Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms
Abstract
Simple Summary
Abstract
1. Introduction
2. miRNA Deregulation in Prostate Cancer: An Overview
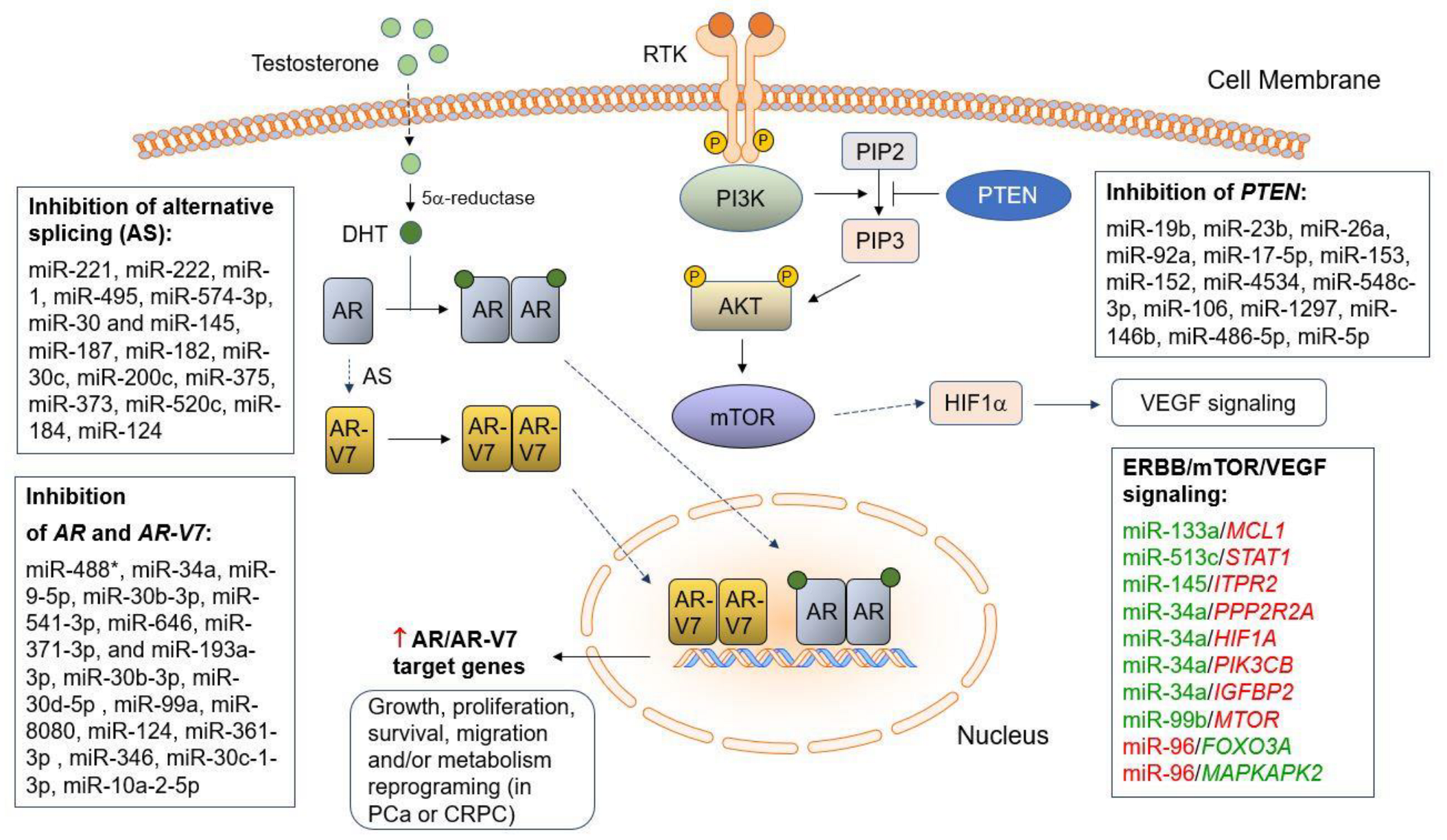
3. miRNAs Involved in AR/AR-V7 Synthesis and Hormone Therapy Resistance in PCa
3.1. miRNAs in AR Synthesis
3.2. Deregulated miRNAs Involved in AR-V7 Synthesis
3.3. miRNAs Involved in Hormone Therapy Resistance of PCa
4. miRNAs Involved in Chemoresistance in PCa
5. miRNAs Involved in PTEN Loss/Downregulation in Aggressive PCa
6. Deregulated miRNAs Regulating Alternative Splicing Mechanisms in PCa
7. Deregulated miRNAs and Differential miRNA–mRNA Regulatory Networks in PCa Disparities
8. Clinical Implications of miRNAs as Diagnostic and Prognostic Biomarkers and Therapeutic Tools/Targets
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-induced changes in miRNA biogenesis and functioning. Cell. Mol. Life Sci. 2018, 75, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Models Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Chen, H.C. Analysis of targets and functions coregulated by microRNAs. Methods Mol. Biol. 2011, 676, 225–241. [Google Scholar] [CrossRef]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Farha, M.W.; Salami, S.S. Biomarkers for prostate cancer detection and risk stratification. Ther. Adv. Urol. 2022, 14, 17562872221103988. [Google Scholar] [CrossRef]
- Ozen, M.; Creighton, C.J.; Ozdemir, M.; Ittmann, M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008, 27, 1788–1793. [Google Scholar] [CrossRef]
- Pang, Y.; Young, C.Y.; Yuan, H. MicroRNAs and prostate cancer. Acta Biochim. Biophys. Sin. 2010, 42, 363–369. [Google Scholar] [CrossRef]
- Bonci, D.; Coppola, V.; Patrizii, M.; Addario, A.; Cannistraci, A.; Francescangeli, F.; Pecci, R.; Muto, G.; Collura, D.; Bedini, R.; et al. A microRNA code for prostate cancer metastasis. Oncogene 2016, 35, 1180–1192. [Google Scholar] [CrossRef]
- Aghdam, S.G.; Ebrazeh, M.; Hemmatzadeh, M.; Seyfizadeh, N.; Shabgah, A.G.; Azizi, G.; Ebrahimi, N.; Babaie, F.; Mohammadi, H. The role of microRNAs in prostate cancer migration, invasion, and metastasis. J. Cell. Physiol. 2019, 234, 9927–9942. [Google Scholar] [CrossRef] [PubMed]
- Andl, T.; Ganapathy, K.; Bossan, A.; Chakrabarti, R. MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology? Int. J. Mol. Sci. 2020, 21, 4796. [Google Scholar] [CrossRef] [PubMed]
- Abramovic, I.; Ulamec, M.; Katusic Bojanac, A.; Bulic-Jakus, F.; Jezek, D.; Sincic, N. miRNA in prostate cancer: Challenges toward translation. Epigenomics 2020, 12, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Baruah, M.M. The microRNA signatures: Aberrantly expressed miRNAs in prostate cancer. Clin. Transl. Oncol. 2019, 21, 126–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, D.; Spring, D.J.; DePinho, R.A. Genetics and biology of prostate cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef]
- Calderon-Aparicio, A.; Wang, B.D. Prostate cancer: Alternatively spliced mRNA transcripts in tumor progression and their uses as therapeutic targets. Int. J. Biochem. Cell Biol. 2021, 141, 106096. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G.; Mukherjee, C.; Shi, Y.; Gelinas, C.; Fan, Y.; et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Loizzo, D.; Pandolfo, S.D.; Rogers, D.; Cerrato, C.; di Meo, N.A.; Autorino, R.; Mirone, V.; Ferro, M.; Porta, C.; Stella, A.; et al. Novel Insights into Autophagy and Prostate Cancer: A Comprehensive Review. Int. J. Mol. Sci. 2022, 23, 3826. [Google Scholar] [CrossRef]
- Takayama, K.I.; Misawa, A.; Inoue, S. Significance of microRNAs in Androgen Signaling and Prostate Cancer Progression. Cancers 2017, 9, 102. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, K.Y.; Liu, Q.; Cao, Q. Androgen Receptor-Related Non-coding RNAs in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 660853. [Google Scholar] [CrossRef]
- Tian, L.; Fang, Y.X.; Xue, J.L.; Chen, J.Z. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS ONE 2013, 8, e75885. [Google Scholar] [CrossRef]
- Amir, S.; Ma, A.H.; Shi, X.B.; Xue, L.; Kung, H.J.; Devere White, R.W. Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PLoS ONE 2013, 8, e61064. [Google Scholar] [CrossRef]
- Cui, Y.; Yang, Y.; Ren, L.; Yang, J.; Wang, B.; Xing, T.; Chen, H.; Chen, M. miR-15a-3p Suppresses Prostate Cancer Cell Proliferation and Invasion by Targeting SLC39A7 Via Downregulating Wnt/beta-Catenin Signaling Pathway. Cancer Biother. Radiopharm. 2019, 34, 472–479. [Google Scholar] [CrossRef]
- Huang, K.; Sun, X.; Wu, H.; Zhao, J.; Jian, Y.; Xu, Z.; Wang, S.; Yang, D. The Regulating Effect of Autophagy-Related MiRNAs in Kidney, Bladder, and Prostate Cancer. J. Oncol. 2021, 2021, 5510318. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, R.; Shen, D.; Cheng, S.; Wang, H.; Lu, Z.; Zheng, Q.; Wang, L.; Xia, L.; Li, G. Role of noncoding RNA in drug resistance of prostate cancer. Cell Death Dis. 2021, 12, 590. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Hashemy, S.I.; Sahebkar, A.; Aghaee Bakhtiari, S.H. MicroRNA Regulation of Androgen Receptor in Castration-Resistant Prostate Cancer: Premises, Promises, and Potentials. Curr. Mol. Pharmacol. 2021, 14, 559–569. [Google Scholar] [CrossRef]
- Balzeau, J.; Menezes, M.R.; Cao, S.; Hagan, J.P. The LIN28/let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef]
- Barh, D.; Malhotra, R.; Ravi, B.; Sindhurani, P. MicroRNA let-7: An emerging next-generation cancer therapeutic. Curr. Oncol. 2010, 17, 70–80. [Google Scholar] [CrossRef]
- Tang, G.; Du, R.; Tang, Z.; Kuang, Y. MiRNALet-7a mediates prostate cancer PC-3 cell invasion, migration by inducing epithelial-mesenchymal transition through CCR7/MAPK pathway. J. Cell. Biochem. 2018, 119, 3725–3731. [Google Scholar] [CrossRef] [PubMed]
- Nadiminty, N.; Tummala, R.; Lou, W.; Zhu, Y.; Shi, X.B.; Zou, J.X.; Chen, H.; Zhang, J.; Chen, X.; Luo, J.; et al. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS ONE 2012, 7, e32832. [Google Scholar] [CrossRef]
- Gao, S.; Zhao, Z.; Wu, R.; Wu, L.; Tian, X.; Zhang, Z. MiR-1 inhibits prostate cancer PC3 cells proliferation through the Akt/mTOR signaling pathway by binding to c-Met. Biomed. Pharmacother. 2019, 109, 1406–1410. [Google Scholar] [CrossRef]
- Chang, J.; Xu, W.; Du, X.; Hou, J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. OncoTargets Ther 2018, 11, 3461–3473. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Hou, X.; Zhu, J.; Jiang, C.; Wei, W. Expression of miR-30c and miR-29b in prostate cancer and its diagnostic significance. Oncol. Lett. 2018, 16, 3140–3144. [Google Scholar] [CrossRef]
- Huang, Y.Q.; Ling, X.H.; Yuan, R.Q.; Chen, Z.Y.; Yang, S.B.; Huang, H.X.; Zhong, W.D.; Qiu, S.P. miR30c suppresses prostate cancer survival by targeting the ASF/SF2 splicing factor oncoprotein. Mol. Med. Rep. 2017, 16, 2431–2438. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.; Steele, R.; Shi, X.; Ray, R.B. miRNA-29b Inhibits Prostate Tumor Growth and Induces Apoptosis by Increasing Bim Expression. Cells 2019, 8, 1455. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Cui, H.; Liang, J.; Su, X. Role of MicroRNA-30c in cancer progression. J. Cancer 2020, 11, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Guo, Q.; Fu, F.J.; Wang, Z.; Yin, Z.; Wei, Y.B.; Yang, J.R. The role of miR-29b in cancer: Regulation, function, and signaling. OncoTargets Ther. 2015, 8, 539–548. [Google Scholar] [CrossRef]
- Gujrati, H.; Ha, S.; Mohamed, A.; Wang, B.D. MicroRNA-mRNA Regulatory Network Mediates Activation of mTOR and VEGF Signaling in African American Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 2926. [Google Scholar] [CrossRef]
- Niture, S.; Tricoli, L.; Qi, Q.; Gadi, S.; Hayes, K.; Kumar, D. MicroRNA-99b-5p targets mTOR/AR axis, induces autophagy and inhibits prostate cancer cell proliferation. Tumor Biol. 2022, 44, 107–127. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gulla, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A new weapon against cancer? Mol. Ther. Nucleic Acids 2014, 3, e194. [Google Scholar] [CrossRef]
- Chen, F.; Hu, S.J. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: A review. J. Biochem. Mol. Toxicol. 2012, 26, 79–86. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef]
- Hermeking, H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat. Rev. Cancer 2012, 12, 613–626. [Google Scholar] [CrossRef]
- Tarasov, V.; Jung, P.; Verdoodt, B.; Lodygin, D.; Epanchintsev, A.; Menssen, A.; Meister, G.; Hermeking, H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 2007, 6, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Chua, C.E.L.; Tang, B.L. miR-34a in Neurophysiology and Neuropathology. J. Mol. Neurosci. 2019, 67, 235–246. [Google Scholar] [CrossRef]
- Lacombe, J.; Zenhausern, F. Emergence of miR-34a in radiation therapy. Crit. Rev. Oncol. Hematol. 2017, 109, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, A.; Zhou, Y.; Zhang, B.; Jiang, Y.; Hong, Z. miR-34a Suppresses Cell Proliferation in Laryngeal Cancer by Targeting Prominin 1. Crit. Rev. Eukaryot. Gene Expr. 2021, 31, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Slabakova, E.; Culig, Z.; Remsik, J.; Soucek, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef]
- Fujita, Y.; Kojima, K.; Hamada, N.; Ohhashi, R.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem. Biophys. Res. Commun. 2008, 377, 114–119. [Google Scholar] [CrossRef]
- Li, W.; Chang, J.; Wang, S.; Liu, X.; Peng, J.; Huang, D.; Sun, M.; Chen, Z.; Zhang, W.; Guo, W.; et al. miRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOR. Oncotarget 2015, 6, 24448–24462. [Google Scholar] [CrossRef]
- Sun, D.; Lee, Y.S.; Malhotra, A.; Kim, H.K.; Matecic, M.; Evans, C.; Jensen, R.V.; Moskaluk, C.A.; Dutta, A. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011, 71, 1313–1324. [Google Scholar] [CrossRef]
- Turcatel, G.; Rubin, N.; El-Hashash, A.; Warburton, D. MIR-99a and MIR-99b modulate TGF-beta induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS ONE 2012, 7, e31032. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Z.; Yang, Y.; Luo, M.; Zhang, M.; Wang, X.; Liu, L.; Hou, N.; Guo, Q.; Song, T.; et al. MiR-99b-5p and miR-203a-3p Function as Tumor Suppressors by Targeting IGF-1R in Gastric Cancer. Sci. Rep. 2018, 8, 10119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, R.; Bai, P.; Li, S.; Zuo, J.; Zhang, Y.; Liu, M.; Wu, L. Down-regulated expression of miR-99a is associated with lymph node metastasis and predicts poor outcome in stage IB cervical squamous cell carcinoma: A case-control study. Ann. Transl. Med. 2022, 10, 663. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.G.; Qin, X.G.; Zhao, B.S.; Qi, B.; Yao, W.J.; Wang, T.Y.; Li, H.C.; Wu, X.N. Differential expression of miRNAs in esophageal cancer tissue. Oncol. Lett. 2013, 5, 1639–1642. [Google Scholar] [CrossRef]
- Gujrati, H.; Ha, S.; Waseem, M.; Wang, B.D. Downregulation of miR-99b-5p and Upregulation of Nuclear mTOR Cooperatively Promotes the Tumor Aggressiveness and Drug Resistance in African American Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 9643. [Google Scholar] [CrossRef]
- Hua, Y.T.; Xu, W.X.; Li, H.; Xia, M. Emerging roles of MiR-133a in human cancers. J. Cancer 2021, 12, 198–206. [Google Scholar] [CrossRef]
- Zheng, L.; Kang, Y.; Zhang, L.; Zou, W. MiR-133a-5p inhibits androgen receptor (AR)-induced proliferation in prostate cancer cells via targeting FUsed in Sarcoma (FUS) and AR. Cancer Biol. Ther. 2020, 21, 34–42. [Google Scholar] [CrossRef]
- Tang, Y.; Pan, J.; Huang, S.; Peng, X.; Zou, X.; Luo, Y.; Ren, D.; Zhang, X.; Li, R.; He, P.; et al. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018, 37, 160. [Google Scholar] [CrossRef]
- Pelka, K.; Klicka, K.; Grzywa, T.M.; Gondek, A.; Marczewska, J.M.; Garbicz, F.; Szczepaniak, K.; Paskal, W.; Wlodarski, P.K. miR-96-5p, miR-134-5p, miR-181b-5p and miR-200b-3p heterogenous expression in sites of prostate cancer versus benign prostate hyperplasia-archival samples study. Histochem. Cell Biol. 2021, 155, 423–433. [Google Scholar] [CrossRef]
- Pan, J.Y.; Zhang, F.; Sun, C.C.; Li, S.J.; Li, G.; Gong, F.Y.; Bo, T.; He, J.; Hua, R.X.; Hu, W.D.; et al. miR-134: A Human Cancer Suppressor? Mol. Ther. Nucleic Acids 2017, 6, 140–149. [Google Scholar] [CrossRef]
- Ngalame, N.N.; Tokar, E.J.; Person, R.J.; Xu, Y.; Waalkes, M.P. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol. Sci. 2014, 138, 268–277. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Chauhan, N.; Dhasmana, A.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. miR-205: A Potential Biomedicine for Cancer Therapy. Cells 2020, 9, 1957. [Google Scholar] [CrossRef] [PubMed]
- Kiener, M.; Chen, L.; Krebs, M.; Grosjean, J.; Klima, I.; Kalogirou, C.; Riedmiller, H.; Kneitz, B.; Thalmann, G.N.; Snaar-Jagalska, E.; et al. miR-221-5p regulates proliferation and migration in human prostate cancer cells and reduces tumor growth in vivo. BMC Cancer 2019, 19, 627. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Ma, G.; Zhang, J.; Zhu, W. miR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancer. BMC Urol. 2018, 18, 14. [Google Scholar] [CrossRef]
- Xuan, H.; Xue, W.; Pan, J.; Sha, J.; Dong, B.; Huang, Y. Downregulation of miR-221, -30d, and -15a contributes to pathogenesis of prostate cancer by targeting Bmi-1. Biochemistry 2015, 80, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, X.; He, H.H.; Sweeney, C.J.; Liu, S.X.; Brown, M.; Balk, S.; Lee, G.S.; Kantoff, P.W. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene 2014, 33, 2790–2800. [Google Scholar] [CrossRef]
- Pillman, K.A.; Phillips, C.A.; Roslan, S.; Toubia, J.; Dredge, B.K.; Bert, A.G.; Lumb, R.; Neumann, D.P.; Li, X.; Conn, S.J.; et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018, 37, e99016. [Google Scholar] [CrossRef]
- Yang, K.; Handorean, A.M.; Iczkowski, K.A. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int. J. Clin. Exp. Pathol. 2009, 2, 361–369. [Google Scholar]
- Abramovic, I.; Vrhovec, B.; Skara, L.; Vrtaric, A.; Nikolac Gabaj, N.; Kulis, T.; Stimac, G.; Ljiljak, D.; Ruzic, B.; Kastelan, Z.; et al. MiR-182-5p and miR-375-3p Have Higher Performance Than PSA in Discriminating Prostate Cancer from Benign Prostate Hyperplasia. Cancers 2021, 13, 2068. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell. Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ning, L.; Wang, L.; Ouyang, T.; Qi, L.; Yang, R.; Wu, Y. miR-21 inhibition reverses doxorubicin-resistance and inhibits PC3 human prostate cancer cells proliferation. Andrologia 2021, 53, e14016. [Google Scholar] [CrossRef]
- Yamada, Y.; Enokida, H.; Kojima, S.; Kawakami, K.; Chiyomaru, T.; Tatarano, S.; Yoshino, H.; Kawahara, K.; Nishiyama, K.; Seki, N.; et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: Correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011, 102, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Haflidadottir, B.S.; Larne, O.; Martin, M.; Persson, M.; Edsjo, A.; Bjartell, A.; Ceder, Y. Upregulation of miR-96 enhances cellular proliferation of prostate cancer cells through FOXO1. PLoS ONE 2013, 8, e72400. [Google Scholar] [CrossRef]
- Sagar, S.K. miR-106b as an emerging therapeutic target in cancer. Genes Dis. 2022, 9, 889–899. [Google Scholar] [CrossRef]
- Yin, W.; Chen, J.; Wang, G.; Zhang, D. MicroRNA-106b functions as an oncogene and regulates tumor viability and metastasis by targeting LARP4B in prostate cancer. Mol. Med. Rep. 2019, 20, 951–958. [Google Scholar] [CrossRef]
- Shi, X.B.; Xue, L.; Ma, A.H.; Tepper, C.G.; Kung, H.J.; White, R.W. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 2011, 71, 538–549. [Google Scholar] [CrossRef]
- Li, J.Z.; Li, J.; Wang, H.Q.; Li, X.; Wen, B.; Wang, Y.J. MiR-141-3p promotes prostate cancer cell proliferation through inhibiting kruppel-like factor-9 expression. Biochem. Biophys. Res. Commun. 2017, 482, 1381–1386. [Google Scholar] [CrossRef]
- Xu, W.; Hua, Y.; Deng, F.; Wang, D.; Wu, Y.; Zhang, W.; Tang, J. MiR-145 in cancer therapy resistance and sensitivity: A comprehensive review. Cancer Sci. 2020, 111, 3122–3131. [Google Scholar] [CrossRef]
- Bai, L.; Luo, L.; Gao, W.; Bu, C.; Huang, J. miR-182 modulates cell proliferation and invasion in prostate cancer via targeting ST6GALNAC5. Braz. J. Med. Biol. Res. 2021, 54, e9695. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef]
- Lin, J.; Lu, Y.; Zhang, X.; Mo, Q.; Yu, L. Effect of miR-200c on proliferation, invasion and apoptosis of prostate cancer LNCaP cells. Oncol. Lett. 2019, 17, 4299–4304. [Google Scholar] [CrossRef]
- Shi, R.; Xiao, H.; Yang, T.; Chang, L.; Tian, Y.; Wu, B.; Xu, H. Effects of miR-200c on the migration and invasion abilities of human prostate cancer Du145 cells and the corresponding mechanism. Front. Med. 2014, 8, 456–463. [Google Scholar] [CrossRef]
- Karimi Mazraehshah, M.; Tavangar, S.M.; Saidijam, M.; Amini, R.; Bahreini, F.; Karimi Dermani, F.; Najafi, R. Anticancer effects of miR-200c in colorectal cancer through BMI1. J. Cell. Biochem. 2018, 119, 10005–10012. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Rossi, M.; Caracciolo, D.; Gulla, A.; Tagliaferri, P.; Tassone, P. Mir-221/222 are promising targets for innovative anticancer therapy. Expert Opin. Ther. Targets 2016, 20, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, N.; Coppola, V.; Bonci, D.; Miele, F.; Costantini, A.; Guadagnoli, M.; Bonanno, E.; Muto, G.; Frajese, G.V.; De Maria, R.; et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE 2008, 3, e4029. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Tian, H. Correlation between miR-222 and uterine cancer and its prognostic value. Oncol. Lett. 2018, 16, 1722–1726. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Xu, L.Y.; Qian, Q.; Zhu, Q.W.; Chen, W.X. MicroRNA-222 promotes drug resistance to doxorubicin in breast cancer via regulation of miR-222/bim pathway. Biosci. Rep. 2019, 39, BSR20190650. [Google Scholar] [CrossRef]
- Lin, T.; Yang, Y.; Ye, X.; Yao, J.; Zhou, H. Low expression of miR-99b promotes progression of clear cell renal cell carcinoma by up-regulating IGF1R/Akt/mTOR signaling. Int. J. Clin. Exp. Pathol. 2020, 13, 3083–3091. [Google Scholar]
- He, M.Q.; Wan, J.F.; Zeng, H.F.; Tang, Y.Y.; He, M.Q. miR-133a-5p suppresses gastric cancer through TCF4 down-regulation. J. Gastrointest. Oncol. 2021, 12, 1007–1019. [Google Scholar] [CrossRef]
- Sun, C.C.; Li, S.J.; Li, D.J. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget 2016, 7, 35960–35978. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sanchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velazquez, I.A.; Gonzalez-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manriquez, R.; Castro-Hernandez, C.; Fragoso-Ontiveros, V.; Alvarez-Gomez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, Y.; Liu, X.; Zeng, Y.; Liu, L. miR-96-5p is the tumor suppressor in osteosarcoma via targeting SYK. Biochem. Biophys. Res. Commun. 2021, 572, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, Y.; Wu, J.; Cui, X.; Zheng, S.; Yan, H.; Wu, Y.; Wang, F. LncRNA FGF14-AS2 represses growth of prostate carcinoma cells via modulating miR-96-5p/AJAP1 axis. J. Clin. Lab. Anal. 2021, 35, e24012. [Google Scholar] [CrossRef]
- Peng, B.; Theng, P.Y.; Le, M.T.N. Essential functions of miR-125b in cancer. Cell Prolif. 2021, 54, e12913. [Google Scholar] [CrossRef]
- Fang, F.; Cheng, L.; Wu, X.; Ye, M.; Zhang, H. miR-141 Promotes Colon Cancer Cell Proliferation by Targeted PHLPP2 Expression Inhibitionn. Cancer Manag. Res. 2020, 12, 11341–11350. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar]
- Chua, F.Y.; Adams, B.D. Androgen receptor and miR-206 regulation in prostate cancer. Transcription 2017, 8, 313–327. [Google Scholar] [CrossRef]
- Sampson, N.; Neuwirt, H.; Puhr, M.; Klocker, H.; Eder, I.E. In vitro model systems to study androgen receptor signaling in prostate cancer. Endocr. Relat. Cancer 2013, 20, R49–R64. [Google Scholar] [CrossRef]
- Fletcher, C.E.; Sulpice, E.; Combe, S.; Shibakawa, A.; Leach, D.A.; Hamilton, M.P.; Chrysostomou, S.L.; Sharp, A.; Welti, J.; Yuan, W.; et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene 2019, 38, 5700–5724. [Google Scholar] [CrossRef]
- Eringyte, I.; Zamarbide Losada, J.N.; Powell, S.M.; Bevan, C.L.; Fletcher, C.E. Coordinated AR and microRNA regulation in prostate cancer. Asian J. Urol. 2020, 7, 233–250. [Google Scholar] [CrossRef]
- Sikand, K.; Slaibi, J.E.; Singh, R.; Slane, S.D.; Shukla, G.C. miR 488* inhibits androgen receptor expression in prostate carcinoma cells. Int. J. Cancer 2011, 129, 810–819. [Google Scholar] [CrossRef]
- Ostling, P.; Leivonen, S.K.; Aakula, A.; Kohonen, P.; Makela, R.; Hagman, Z.; Edsjo, A.; Kangaspeska, S.; Edgren, H.; Nicorici, D.; et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011, 71, 1956–1967. [Google Scholar] [CrossRef] [PubMed]
- Kashat, M.; Azzouz, L.; Sarkar, S.H.; Kong, D.; Li, Y.; Sarkar, F.H. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 2012, 4, 432–442. [Google Scholar] [PubMed]
- Aakula, A.; Leivonen, S.K.; Hintsanen, P.; Aittokallio, T.; Ceder, Y.; Borresen-Dale, A.L.; Perala, M.; Ostling, P.; Kallioniemi, O. MicroRNA-135b regulates ERalpha, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol. Oncol. 2015, 9, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Khaleghzadegan, S.; Mears, B.; Hatano, K.; Kudrolli, T.A.; Chowdhury, W.H.; Yeater, D.B.; Ewing, C.M.; Luo, J.; Isaacs, W.B.; et al. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 2016, 7, 72593–72607. [Google Scholar] [CrossRef]
- Hu, R.; Isaacs, W.B.; Luo, J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate 2011, 71, 1656–1667. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Armstrong, A.J.; Dehm, S.M.; Luo, J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016, 19, 231–241. [Google Scholar] [CrossRef]
- Shi, X.B.; Ma, A.H.; Xue, L.; Li, M.; Nguyen, H.G.; Yang, J.C.; Tepper, C.G.; Gandour-Edwards, R.; Evans, C.P.; Kung, H.J.; et al. miR-124 and Androgen Receptor Signaling Inhibitors Repress Prostate Cancer Growth by Downregulating Androgen Receptor Splice Variants, EZH2, and Src. Cancer Res. 2015, 75, 5309–5317. [Google Scholar] [CrossRef]
- Chen, W.; Yao, G.; Zhou, K. miR-103a-2-5p/miR-30c-1-3p inhibits the progression of prostate cancer resistance to androgen ablation therapy via targeting androgen receptor variant 7. J. Cell. Biochem. 2019, 120, 14055–14064. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Kojima, K.; Ohhashi, R.; Hamada, N.; Nozawa, Y.; Kitamoto, A.; Sato, A.; Kondo, S.; Kojima, T.; Deguchi, T.; et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J. Biol. Chem. 2010, 285, 19076–19084. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Mahato, R.I. MicroRNAs and drug resistance in prostate cancers. Mol. Pharm. 2014, 11, 2539–2552. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Ikeda, K.; Sato, W.; Horie-Inoue, K.; Okamoto, K.; Inoue, S. MicroRNA Library-Based Functional Screening Identified Androgen-Sensitive miR-216a as a Player in Bicalutamide Resistance in Prostate Cancer. J. Clin. Med. 2015, 4, 1853–1865. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Wu, Z.; Ding, W.; Gao, S.; Gao, Y.; Xu, C. Mechanisms of enzalutamide resistance in castration-resistant prostate cancer and therapeutic strategies to overcome it. Br. J. Pharmacol. 2021, 178, 239–261. [Google Scholar] [CrossRef]
- Lin, S.C.; Kao, C.Y.; Lee, H.J.; Creighton, C.J.; Ittmann, M.M.; Tsai, S.J.; Tsai, S.Y.; Tsai, M.J. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nat. Commun. 2016, 7, 11418. [Google Scholar] [CrossRef]
- Ebron, J.S.; Shankar, E.; Singh, J.; Sikand, K.; Weyman, C.M.; Gupta, S.; Lindner, D.J.; Liu, X.; Campbell, M.J.; Shukla, G.C. MiR-644a Disrupts Oncogenic Transformation and Warburg Effect by Direct Modulation of Multiple Genes of Tumor-Promoting Pathways. Cancer Res. 2019, 79, 1844–1856. [Google Scholar] [CrossRef]
- Zoni, E.; Karkampouna, S.; Thalmann, G.N.; Kruithof-de Julio, M.; Spahn, M. Emerging aspects of microRNA interaction with TMPRSS2-ERG and endocrine therapy. Mol. Cell. Endocrinol. 2018, 462, 9–16. [Google Scholar] [CrossRef]
- Visakorpi, T. Novel endocrine aspects of prostate cancer. Mol. Cell. Endocrinol. 2012, 360, 1–2. [Google Scholar] [CrossRef]
- Gordanpour, A.; Stanimirovic, A.; Nam, R.K.; Moreno, C.S.; Sherman, C.; Sugar, L.; Seth, A. miR-221 Is down-regulated in TMPRSS2:ERG fusion-positive prostate cancer. Anticancer Res. 2011, 31, 403–410. [Google Scholar]
- Kao, C.J.; Martiniez, A.; Shi, X.B.; Yang, J.; Evans, C.P.; Dobi, A.; deVere White, R.W.; Kung, H.J. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014, 33, 2495–2503. [Google Scholar] [CrossRef]
- Hart, M.; Wach, S.; Nolte, E.; Szczyrba, J.; Menon, R.; Taubert, H.; Hartmann, A.; Stoehr, R.; Wieland, W.; Grasser, F.A.; et al. The proto-oncogene ERG is a target of microRNA miR-145 in prostate cancer. FEBS J. 2013, 280, 2105–2116. [Google Scholar] [CrossRef]
- Casanova-Salas, I.; Rubio-Briones, J.; Calatrava, A.; Mancarella, C.; Masia, E.; Casanova, J.; Fernandez-Serra, A.; Rubio, L.; Ramirez-Backhaus, M.; Arminan, A.; et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J. Urol. 2014, 192, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.B.; Fu, G.B.; Wang, L.; Ge, X.; Liu, W.T.; Wen, Y.Y.; Sun, H.R.; Liu, L.Z.; Wang, Z.J.; Jiang, B.H. Insulin-like growth factor-I induces chemoresistence to docetaxel by inhibiting miR-143 in human prostate cancer. Oncotarget 2017, 8, 107157–107166. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Niu, X.; Zhang, X.; Tao, J.; Wu, D.; Wang, Z.; Li, P.; Zhang, W.; Wu, H.; Feng, N.; et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol. Cell. Biochem. 2011, 350, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Lin, S.; Cheng, C.; Zhu, A.; Hu, Y.; Shi, Z.; Zhang, X.; Hong, Z. Long non-coding RNA CASC2 regulates Sprouty2 via functioning as a competing endogenous RNA for miR-183 to modulate the sensitivity of prostate cancer cells to docetaxel. Arch. Biochem. Biophys. 2019, 665, 69–78. [Google Scholar] [CrossRef]
- Dong, B.; Shi, Z.; Wang, J.; Wu, J.; Yang, Z.; Fang, K. IL-6 Inhibits the Targeted Modulation of PDCD4 by miR-21 in Prostate Cancer. PLoS ONE 2015, 10, e0134366. [Google Scholar] [CrossRef]
- Shi, G.H.; Ye, D.W.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhang, H.L.; Shen, Y.J.; Zhu, Y.; Zhu, Y.P.; Xiao, W.J.; et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol. Sin. 2010, 31, 867–873. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Li, X.; Padi, S.K.; Zhang, Q.; Tang, M.S.; Guo, B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010, 1, e105. [Google Scholar] [CrossRef]
- Zhang, Q.; Padi, S.K.; Tindall, D.J.; Guo, B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis. 2014, 5, e1486. [Google Scholar] [CrossRef]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zou, L.; Li, X.; Chen, Z.; Lin, Q.; Wu, X. MicroRNA-195 regulates docetaxel resistance by targeting clusterin in prostate cancer. Biomed. Pharmacother. 2018, 99, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; He, P.; Wang, Y. MicroRNA-223-3p regulates cell chemo-sensitivity by targeting FOXO3 in prostatic cancer. Gene 2018, 658, 152–158. [Google Scholar] [CrossRef]
- Zhou, W.; Huang, S.; Jiang, Q.; Yuan, T. Suppression of miR-4735-3p in androgen receptor-expressing prostate cancer cells increases cell death during chemotherapy. Am. J. Transl. Res. 2017, 9, 3714–3722. [Google Scholar] [PubMed]
- Samli, H.; Samli, M.; Vatansever, B.; Ardicli, S.; Aztopal, N.; Dincel, D.; Sahin, A.; Balci, F. Paclitaxel resistance and the role of miRNAs in prostate cancer cell lines. World J. Urol. 2019, 37, 1117–1126. [Google Scholar] [CrossRef]
- Razdan, A.; de Souza, P.; Roberts, T.L. Role of MicroRNAs in Treatment Response in Prostate Cancer. Curr. Cancer Drug Targets 2018, 18, 929–944. [Google Scholar] [CrossRef] [PubMed]
- McKie, A.B.; Douglas, D.A.; Olijslagers, S.; Graham, J.; Omar, M.M.; Heer, R.; Gnanapragasam, V.J.; Robson, C.N.; Leung, H.Y. Epigenetic inactivation of the human sprouty2 (hSPRY2) homologue in prostate cancer. Oncogene 2005, 24, 2166–2174. [Google Scholar] [CrossRef]
- Sun, X.; Yang, Z.; Zhang, Y.; He, J.; Wang, F.; Su, P.; Han, J.; Song, Z.; Fei, Y. Prognostic implications of tissue and serum levels of microRNA-128 in human prostate cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 8394–8401. [Google Scholar]
- Jin, M.; Zhang, T.; Liu, C.; Badeaux, M.A.; Liu, B.; Liu, R.; Jeter, C.; Chen, X.; Vlassov, A.V.; Tang, D.G. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014, 74, 4183–4195. [Google Scholar] [CrossRef]
- Nip, H.; Dar, A.A.; Saini, S.; Colden, M.; Varahram, S.; Chowdhary, H.; Yamamura, S.; Mitsui, Y.; Tanaka, Y.; Kato, T.; et al. Oncogenic microRNA-4534 regulates PTEN pathway in prostate cancer. Oncotarget 2016, 7, 68371–68384. [Google Scholar] [CrossRef]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef]
- Stoen, M.J.; Andersen, S.; Rakaee, M.; Pedersen, M.I.; Ingebriktsen, L.M.; Bremnes, R.M.; Donnem, T.; Lombardi, A.P.G.; Kilvaer, T.K.; Busund, L.T.; et al. High expression of miR-17-5p in tumor epithelium is a predictor for poor prognosis for prostate cancer patients. Sci. Rep. 2021, 11, 13864. [Google Scholar] [CrossRef] [PubMed]
- Turnham, D.J.; Bullock, N.; Dass, M.S.; Staffurth, J.N.; Pearson, H.B. The PTEN Conundrum: How to Target PTEN-Deficient Prostate Cancer. Cells 2020, 9, 2342. [Google Scholar] [CrossRef]
- Hoey, C.; Ahmed, M.; Fotouhi Ghiam, A.; Vesprini, D.; Huang, X.; Commisso, K.; Commisso, A.; Ray, J.; Fokas, E.; Loblaw, D.A.; et al. Circulating miRNAs as non-invasive biomarkers to predict aggressive prostate cancer after radical prostatectomy. J. Transl. Med. 2019, 17, 173. [Google Scholar] [CrossRef] [PubMed]
- Aboushousha, T.; El-Nahas, E.I.; El-Hindawi, A.; Elwi, D.; El-Ganzouri, H.; Hammam, O.; Magdy, M.; Helal, N.S. Implication of miRNA-153 on PTEN expression in prostatic adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 6834–6843. [Google Scholar] [CrossRef] [PubMed]
- Gurbuz, V.; Sozen, S.; Bilen, C.Y.; Konac, E. miR-148a, miR-152 and miR-200b promote prostate cancer metastasis by targeting DNMT1 and PTEN expression. Oncol. Lett. 2021, 22, 805. [Google Scholar] [CrossRef]
- Huang, S.; Li, X.; Zhu, H. MicroRNA-152 Targets Phosphatase and Tensin Homolog to Inhibit Apoptosis and Promote Cell Migration of Nasopharyngeal Carcinoma Cells. Med. Sci. Monit. 2016, 22, 4330–4337. [Google Scholar] [CrossRef]
- Saffari, M.; Ghaderian, S.M.H.; Omrani, M.D.; Afsharpad, M.; Shankaie, K.; Samadaian, N. The Association of miR-let 7b and miR-548 with PTEN in Prostate Cancer. Urol. J. 2019, 16, 267–273. [Google Scholar] [CrossRef]
- Lu, J.; Mu, X.; Yin, Q.; Hu, K. miR-106a contributes to prostate carcinoma progression through PTEN. Oncol. Lett. 2019, 17, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, J.; Zhang, Y.; Kang, S. Silencing miRNA-1297 suppresses the invasion and migration of prostate cancer cells via targeting modulation of PTEN and blocking of the AKT/ERK pathway. Exp. Ther. Med. 2021, 22, 768. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhao, Z.; Wu, R.; Wu, L.; Tian, X.; Zhang, Z. MiR-146b inhibits autophagy in prostate cancer by targeting the PTEN/Akt/mTOR signaling pathway. Aging 2018, 10, 2113–2121. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ji, C.; Guo, S.; Su, X.; Zhao, X.; Zhang, S.; Liu, G.; Qiu, X.; Zhang, Q.; Guo, H.; et al. The miR-486-5p plays a causative role in prostate cancer through negative regulation of multiple tumor suppressor pathways. Oncotarget 2017, 8, 72835–72846. [Google Scholar] [CrossRef]
- Ghorbanmehr, N.; Gharbi, S.; Korsching, E.; Tavallaei, M.; Einollahi, B.; Mowla, S.J. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate 2019, 79, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, J.X.; Shao, Z.Q. miR-21 targets and inhibits tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion: An experimental study. Asian Pac. J. Trop. Med. 2017, 10, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Anczukow, O.; Akerman, M.; Krainer, A.R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012, 1, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Krainer, A.R. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014, 12, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Q.; Balk, S.; Brown, M.; Lee, G.S.; Kantoff, P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009, 69, 3356–3363. [Google Scholar] [CrossRef] [PubMed]
- Ratnadiwakara, M.; Mohenska, M.; Anko, M.L. Splicing factors as regulators of miRNA biogenesis—links to human disease. Semin. Cell Dev. Biol. 2018, 79, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Gong, J.Y.; Goodman, O.B., Jr.; Cartegni, L.; Nanus, D.M.; Shen, R. Bombesin attenuates pre-mRNA splicing of glucocorticoid receptor by regulating the expression of serine-arginine protein p30c (SRp30c) in prostate cancer cells. Biochim. Biophys. Acta 2007, 1773, 1087–1094. [Google Scholar] [CrossRef]
- van Alphen, R.J.; Wiemer, E.A.; Burger, H.; Eskens, F.A. The spliceosome as target for anticancer treatment. Br. J. Cancer 2009, 100, 228–232. [Google Scholar] [CrossRef]
- Yoshino, H.; Enokida, H.; Chiyomaru, T.; Tatarano, S.; Hidaka, H.; Yamasaki, T.; Gotannda, T.; Tachiwada, T.; Nohata, N.; Yamane, T.; et al. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem. Biophys. Res. Commun. 2012, 417, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hu, H. Identification of prognosis biomarkers of prostatic cancer in a cohort of 498 patients from TCGA. Curr. Probl. Cancer 2019, 43, 100503. [Google Scholar] [CrossRef] [PubMed]
- Pewzner-Jung, Y.; Ben-Dor, S.; Futerman, A.H. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J. Biol. Chem. 2006, 281, 25001–25005. [Google Scholar] [CrossRef]
- Eto, M.; Bennouna, J.; Hunter, O.C.; Hershberger, P.A.; Kanto, T.; Johnson, C.S.; Lotze, M.T.; Amoscato, A.A. C16 ceramide accumulates following androgen ablation in LNCaP prostate cancer cells. Prostate 2003, 57, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Eto, M.; Bennouna, J.; Hunter, O.C.; Lotze, M.T.; Amoscato, A.A. Importance of C16 ceramide accumulation during apoptosis in prostate cancer cells. Int. J. Urol. 2006, 13, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Mak, B.; Yeung, N.; Huynh, K.; Meikle, T.G.; Mellett, N.A.; Kwan, E.M.; Fettke, H.; Tran, B.; Davis, I.D.; et al. Overcoming enzalutamide resistance in metastatic prostate cancer by targeting sphingosine kinase. EBioMedicine 2021, 72, 103625. [Google Scholar] [CrossRef]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Hidaka, H.; Majid, S.; Saini, S.; Arora, S.; Deng, G.; Shahryari, V.; Chang, I.; et al. Genistein up-regulates tumor suppressor microRNA-574-3p in prostate cancer. PLoS ONE 2013, 8, e58929. [Google Scholar] [CrossRef]
- Marima, R.; Francies, F.Z.; Hull, R.; Molefi, T.; Oyomno, M.; Khanyile, R.; Mbatha, S.; Mabongo, M.; Owen Bates, D.; Dlamini, Z. MicroRNA and Alternative mRNA Splicing Events in Cancer Drug Response/Resistance: Potent Therapeutic Targets. Biomedicines 2021, 9, 1818. [Google Scholar] [CrossRef] [PubMed]
- Meyers-Needham, M.; Ponnusamy, S.; Gencer, S.; Jiang, W.; Thomas, R.J.; Senkal, C.E.; Ogretmen, B. Concerted functions of HDAC1 and microRNA-574-5p repress alternatively spliced ceramide synthase 1 expression in human cancer cells. EMBO Mol. Med. 2012, 4, 78–92. [Google Scholar] [CrossRef]
- Mavrou, A.; Brakspear, K.; Hamdollah-Zadeh, M.; Damodaran, G.; Babaei-Jadidi, R.; Oxley, J.; Gillatt, D.A.; Ladomery, M.R.; Harper, S.J.; Bates, D.O.; et al. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene 2015, 34, 4311–4319. [Google Scholar] [CrossRef]
- Oltean, S.; Gammons, M.; Hulse, R.; Hamdollah-Zadeh, M.; Mavrou, A.; Donaldson, L.; Salmon, A.H.; Harper, S.J.; Ladomery, M.R.; Bates, D.O. SRPK1 inhibition in vivo: Modulation of VEGF splicing and potential treatment for multiple diseases. Biochem. Soc. Trans. 2012, 40, 831–835. [Google Scholar] [CrossRef]
- Amin, E.M.; Oltean, S.; Hua, J.; Gammons, M.V.; Hamdollah-Zadeh, M.; Welsh, G.I.; Cheung, M.K.; Ni, L.; Kase, S.; Rennel, E.S.; et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell 2011, 20, 768–780. [Google Scholar] [CrossRef]
- Gao, X.; Dai, C.; Huang, S.; Tang, J.; Chen, G.; Li, J.; Zhu, Z.; Zhu, X.; Zhou, S.; Gao, Y.; et al. Functional Silencing of HSD17B2 in Prostate Cancer Promotes Disease Progression. Clin. Cancer Res. 2019, 25, 1291–1301. [Google Scholar] [CrossRef]
- Lin, S.L.; Chiang, A.; Chang, D.; Ying, S.Y. Loss of mir-146a function in hormone-refractory prostate cancer. RNA 2008, 14, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.I.; Suzuki, T.; Fujimura, T.; Yamada, Y.; Takahashi, S.; Homma, Y.; Suzuki, Y.; Inoue, S. Dysregulation of spliceosome gene expression in advanced prostate cancer by RNA-binding protein PSF. Proc. Natl. Acad. Sci. USA 2017, 114, 10461–10466. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lan, T. Molecular Origin, Expression Regulation, and Biological Function of Androgen Receptor Splicing Variant 7 in Prostate Cancer. Urol. Int. 2021, 105, 337–353. [Google Scholar] [CrossRef]
- Naiki-Ito, A.; Naiki, T.; Kato, H.; Iida, K.; Etani, T.; Nagayasu, Y.; Suzuki, S.; Yamashita, Y.; Inaguma, S.; Onishi, M.; et al. Recruitment of miR-8080 by luteolin inhibits androgen receptor splice variant 7 expression in castration-resistant prostate cancer. Carcinogenesis 2020, 41, 1145–1157. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.B.; Xue, L.; Ma, A.H.; Tepper, C.G.; Gandour-Edwards, R.; Kung, H.J.; deVere White, R.W. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene 2013, 32, 4130–4138. [Google Scholar] [CrossRef]
- Marcias, G.; Erdmann, E.; Lapouge, G.; Siebert, C.; Barthelemy, P.; Duclos, B.; Bergerat, J.P.; Ceraline, J.; Kurtz, J.E. Identification of novel truncated androgen receptor (AR) mutants including unreported pre-mRNA splicing variants in the 22Rv1 hormone-refractory prostate cancer (PCa) cell line. Hum. Mutat. 2010, 31, 74–80. [Google Scholar] [CrossRef]
- Ma, J.; Jemal, A.; Fedewa, S.A.; Islami, F.; Lichtenfeld, J.L.; Wender, R.C.; Cullen, K.J.; Brawley, O.W. The American Cancer Society 2035 challenge goal on cancer mortality reduction. CA Cancer J. Clin. 2019, 69, 351–362. [Google Scholar] [CrossRef]
- A summary of the American Cancer Society Report to the Nation: Cancer in the poor. CA Cancer J. Clin. 1989, 39, 263–265. [CrossRef]
- Freeman, H.P. Cancer in the socioeconomically disadvantaged. CA Cancer J. Clin. 1989, 39, 266–288. [Google Scholar] [CrossRef] [PubMed]
- Freeman, H.P. Cancer in the economically disadvantaged. Cancer 1989, 64, 324–334; discussion 342–345. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Thun, M.J. Cancer statistics, 2007. CA Cancer J. Clin. 2007, 57, 43–66. [Google Scholar] [CrossRef]
- Powell, I.J. Epidemiology and pathophysiology of prostate cancer in African-American men. J. Urol. 2007, 177, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.; Shapiro, M.; Morton, R., Jr.; Brawley, O.W. Prostate cancer in black and white Americans. Cancer Metastasis. Rev. 2003, 22, 83–86. [Google Scholar] [CrossRef]
- Robbins, A.S.; Whittemore, A.S.; Thom, D.H. Differences in socioeconomic status and survival among white and black men with prostate cancer. Am. J. Epidemiol. 2000, 151, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.; Metcalfe, C.; Ibrahim, F.; Persad, R.; Ben-Shlomo, Y. Investigating Black-White differences in prostate cancer prognosis: A systematic review and meta-analysis. Int. J. Cancer 2008, 123, 430–435. [Google Scholar] [CrossRef]
- Conti, D.V.; Darst, B.F.; Moss, L.C.; Saunders, E.J.; Sheng, X.; Chou, A.; Schumacher, F.R.; Olama, A.A.A.; Benlloch, S.; Dadaev, T.; et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021, 53, 65–75. [Google Scholar] [CrossRef]
- Johnson, J.R.; Woods-Burnham, L.; Hooker, S.E., Jr.; Batai, K.; Kittles, R.A. Genetic Contributions to Prostate Cancer Disparities in Men of West African Descent. Front. Oncol. 2021, 11, 770500. [Google Scholar] [CrossRef]
- Rayford, W.; Beksac, A.T.; Alger, J.; Alshalalfa, M.; Ahmed, M.; Khan, I.; Falagario, U.G.; Liu, Y.; Davicioni, E.; Spratt, D.E.; et al. Comparative analysis of 1152 African-American and European-American men with prostate cancer identifies distinct genomic and immunological differences. Commun. Biol. 2021, 4, 670. [Google Scholar] [CrossRef]
- Awasthi, S.; Berglund, A.; Abraham-Miranda, J.; Rounbehler, R.J.; Kensler, K.; Serna, A.; Vidal, A.; You, S.; Freeman, M.R.; Davicioni, E.; et al. Comparative Genomics Reveals Distinct Immune-oncologic Pathways in African American Men with Prostate Cancer. Clin. Cancer Res. 2021, 27, 320–329. [Google Scholar] [CrossRef]
- Theodore, S.C.; Rhim, J.S.; Turner, T.; Yates, C. MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn. Dis. 2010, 20 (Suppl. S1), S96–S100. [Google Scholar]
- Shiina, M.; Hashimoto, Y.; Kulkarni, P.; Dasgupta, P.; Shahryari, V.; Yamamura, S.; Tanaka, Y.; Dahiya, R. Role of miR-182/PDCD4 axis in aggressive behavior of prostate cancer in the African Americans. BMC Cancer 2021, 21, 1028. [Google Scholar] [CrossRef]
- Theodore, S.C.; Davis, M.; Zhao, F.; Wang, H.; Chen, D.; Rhim, J.; Dean-Colomb, W.; Turner, T.; Ji, W.; Zeng, G.; et al. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget 2014, 5, 3512–3525. [Google Scholar] [CrossRef] [PubMed]
- Cagle, P.; Niture, S.; Srivastava, A.; Ramalinga, M.; Aqeel, R.; Rios-Colon, L.; Chimeh, U.; Suy, S.; Collins, S.P.; Dahiya, R.; et al. MicroRNA-214 targets PTK6 to inhibit tumorigenic potential and increase drug sensitivity of prostate cancer cells. Sci. Rep. 2019, 9, 9776. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Shiina, M.; Dasgupta, P.; Kulkarni, P.; Kato, T.; Wong, R.K.; Tanaka, Y.; Shahryari, V.; Maekawa, S.; Yamamura, S.; et al. Upregulation of miR-130b Contributes to Risk of Poor Prognosis and Racial Disparity in African-American Prostate Cancer. Cancer Prev. Res. 2019, 12, 585–598. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Shiina, M.; Kato, T.; Yamamura, S.; Tanaka, Y.; Majid, S.; Saini, S.; Shahryari, V.; Kulkarni, P.; Dasgupta, P.; et al. The role of miR-24 as a race related genetic factor in prostate cancer. Oncotarget 2017, 8, 16581–16593. [Google Scholar] [CrossRef]
- Parra-Medina, R.; Lopez-Kleine, L.; Ramirez-Clavijo, S.; Payan-Gomez, C. Identification of candidate miRNAs in early-onset and late-onset prostate cancer by network analysis. Sci. Rep. 2020, 10, 12345. [Google Scholar] [CrossRef]
- Ren, Q.; Liang, J.; Wei, J.; Basturk, O.; Wang, J.; Daniels, G.; Gellert, L.L.; Li, Y.; Shen, Y.; Osman, I.; et al. Epithelial and stromal expression of miRNAs during prostate cancer progression. Am. J. Transl. Res. 2014, 6, 329–339. [Google Scholar] [PubMed]
- Shiina, M.; Hashimoto, Y.; Kato, T.; Yamamura, S.; Tanaka, Y.; Majid, S.; Saini, S.; Varahram, S.; Kulkarni, P.; Dasgupta, P.; et al. Differential expression of miR-34b and androgen receptor pathway regulate prostate cancer aggressiveness between African-Americans and Caucasians. Oncotarget 2017, 8, 8356–8368. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Ramalinga, M.; Chijioke, J.; Marian, C.; Oermann, E.K.; Uhm, S.; Kim, J.S.; Chen, L.N.; et al. MicroRNA profiling in prostate cancer--the diagnostic potential of urinary miR-205 and miR-214. PLoS ONE 2013, 8, e76994. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jia, D.; Kim, H.; Abd Elmageed, Z.Y.; Datta, A.; Davis, R.; Srivastav, S.; Moroz, K.; Crawford, B.E.; Moparty, K.; et al. Dysregulation of miR-212 Promotes Castration Resistance through hnRNPH1-Mediated Regulation of AR and AR-V7: Implications for Racial Disparity of Prostate Cancer. Clin. Cancer Res. 2016, 22, 1744–1756. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Ha, J.W.; Zhang, B.T. Constructing higher-order miRNA-mRNA interaction networks in prostate cancer via hypergraph-based learning. BMC Syst. Biol. 2013, 7, 47. [Google Scholar] [CrossRef]
- Yu, J.; Sun, S.; Mao, W.; Xu, B.; Chen, M. Identification of Enzalutamide Resistance-Related circRNA-miRNA-mRNA Regulatory Networks in Patients with Prostate Cancer. OncoTargets Ther. 2021, 14, 3833–3848. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Wang, P.; Yang, C.; Li, S. Exploration of the molecular mechanism of prostate cancer based on mRNA and miRNA expression profiles. OncoTargets Ther. 2017, 10, 3225–3232. [Google Scholar] [CrossRef]
- Wang, B.D.; Ceniccola, K.; Yang, Q.; Andrawis, R.; Patel, V.; Ji, Y.; Rhim, J.; Olender, J.; Popratiloff, A.; Latham, P.; et al. Identification and Functional Validation of Reciprocal microRNA-mRNA Pairings in African American Prostate Cancer Disparities. Clin. Cancer Res. 2015, 21, 4970–4984. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Benoist, G.E.; van Oort, I.M.; Boerrigter, E.; Verhaegh, G.W.; van Hooij, O.; Groen, L.; Smit, F.; de Mol, P.; Hamberg, P.; Dezentje, V.O.; et al. Prognostic Value of Novel Liquid Biomarkers in Patients with Metastatic Castration-Resistant Prostate Cancer Treated with Enzalutamide: A Prospective Observational Study. Clin. Chem. 2020, 66, 842–851. [Google Scholar] [CrossRef]
- Crocetto, F.; Russo, G.; Di Zazzo, E.; Pisapia, P.; Mirto, B.F.; Palmieri, A.; Pepe, F.; Bellevicine, C.; Russo, A.; La Civita, E.; et al. Liquid Biopsy in Prostate Cancer Management-Current Challenges and Future Perspectives. Cancers 2022, 14, 3272. [Google Scholar] [CrossRef]
- Rana, S.; Valbuena, G.N.; Curry, E.; Bevan, C.L.; Keun, H.C. MicroRNAs as biomarkers for prostate cancer prognosis: A systematic review and a systematic reanalysis of public data. Br. J. Cancer 2022, 126, 502–513. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.M.; Engelman, D.M.; et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015, 518, 107–110. [Google Scholar] [CrossRef]
- Hu, C.M.; Zhang, L. Nanoparticle-based combination therapy toward overcoming drug resistance in cancer. Biochem. Pharmacol. 2012, 83, 1104–1111. [Google Scholar] [CrossRef]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef]
- Ji, J.; Shi, J.; Budhu, A.; Yu, Z.; Forgues, M.; Roessler, S.; Ambs, S.; Chen, Y.; Meltzer, P.S.; Croce, C.M.; et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 2009, 361, 1437–1447. [Google Scholar] [CrossRef]
- Tang, X.; Jin, L.; Cao, P.; Cao, K.; Huang, C.; Luo, Y.; Ma, J.; Shen, S.; Tan, M.; Li, X.; et al. MicroRNA-16 sensitizes breast cancer cells to paclitaxel through suppression of IKBKB expression. Oncotarget 2016, 7, 23668–23683. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Fava, P.; Bergallo, M.; Astrua, C.; Brizio, M.; Galliano, I.; Montanari, P.; Dapra, V.; Novelli, M.; Savoia, P.; Quaglino, P.; et al. miR-155 expression in Primary Cutaneous T-Cell Lymphomas (CTCL). J. Eur. Acad. Dermatol. Venereol. 2017, 31, e27–e29. [Google Scholar] [CrossRef] [PubMed]
- Kopp, K.L.; Ralfkiaer, U.; Gjerdrum, L.M.; Helvad, R.; Pedersen, I.H.; Litman, T.; Jonson, L.; Hagedorn, P.H.; Krejsgaard, T.; Gniadecki, R.; et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle 2013, 12, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Pel, M.E.; Kirschner, M.B.; Cheng, Y.Y.; Mugridge, N.; Weiss, J.; Williams, M.; Wright, C.; Edelman, J.J.; Vallely, M.P.; et al. Restoring expression of miR-16: A novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. 2013, 24, 3128–3135. [Google Scholar] [CrossRef] [PubMed]
- Gaur, S.; Wen, Y.; Song, J.H.; Parikh, N.U.; Mangala, L.S.; Blessing, A.M.; Ivan, C.; Wu, S.Y.; Varkaris, A.; Shi, Y.; et al. Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget 2015, 6, 29161–29177. [Google Scholar] [CrossRef]
- Wen, D.; Peng, Y.; Lin, F.; Singh, R.K.; Mahato, R.I. Micellar Delivery of miR-34a Modulator Rubone and Paclitaxel in Resistant Prostate Cancer. Cancer Res. 2017, 77, 3244–3254. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, C.; Tao, Y.; Zou, P.; Gao, F.; Jia, C.; Liu, L.; Li, G.; Zhang, G.; Duan, Y.; et al. Ultrasound-Induced Microbubble Cavitation Combined with miR-34a-Loaded Nanoparticles for the Treatment of Castration-Resistant Prostate Cancer. J. Biomed. Nanotechnol. 2021, 17, 78–89. [Google Scholar] [CrossRef]
- Yao, C.; Liu, J.; Wu, X.; Tai, Z.; Gao, Y.; Zhu, Q.; Li, J.; Zhang, L.; Hu, C.; Gu, F.; et al. Reducible self-assembling cationic polypeptide-based micelles mediate co-delivery of doxorubicin and microRNA-34a for androgen-independent prostate cancer therapy. J. Control. Release 2016, 232, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Wen, D.; Wang, X.; Mahato, R.I. Dual responsive micelles capable of modulating miRNA-34a to combat taxane resistance in prostate cancer. Biomaterials 2019, 192, 95–108. [Google Scholar] [CrossRef]
- Takeshita, F.; Patrawala, L.; Osaki, M.; Takahashi, R.U.; Yamamoto, Y.; Kosaka, N.; Kawamata, M.; Kelnar, K.; Bader, A.G.; Brown, D.; et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. 2010, 18, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Fan, W.; Hao, J.; Wu, X.; Zeng, G.Q.; Zhang, L.J.; Nie, S.F.; Wang, X.D. Efficient delivery of micro RNA to bone-metastatic prostate tumors by using aptamer-conjugated atelocollagen in vitro and in vivo. Drug Deliv. 2016, 23, 874–881. [Google Scholar] [CrossRef] [PubMed]
| MiRNAs | Regulation in PCa | MiRNA Target Genes and Regulated Pathways in PCa | References |
|---|---|---|---|
| let-7/let-7a | Downregulation | CCR7 MAPK/EMT signaling pathways | [35,36] |
| miR-1 | Downregulation | MET, KRAS c-Met/AKT/mTOR signaling | [38,39] |
| miR-29b | Downregulation | AKT2, MCL1 | [42,44] |
| miR-30c | Downregulation | ASF/SF2, E2F7 | [40,41] |
| miR-34a | Downregulation | SIRT1, E2F1, CDK6, Cyclin D1, E2F3, BCL2 | [55,57,58] |
| miR-99b | Downregulation | MTOR, SMARCA5, SMARCD1, PSA, AR | [59,60,62,100] |
| miR-133a | Downregulation | FUS, AR, EGFR, FGFR1, IGF1R, and MET | [67,68,101] |
| miR-134 | Downregulation | EGFR, cyclin D/cyclin D2/CDK4, KRAS, EGFR, POGLUT1, STAT5B, KRAS, NANOG, HNF4A, EGFR, ITGB1, FOXM1, HER2, PIK3CA, ITGB1, POGLUT1 | [69,70,102] |
| miR-205 | Downregulation | AR, MAPK | [73] |
| miR-221 | Downregulation Upregulation | SOCS1, BMI1 Ras/Raf/MEK/ERK signaling HECTD2, RAB1A, AR reprograming in CRPC | [74,75,76,77] |
| miR-375 | Downregulation | QKI5, CD44 | [78,79,80] |
| miR-21 | Upregulation | PTEN, RECK, MARCKS, PDCD4, TPM1 JAK/STAT3 cascades | [82,83,103] |
| miR-96 | Upregulation | FOXO1, MTSS1, EGFR, MTOR, ATG7 | [69,104,105] |
| miR-106b | Upregulation | TEN, AKT, CNN1, LARP4B, RUNX3, DAB2, DLC1, FOG2, REST1, FUT6, ALEX1 and BTG3, CDKN1A (p21 gene), E2F1, Ki67, MMP2, CD44, and SMAD2 | [86] |
| miR-125b | Upregulation | BAK1, TP53, PUMA | [88,106] |
| miR-141 | Upregulation | AKT and Rb/E2F signaling | [89,107] |
| miR-145 | Upregulation | SP1, CDK6, AKAP12, RAD51, MCL1, PAR4, PARP1 | [90] |
| miR-182 | Upregulation | NDRG1, GNA13, BRCA1, PDCD4, ST6GALNAC5 | [91] |
| miR-200c | Upregulation | BRD7, BMI1 EMT, AKT signaling pathways | [93,94,95] |
| miR-222 | Upregulation | CDKN1B, CDKN1C | [97] |
| MiRNA (Up or Down in PCa) | Target Gene(s) | Treatment Resistance | References |
|---|---|---|---|
| miR-148a ↓ | MSK1 | ADT resistance | [122] |
| miR-125b ↓ | TP53, PUMA, BAK11 | ADT resistance | [88] |
| miR-216a ↑ | PTEN, TGFBR2 | ADT resistance | [124] |
| miR-194 ↓ | FOXA1 | Enz resistance | [125] |
| miR-27a, miR-101 ↓ | NR2F2 | Enz resistance | [126] |
| miR-30c-1/miR-103a-2 ↑ | AR-V7 | Enz resistance | [121] |
| miR-644a ↓ | MYC, BCLXL, BCL2, AKT, IGF1R | Enz resistance | [125] |
| miR-221 ↓ | ERG | Androgen-independent | [130,131] |
| miR-145 ↓ | ERG | Enz resistance | [132] |
| miR-187 ↓, miR-182 ↑ | TMPRSS2-ERG | Enz resistance | [133] |
| miR-143 ↓ | IGF1R, IRS1 | DCT resistance | [135] |
| miR-183 | DCT resistance | [147] | |
| miR-21 ↑ | PDCD4 | DCT resistance | [137,138] |
| miR-205, miR-31 ↓ | BCLW, E2F6 | DCT resistance | [139,140] |
| miR-195 ↓ | CUL | DCT resistance | [142] |
| miR-223-3p ↑ | FOXO3 | DCT resistance | [143] |
| miR-4735-3p ↓ | MEKK1 | DCT resistance | [144] |
| miR-19b-3p, miR-26b-5p, miR-374b-5p ↓ | LARP1, CCND1 | Paclitaxel resistance | [145] |
| miR-15a, miR-16 ↓ | CCND1, WNT3A | Cisplatin resistance | [146] |
| miR-128 ↓ | ZEB1, BIM1 | Cisplatin resistance | [148,149] |
| miR-205 ↓ | RAB27A, LAMP3 | Cisplatin resistance | [32] |
| MiRNA | Regulation of PTEN Expression | Function | Reference |
|---|---|---|---|
| miR-19b | Downregulates PTEN | Cell proliferation ↑ | [28] |
| miR-23b | Downregulates PTEN | Cell proliferation ↑ | [28] |
| miR-26a | Downregulates PTEN | Cell proliferation ↑ | [28] |
| miR-92a | Downregulates PTEN | Cell proliferation ↑ | [28] |
| miR-17-5p | Downregulates PTEN | Tumor growth ↑/cell proliferation ↑/invasion ↑ | [152,154] |
| miR-153 | Downregulates PTEN | Higher Gleason scores | [155] |
| miR-152 | Downregulates PTEN | Tumor growth ↑ | [156,157] |
| miR-4534 | Downregulates PTEN | Cell growth ↑, proliferation ↑, and progression ↑ | [150] |
| miR-548c-3p | Downregulates PTEN | Higher Gleason scores | [158] |
| miR-106 | Downregulates PTEN | Cell growth and proliferation ↑ | [159] |
| miR-1297 | Downregulates PTEN | Migration ↑, invasion ↑ | [160] |
| miR-146b | Downregulates PTEN | Tumorigenesis ↑, survival ↑, proliferation ↑, AKT/mTOR signaling↑ | [161] |
| miR-486-5p | Downregulates PTEN | Migration ↑, invasion ↑, motility ↑ | [162] |
| miR-21-5p | Downregulates PTEN | Cell proliferation ↑, invasion ↑ | [163,164] |
| MiRNA | Clinical Trial | Biomarker/Therapy | Status |
|---|---|---|---|
| miR-141, miR-375 | NCT02391051 | Prognostic biomarker for low-risk PCa | Phase II |
| miRNA profiling | NCT01220427 | Diagnostic biomarker for high-grade PCa | Phase II |
| miRNA profiling | NCT02964351 | Biomarker for high-PSA PCa | Recruiting |
| Exosomal miRNA signature | NCT02366494 | Prognostic biomarker for PCa under ADT treatment | Active, not recruiting |
| miRNA panel | NCT02471469 | Prognostic biomarker for mCRPC under Enz | Complete recruiting |
| miRNA profiling | NCT01503229 | Prognostic biomarker for mCRPC under Abi | Phase II |
| miRNA profiling | NCT01120236 | Prognostic biomarker for PCa under ADT/cixutumumab | Phase II |
| MRX34 | NCT01829971 | miR-34a-based therapy for multiple solid tumors | Phase I, terminated |
| MRG-106 | NCT03713320 | miR-155 inhibitor for treating CTCL | Phase II |
| TargomiRs | NCT02369198 | miR-16-based therapy for NSCLC | Phase I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gujrati, H.; Ha, S.; Wang, B.-D. Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms. Cancers 2023, 15, 3140. https://doi.org/10.3390/cancers15123140
Gujrati H, Ha S, Wang B-D. Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms. Cancers. 2023; 15(12):3140. https://doi.org/10.3390/cancers15123140
Chicago/Turabian StyleGujrati, Himali, Siyoung Ha, and Bi-Dar Wang. 2023. "Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms" Cancers 15, no. 12: 3140. https://doi.org/10.3390/cancers15123140
APA StyleGujrati, H., Ha, S., & Wang, B.-D. (2023). Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms. Cancers, 15(12), 3140. https://doi.org/10.3390/cancers15123140






