du, Y.; Yang, S.; Zhang, J.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. Retccl: Clustering-guided contrastive learning for whole-slide image retrieval. Med. Image Anal. 2023, 83, 102645. [Google Scholar] [CrossRef]
- Chanchal, A.K.; Lal, S.; Kumar, R.; Kwak, J.T.; Kini, J. A novel dataset and efficient deep learning framework for automated grading of renal cell carcinoma from kidney histopathology images. Sci. Rep. 2023, 13, 5728. [Google Scholar] [CrossRef]
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696. [Google Scholar] [CrossRef]
- Cao, R.; Yang, F.; Ma, S.C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.H.; Wang, T.; Lu, W.J.; Cai, W.J.; et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in colorectal cancer. Theranostics 2020, 10, 11080–11091. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chen, L.; Cheng, Z.; Yang, M.; Wang, J.; Lin, C.; Wang, Y.; Huang, L.; Chen, Y.; Peng, S.; et al. Deep learning-based six-type classifier for lung cancer and mimics from histopathological whole slide images: A retrospective study. BMC Med. 2021, 19, 80. [Google Scholar] [CrossRef] [PubMed]
- Al-Aynati, M.; Chen, V.; Salama, S.; Shuhaibar, H.; Treleaven, D.; Vincic, L. Interobserver and intraobserver variability using the fuhrman grading system for renal cell carcinoma. Arch. Pathol. Lab. Med. 2003, 127, 593–596. [Google Scholar] [CrossRef]
- Hong, S.K.; Jeong, C.W.; Park, J.H.; Kim, H.S.; Kwak, C.; Choe, G.; Kim, H.H.; Lee, S.E. Application of simplified fuhrman grading system in clear-cell renal cell carcinoma. BJU Int. 2011, 107, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lughezzani, G.; Jeldres, C.; Isbarn, H.; Shariat, S.F.; Arjane, P.; Widmer, H.; Pharand, D.; Latour, M.; Perrotte, P.; et al. A proposal for reclassification of the fuhrman grading system in patients with clear cell renal cell carcinoma. Eur. Urol. 2009, 56, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.; Lindner, V.; de Fromont, M.; Molinié, V.; Letourneux, H.; Meyer, N.; Martin, M.; Jacqmin, D. Multicenter determination of optimal interobserver agreement using the fuhrman grading system for renal cell carcinoma: Assessment of 241 patients with >15-year follow-up. Cancer 2005, 103, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, S.A.; Lasky, L.C.; Limas, C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 1982, 6, 655–663. [Google Scholar] [CrossRef] [PubMed]

| TCGA | CPTAC | |
|---|---|---|
| Number of patients | 504 | 188 |
| WSI format | SVS | SVS |
| Age (years) | 60.57 (±12.20) | 60.92 (±12.05) |
| Gender | ||
| Female | 177 (35.12%) | 64 (34.04%) |
| Male | 327 (64.88%) | 124 (65.96%) |
| pT stage | ||
| pT1 | 257 (50.99%) | 58 (30.85%) |
| pT2 | 66 (13.10%) | 14 (7.45%) |
| pT3 | 170 (33.73%) | 41 (21.8%) |
| pT4 | 11 (2.18%) | 3 (1.6%) |
| pTx | 0 (0%) | 72 (38.3%) |
| pN stage | ||
| pN0 | 230 (45.63%) | 23 (12.23%) |
| pN1 | 14 (2.78%) | 4 (2.13%) |
| pNx | 260 (51.59%) | 161 (85.64%) |
| pM stage | ||
| pM0 | 402 (79.76%) | 33 (17.55%) |
| pM1 | 74 (14.68%) | 3 (1.6%) |
| pMx | 28 (5.56%) | 152 (80.85%) |
| pTNM stage | ||
| Stage I | 251 (49.81%) | 84 (44.68%) |
| Stage II | 54 (10.71%) | 20 (10.64%) |
| Stage III | 118 (23.41%) | 47 (25.00%) |
| Stage IV | 80 (15.87%) | 21 (11.17%) |
| Missing | 1 (0.20%) | 16 (8.51%) |
| Fuhrman grade | ||
| G1 | 12 (2.38%) | 13 (6.92%) |
| G2 | 216 (42.86%) | 95 (50.53%) |
| G3 | 202 (40.08%) | 60 (31.91%) |
| G4 | 74 (14.68%) | 20 (10.64%) |
| Survival status | ||
| Alive | 334 (66.27%) | 145 (77.13%) |
| Dead | 170 (33.73%) | 27 (14.36%) |
| Not reported | 0 (0%) | 16 (8.51%) |
| Overall survival (years) | 3.64 (± 2.67) | 2.43 (± 1.83) |
| a. Diagnostic Performance in Five-Class Fuhrman Grade (Grade-0, 1, 2, 3, 4) | ||
| Accuracy (95% CI) | AUC (95% CI) | |
| Training set | 0.818 (0.805, 0.831) | 0.947 (0.938, 0.956) |
| Internal validation set | 0.776 (0.742, 0.812) | 0.917 (0.905, 0.928) |
| External validation set | 0.771 (0.739, 0.803) | 0.887 (0.872, 0.904) |
| b. Diagnostic performance in normal/tumor classification (Grade-0, Grade-1/2/3/4) | ||
| Accuracy (95% CI) | AUC (95% CI) | |
| Internal validation set | 0.997 (0.992, 1.000) | 0.999 (0.999, 1.000) |
| External validation set | 0.989 (0.986, 0.992) | 0.991 (0.989, 0.994) |
| c. Diagnostic performance in two-tiered Fuhrman grading (Grade-0, Grade-1/2, Grade-3/4) | ||
| Accuracy (95% CI) | AUC (95% CI) | |
| Internal validation set | 0.872 (0.845, 0.899) | 0.936 (0.906, 0.962) |
| External validation set | 0.838 (0.829, 0.847) | 0.915 (0.907, 0.922) |
| Accuracy (95% CI) | Precision (95% CI) | p-Value * | Kappa # | |
|---|---|---|---|---|
| SSL-CLAM model | 0.771 (0.739, 0.803) | 0.786 (0.762, 0.811) | - | - |
| Junior Pathologist A | 0.737 (0.721, 0.753) | 0.695 (0.678, 0.712) | 0.002 | 0.837 |
| Expert Uropathologist B | 0.824 (0.808, 0.839) | 0.800 (0.779, 0.821) | 0.336 | 0.889 |
| Junior A—SSL-CLAM fusion | 0.787 (0.772, 0.801) | 0.773 (0.758, 0.788) | 0.902 | 0.904 |
| Expert B—SSL-CLAM fusion | 0.856 (0.843, 0.867) | 0.839 (0.819, 0.858) | <0.001 | 0.906 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Yang, R.; Xu, H.; Fan, J.; Jiao, P.; Ni, X.; Yuan, J.; Wang, L.; Chen, Z.; Liu, X. A Weakly Supervised Deep Learning Model and Human–Machine Fusion for Accurate Grading of Renal Cell Carcinoma from Histopathology Slides. Cancers 2023, 15, 3198. https://doi.org/10.3390/cancers15123198
Zheng Q, Yang R, Xu H, Fan J, Jiao P, Ni X, Yuan J, Wang L, Chen Z, Liu X. A Weakly Supervised Deep Learning Model and Human–Machine Fusion for Accurate Grading of Renal Cell Carcinoma from Histopathology Slides. Cancers. 2023; 15(12):3198. https://doi.org/10.3390/cancers15123198
Chicago/Turabian StyleZheng, Qingyuan, Rui Yang, Huazhen Xu, Junjie Fan, Panpan Jiao, Xinmiao Ni, Jingping Yuan, Lei Wang, Zhiyuan Chen, and Xiuheng Liu. 2023. "A Weakly Supervised Deep Learning Model and Human–Machine Fusion for Accurate Grading of Renal Cell Carcinoma from Histopathology Slides" Cancers 15, no. 12: 3198. https://doi.org/10.3390/cancers15123198
APA StyleZheng, Q., Yang, R., Xu, H., Fan, J., Jiao, P., Ni, X., Yuan, J., Wang, L., Chen, Z., & Liu, X. (2023). A Weakly Supervised Deep Learning Model and Human–Machine Fusion for Accurate Grading of Renal Cell Carcinoma from Histopathology Slides. Cancers, 15(12), 3198. https://doi.org/10.3390/cancers15123198






