B7H3 Role in Reshaping Immunosuppressive Landscape in MSI and MSS Colorectal Cancer Tumours
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of the Patient Group
2.2. Evaluation of the B7H3 Concentrations with ELISA
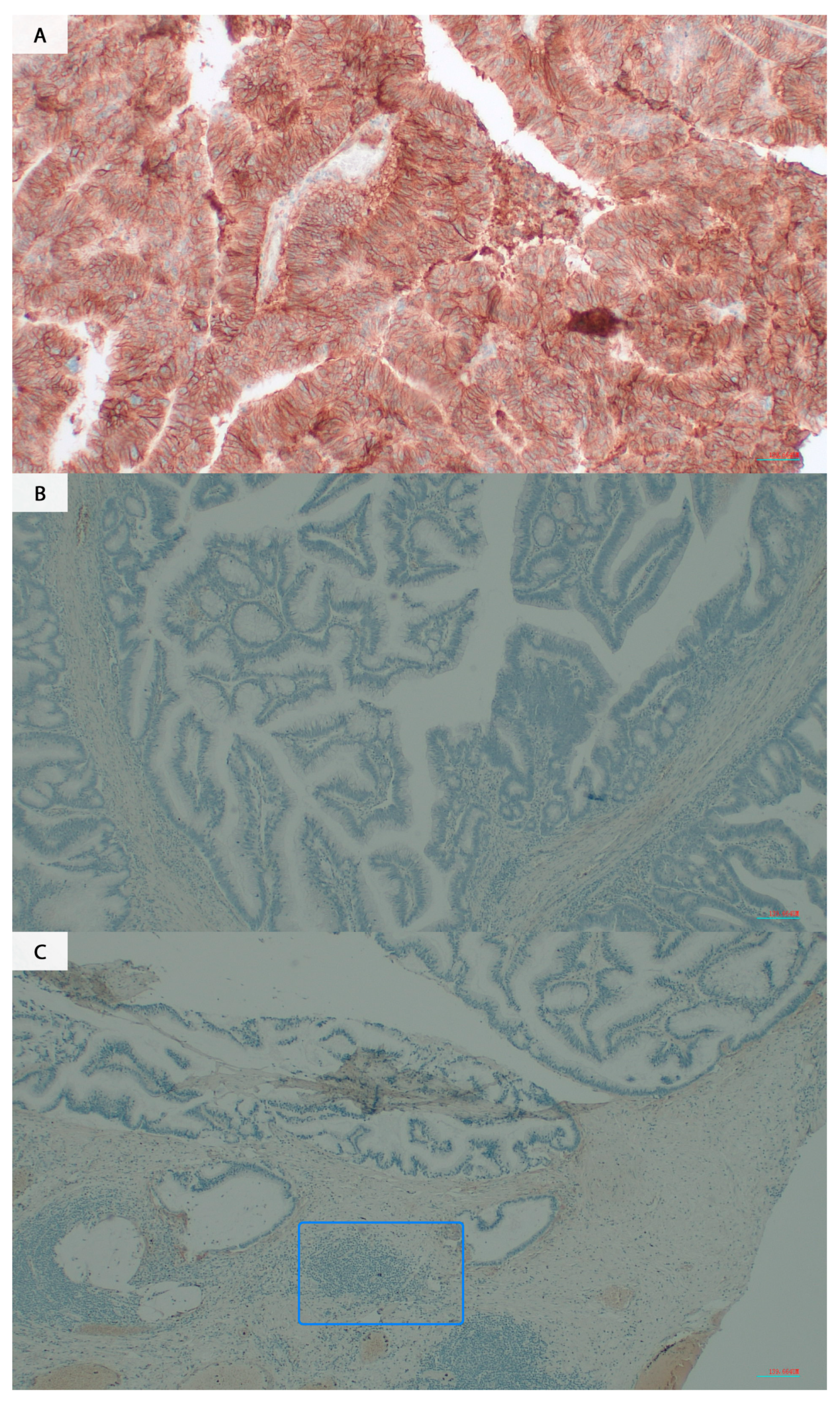
2.3. Evaluation of the B7H3 Expression by IHC
2.4. Assessment of the MSI/MSS Status
2.5. Evaluation of the Tumour-Infiltrating CD8+ T-cells
2.6. Assessment of the TILs and Budding
2.7. Evaluation of the Cytokines Screening Panel
2.8. Exploration of B7H3 Biological Functions
2.9. Statistical Analysis
3. Results
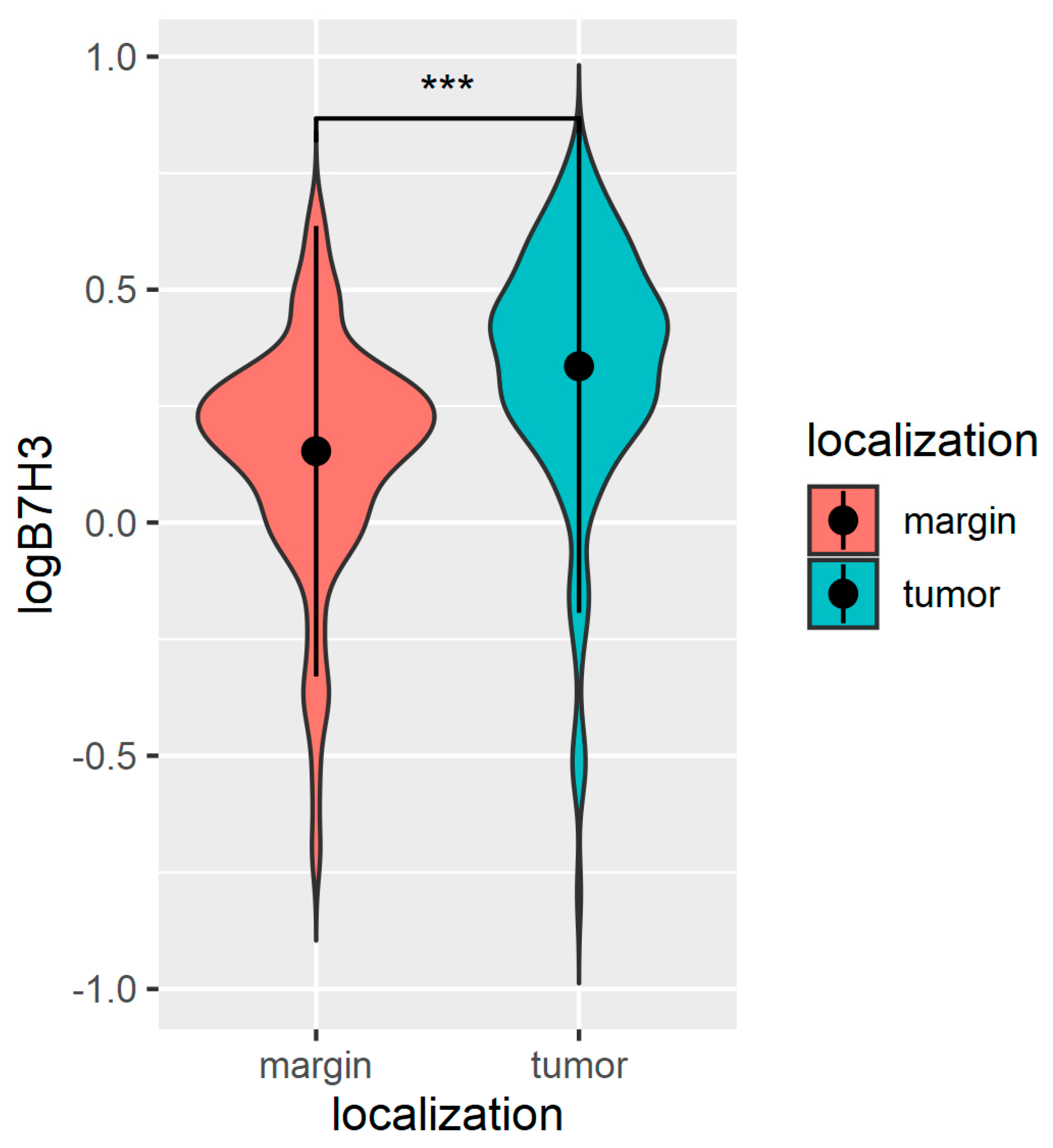
3.1. The B7H3 Expression Is Upregulated in Tumour Tissues
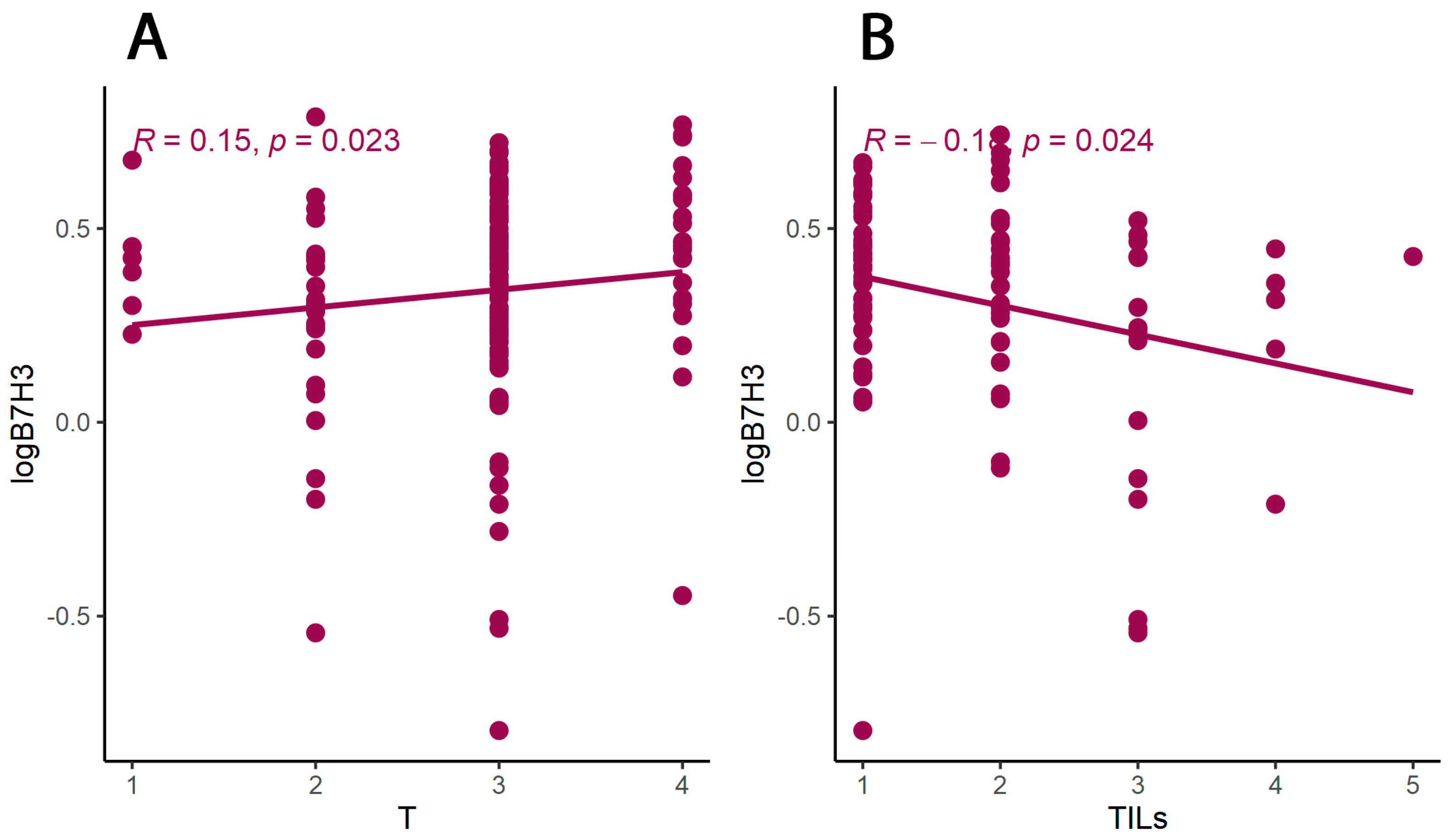
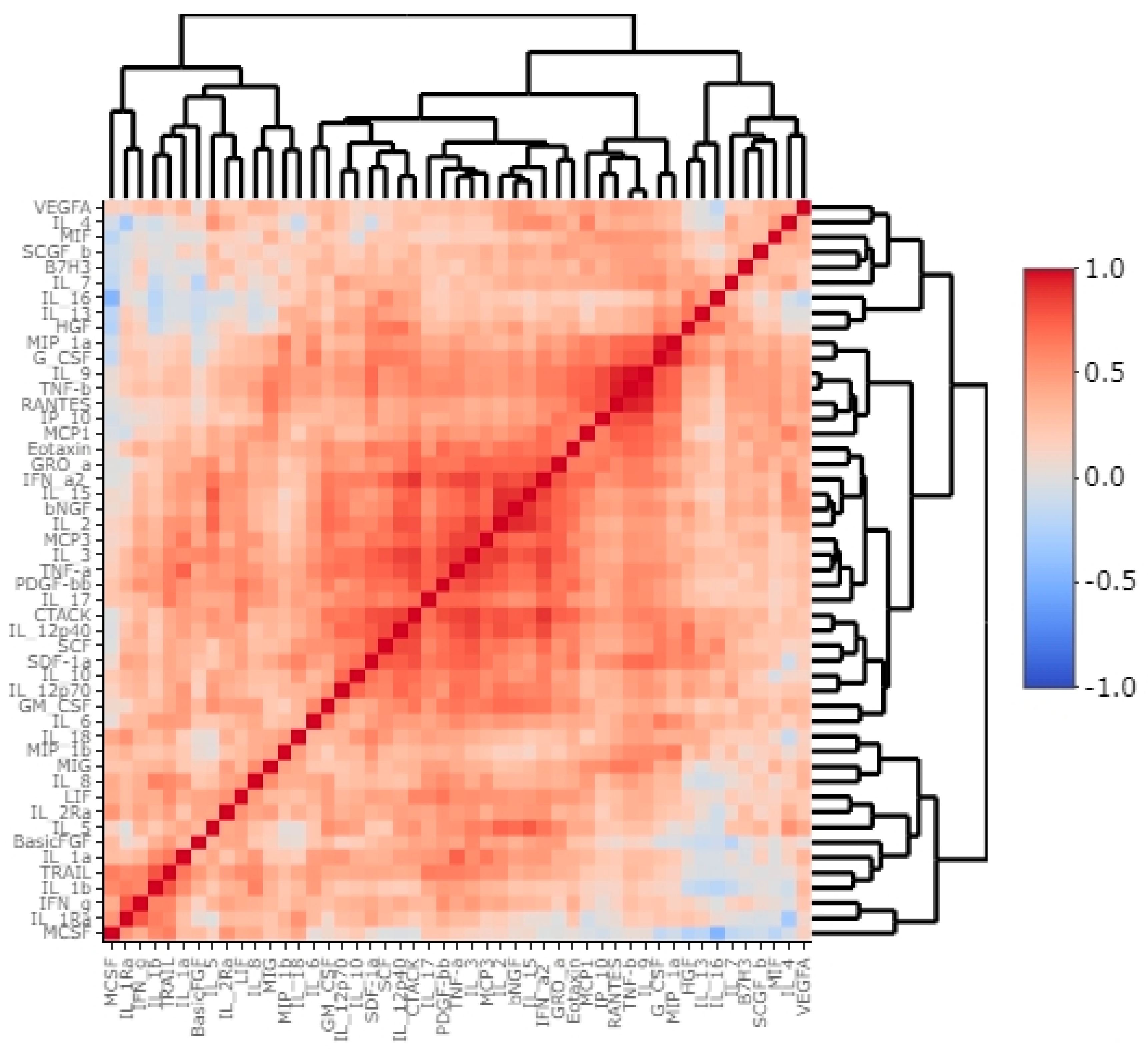
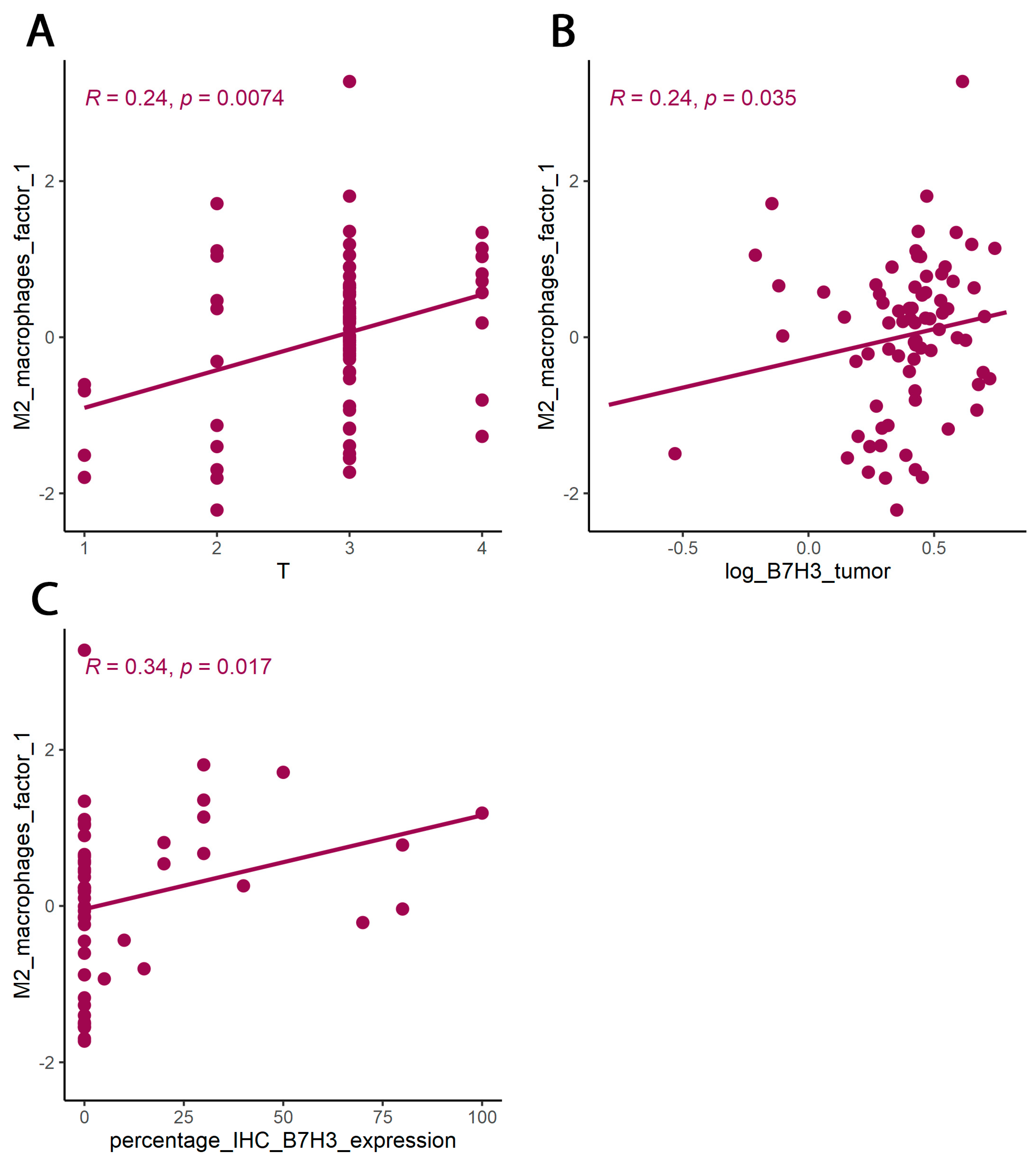
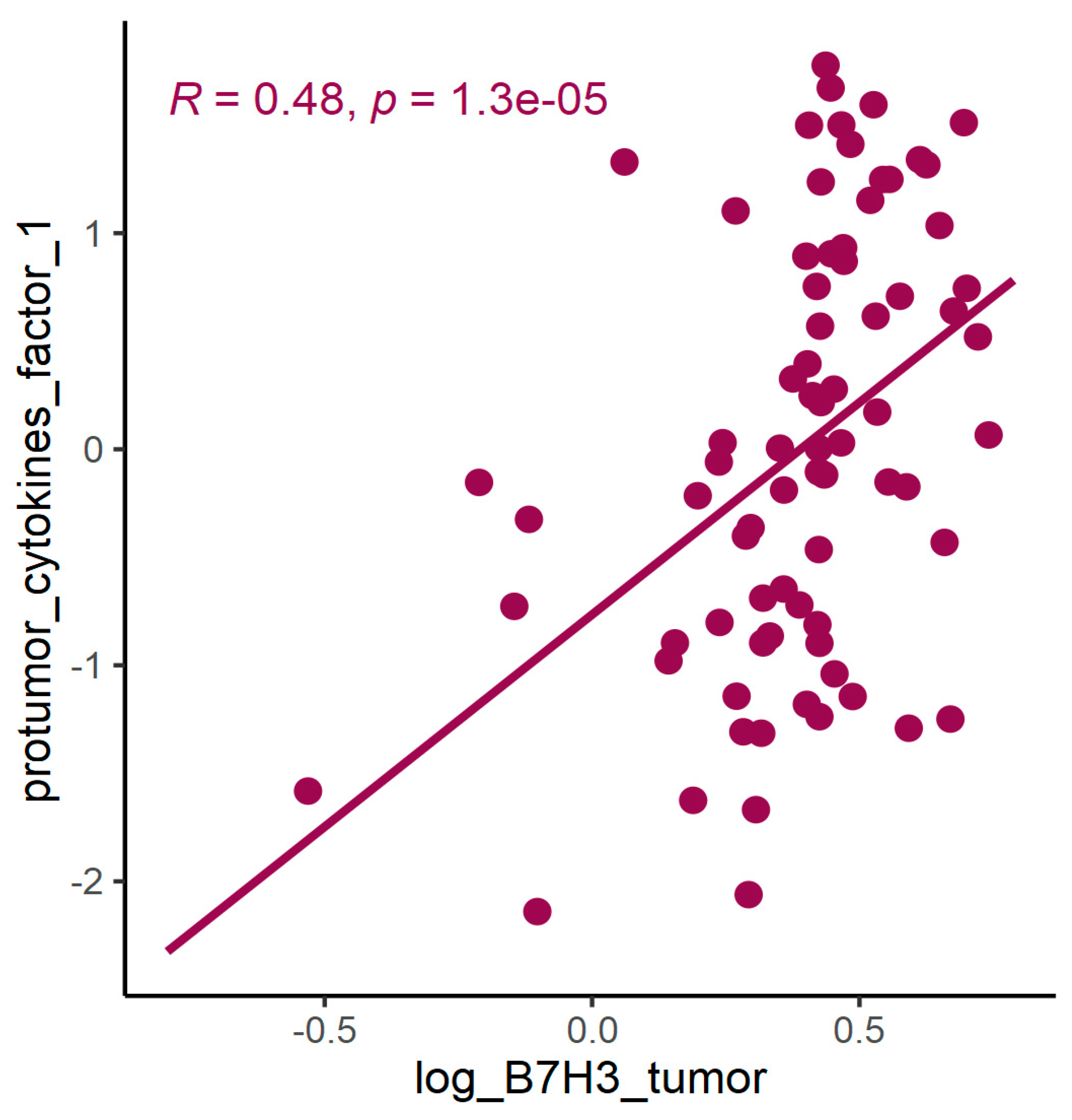
3.2. B7H3 Expression and Cytokines Landscape in CRC Tumours
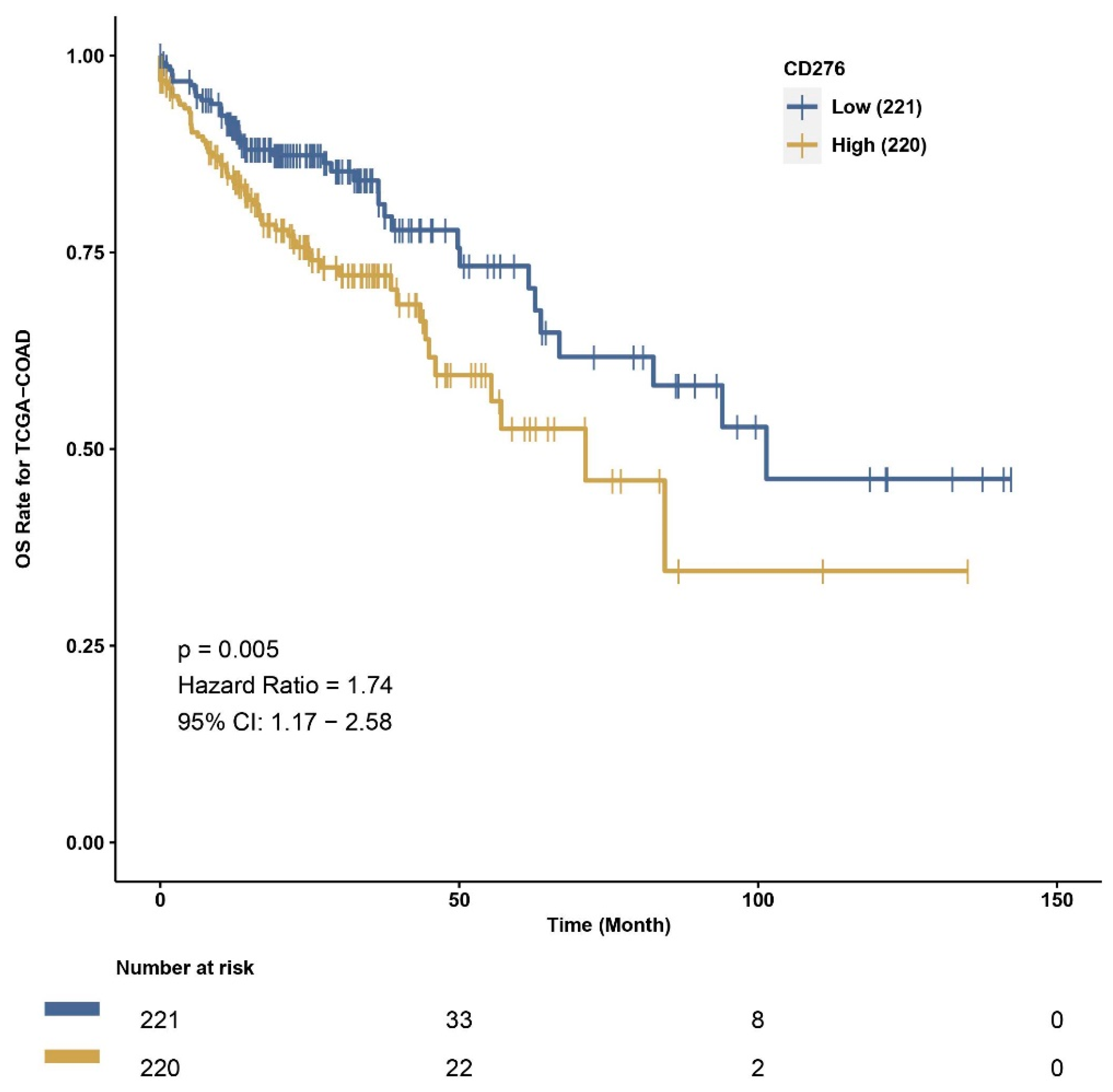
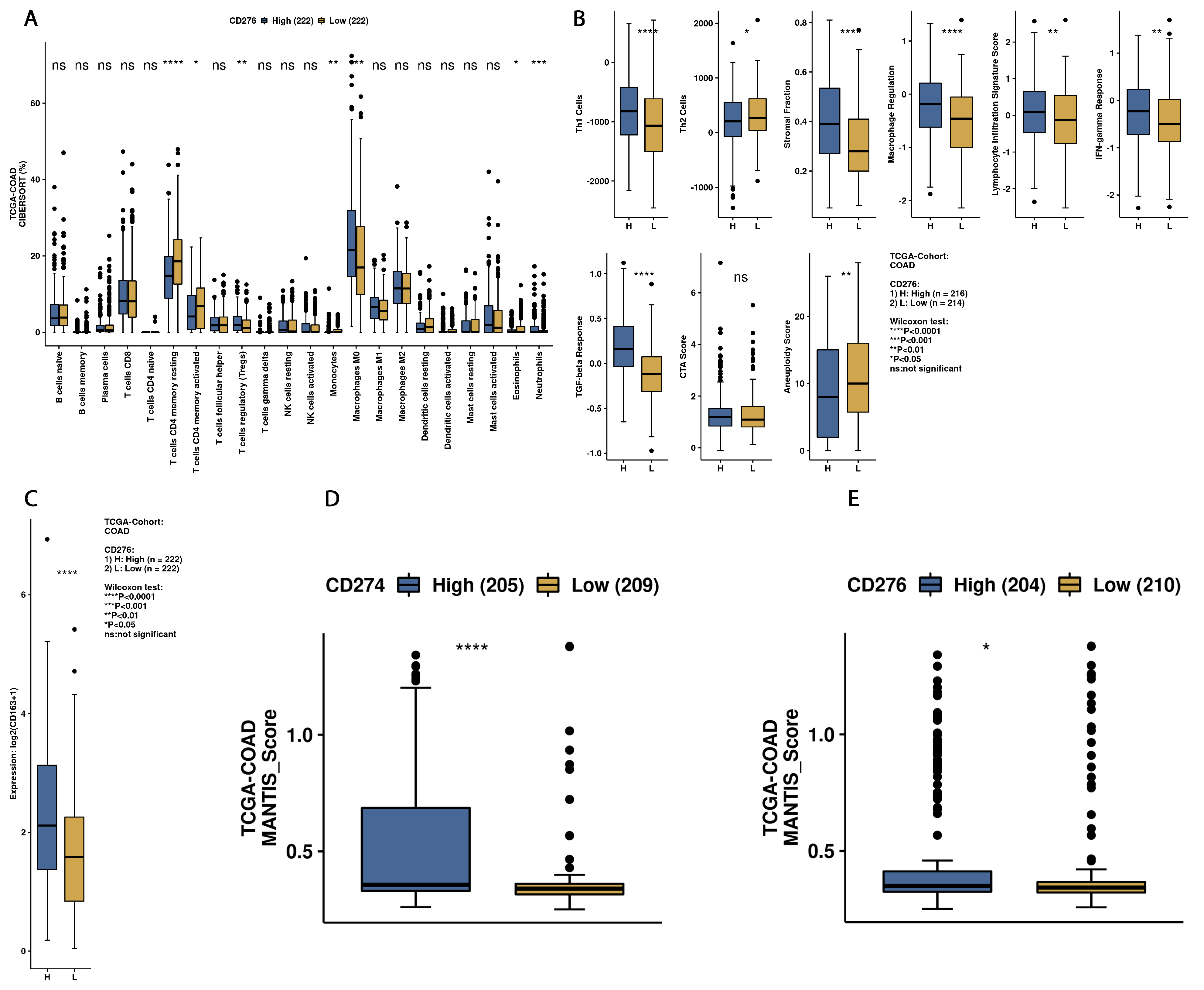
3.3. Exploration of Survival Data and B7H3 Immune Infiltration Landscape Based on TCGA-COAD Data
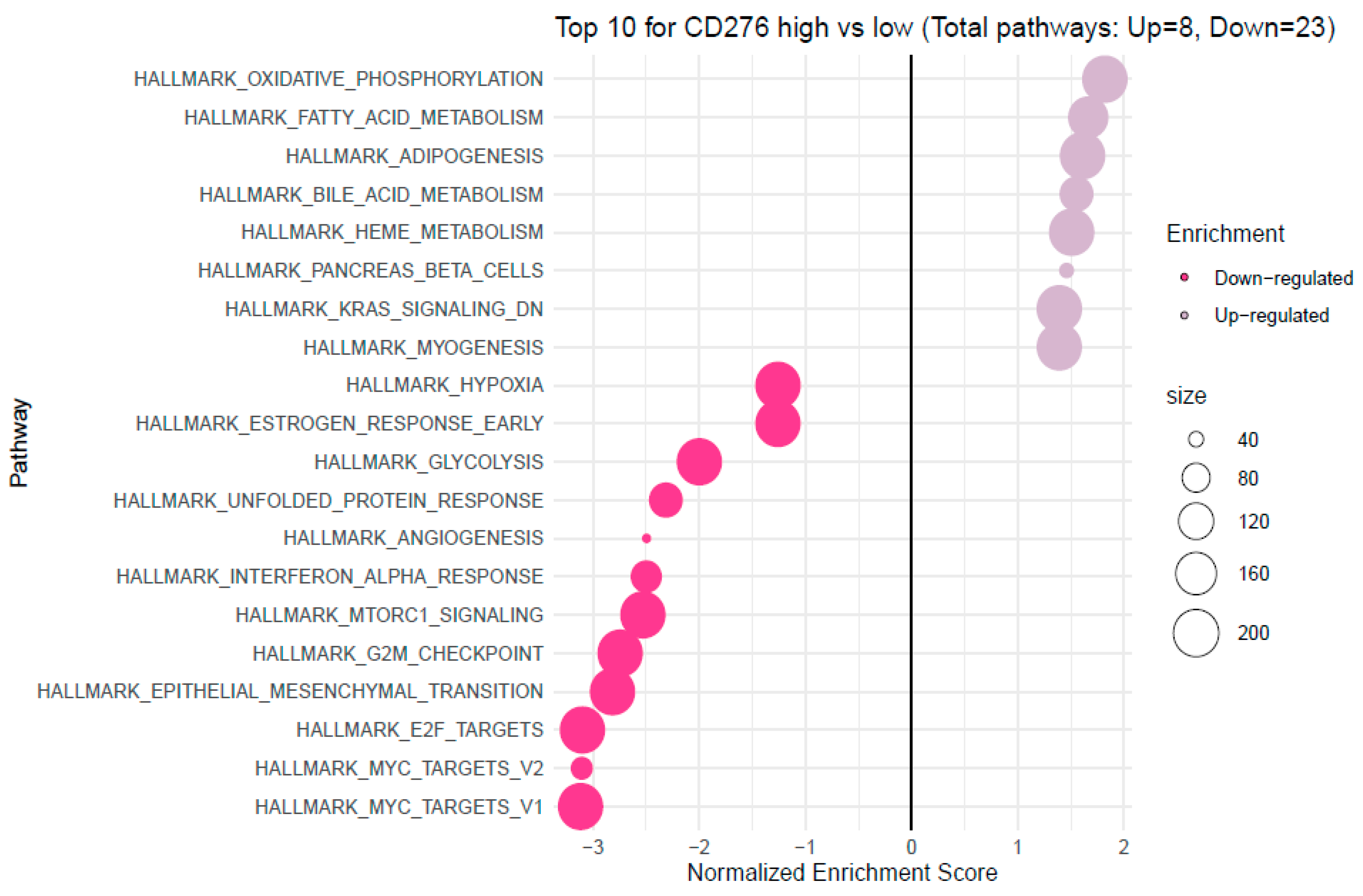
3.4. Analysis of Functional Annotations and B7H3-Induced Signaling Pathways in CRC
4. Discussion
5. Study Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bondhopadhyay, B.; Sisodiya, S.; Chikara, A.; Khan, A.; Tanwar, P.; Afroze, D.; Singh, N.; Agrawal, U.; Mehrotra, R.; Hussain, S. Cancer immunotherapy: A promising dawn in cancer research. Am. J. Blood Res. 2020, 10, 375–385. [Google Scholar] [PubMed]
- Sahin, I.H.; Akce, M.; Alese, O.; Shaib, W.; Lesinski, G.B.; El-Rayes, B.; Wu, C. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br. J. Cancer 2019, 121, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite Instability in Colorectal Cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Lenschow, D.J.; Walunas, T.L.; Bluestone, J.A. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 1996, 14, 233–258. [Google Scholar] [CrossRef]
- Linsley, P.S.; Greene, J.L.; Brady, W.; Bajorath, J.; Ledbetter, J.A.; Peach, R. Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity 1994, 1, 793–801. [Google Scholar] [CrossRef]
- Steinberger, P.; Majdic, O.; Derdak, S.V.; Pfistershammer, K.; Kirchberger, S.; Klauser, C.; Zlabinger, G.; Pickl, W.F.; Stöckl, J.; Knapp, W. Molecular characterization of human 4Ig-B7-H3, a member of the B7 family with four Ig-like domains. J. Immunol. 2004, 172, 2352–2359. [Google Scholar] [CrossRef]
- Chapoval, A.I.; Ni, J.; Lau, J.S.; Wilcox, R.A.; Flies, D.B.; Liu, D.; Dong, H.; Sica, G.L.; Zhu, G.; Tamada, K.; et al. B7-H3: A costimulatory molecule for T cell activation and IFN-γ production. Nat. Immunol. 2001, 2, 269–274. [Google Scholar] [CrossRef]
- Zhou, Y.-H.; Chen, Y.-J.; Ma, Z.-Y.; Xu, L.; Wang, Q.; Zhang, G.-B.; Xie, F.; Ge, Y.; Wang, X.-F.; Zhang, X.-G. 4IgB7-H3 is the major isoform expressed on immunocytes as well as malignant cells. Tissue Antigens 2007, 70, 96–104. [Google Scholar] [CrossRef]
- Sun, M.; Richards, S.; Prasad, D.V.R.; Mai, X.M.; Rudensky, A.; Dong, C. Characterization of Mouse and Human B7-H3 Genes. J. Immunol. 2002, 168, 6294–6297. [Google Scholar] [CrossRef]
- Vigdorovich, V.; Ramagopal, U.A.; Lázár-Molnár, E.; Sylvestre, E.; Lee, J.S.; Hofmeyer, K.A.; Zang, X.; Nathenson, S.G.; Almo, S.C. Structure and T Cell Inhibition Properties of B7 Family Member, B7-H3. Structure 2013, 21, 707–717. [Google Scholar] [CrossRef]
- Kontos, F.; Michelakos, T.; Kurokawa, T.; Sadagopan, A.; Schwab, J.H.; Ferrone, C.R.; Ferrone, S. B7-H3: An Attractive Target for Antibody-based Immunotherapy. Clin. Cancer Res. 2021, 27, 1227–1235. [Google Scholar] [CrossRef]
- Getu, A.A.; Tigabu, A.; Zhou, M.; Lu, J.; Fodstad, Ø.; Tan, M. New frontiers in immune checkpoint B7-H3 (CD276) research and drug development. Mol. Cancer 2023, 22, 43. [Google Scholar] [CrossRef]
- Bin, Z.; Guangbo, Z.; Yan, G.; Huan, Z.; Desheng, L.; Xueguang, Z. Overexpression of B7-H3 in CD133+ colorectal cancer cells is associated with cancer progression and survival in human patients. J. Surg. Res. 2014, 188, 396–403. [Google Scholar] [CrossRef]
- Ingebrigtsen, V.A.; Boye, K.; Tekle, C.; Nesland, J.M.; Flatmark, K.; Fodstad, Ø. B7-H3 expression in colorectal cancer: Nuclear localization strongly predicts poor outcome in colon cancer. Int. J. Cancer 2012, 131, 2528–2536. [Google Scholar] [CrossRef]
- Lu, Z.; Zhao, Z.-X.; Cheng, P.; Huang, F.; Guan, X.; Zhang, M.-G.; Chen, H.-P.; Liu, Z.; Jiang, Z.; Zheng, Z.-X.; et al. B7-H3 immune checkpoint expression is a poor prognostic factor in colorectal carcinoma. Mod. Pathol. 2020, 33, 2330–2340. [Google Scholar] [CrossRef]
- Mielcarska, S.; Kula, A.; Dawidowicz, M.; Kiczmer, P.; Chrabańska, M.; Rynkiewicz, M.; Wziątek-Kuczmik, D.; Świętochowska, E.; Waniczek, D. Assessment of the RANTES Level Correlation and Selected Inflammatory and Pro-Angiogenic Molecules Evaluation of Their Influence on CRC Clinical Features: A Preliminary Observational Study. Medicina 2022, 58, 203. [Google Scholar] [CrossRef]
- Kiczmer, P.; Seńkowska, A.P.; Kula, A.; Dawidowicz, M.; Strzelczyk, J.K.; Zajdel, E.N.; Walkiewicz, K.; Waniczek, D.; Ostrowska, Z.; Świętochowska, E. Assessment of CMKLR1 level in colorectal cancer and its correlation with angiogenic markers. Exp. Mol. Pathol. 2020, 113, 104377. [Google Scholar] [CrossRef]
- Mielcarska, S.; Stopińska, K.; Dawidowicz, M.; Kula, A.; Kiczmer, P.; Seńkowska, A.P.; Zajdel, E.N.; Walkiewicz, K.; Waniczek, D.; Świętochowska, E. GDF-15 Level Correlates with CMKLR1 and VEGF-A in Tumor-free Margin in Colorectal Cancer. Curr. Med. Sci. 2021, 41, 522–528. [Google Scholar] [CrossRef]
- Dawidowicz, M.; Kula, A.; Mielcarska, S.; Kiczmer, P.; Skiba, H.; Krygier, M.; Chrabańska, M.; Piecuch, J.; Szrot, M.; Robotycka, J.; et al. B7H4 Expression Is More Frequent in MSS Status Colorectal Cancer and Is Negatively Associated with Tumour Infiltrating Lymphocytes. Cells 2023, 12, 861. [Google Scholar] [CrossRef]
- Kula, A.; Dawidowicz, M.; Mielcarska, S.; Kiczmer, P.; Chrabańska, M.; Rynkiewicz, M.; Świętochowska, E.; Waniczek, D. Periostin in Angiogenesis and Inflammation in CRC—A Preliminary Observational Study. Medicina 2022, 58, 96. [Google Scholar] [CrossRef] [PubMed]
- Olave, M.C.; Graham, R.P. Mismatch repair deficiency: The what, how and why it is important. Genes Chromosom. Cancer 2022, 61, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Kula, A.; Dawidowicz, M.; Mielcarska, S.; Kiczmer, P.; Skiba, H.; Krygier, M.; Chrabańska, M.; Piecuch, J.; Szrot, M.; Robotycka, J.; et al. Overexpression and Role of HHLA2, a Novel Immune Checkpoint, in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 5876. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Dampier, C.H.; Devall, M.; Jennelle, L.T.; Díez-Obrero, V.; Plummer, S.J.; Moreno, V.; Casey, G. Oncogenic Features in Histologically Normal Mucosa: Novel Insights Into Field Effect from a Mega-Analysis of Colorectal Transcriptomes. Clin. Transl. Gastroenterol. 2020, 11, e00210. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Liu, S.; Liang, J.; Liu, Z.; Zhang, C.; Wang, Y.; Watson, A.H.; Zhou, C.; Zhang, F.; Wu, K.; Zhang, F.; et al. The Role of CD276 in Cancers. Front. Oncol. 2021, 11, 654684. [Google Scholar] [CrossRef]
- Hashiguchi, M.; Inamochi, Y.; Nagai, S.; Otsuki, N.; Piao, J.; Kobori, H.; Kanno, Y.; Kojima, H.; Kobata, T.; Azuma, M. Human B7-H3 binds to Triggering receptor expressed on myeloid cells-like transcript 2 (TLT-2) and enhances T cell responses. Open J. Immunol. 2012, 2, 9–16. [Google Scholar] [CrossRef]
- Hashiguchi, M.; Kobori, H.; Ritprajak, P.; Kamimura, Y.; Kozono, H.; Azuma, M. Triggering receptor expressed on myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for B7-H3 and enhances T cell responses. Proc. Natl. Acad. Sci. USA 2008, 105, 10495–10500. [Google Scholar] [CrossRef]
- Husain, B.; Ramani, S.R.; Chiang, E.; Lehoux, I.; Paduchuri, S.; Arena, T.A.; Patel, A.; Wilson, B.; Chan, P.; Franke, Y.; et al. A Platform for Extracellular Interactome Discovery Identifies Novel Functional Binding Partners for the Immune Receptors B7-H3/CD276 and PVR/CD155. Mol. Cell. Proteom. 2019, 18, 2310–2323. [Google Scholar] [CrossRef]
- Zhou, W.-T.; Jin, W.-L. B7-H3/CD276: An Emerging Cancer Immunotherapy. Front. Immunol. 2021, 12, 701006. [Google Scholar] [CrossRef]
- Ingebrigtsen, V.A.; Boye, K.; Nesland, J.M.; Nesbakken, A.; Flatmark, K.; Fodstad, Ø. B7-H3 expression in colorectal cancer: Associations with clinicopathological parameters and patient outcome. BMC Cancer 2014, 14, 602. [Google Scholar] [CrossRef]
- Fan, H.; Zhu, J.-H.; Yao, X.-Q. Prognostic significance of B7-H3 expression in patients with colorectal cancer: A meta-analysis. Pak. J. Med. Sci. 2016, 32, 1568–1573. [Google Scholar] [CrossRef]
- Zhang, T. Overexpression of B7-H3 augments anti-apoptosis of colorectal cancer cells by Jak2-STAT3. World J. Gastroenterol. 2015, 21, 1804. [Google Scholar] [CrossRef]
- Bai, Z.; Zhou, Y.; Ye, Z.; Xiong, J.; Lan, H.; Wang, F. Tumor-Infiltrating Lymphocytes in Colorectal Cancer: The Fundamental Indication and Application on Immunotherapy. Front. Immunol. 2022, 12, 808964. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Zhao, J.; Gu, M.; Giscombe, R.; Lefvert, A.K.; Wang, X. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer 2006, 53, 143–151. [Google Scholar] [CrossRef]
- Li, F.; Guo, H.; Wang, Y.; Liu, B.; Zhou, H. Profiles of tumor-infiltrating immune cells and prognostic genes associated with the microenvironment of bladder cancer. Int. Immunopharmacol. 2020, 85, 106641. [Google Scholar] [CrossRef]
- Wang, C.; Feng, H.; Cheng, X.; Liu, K.; Cai, D.; Zhao, R. Potential Therapeutic Targets of B7 Family in Colorectal Cancer. Front. Immunol. 2020, 11, 681. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Q.; Chen, W.; Shan, B.; Ding, Y.; Zhang, G.; Cao, N.; Liu, L.; Zhang, Y. B7-H3 is Overexpressed in Patients Suffering Osteosarcoma and Associated with Tumor Aggressiveness and Metastasis. PLoS ONE 2013, 8, e70689. [Google Scholar] [CrossRef]
- Brunner, A.; Hinterholzer, S.; Riss, P.; Heinze, G.; Brustmann, H. Immunoexpression of B7-H3 in endometrial cancer: Relation to tumor T-cell infiltration and prognosis. Gynecol. Oncol. 2012, 124, 105–111. [Google Scholar] [CrossRef]
- Mao, Y.; Chen, L.; Wang, F.; Zhu, D.; Ge, X.; Hua, D.; Sun, J. Cancer cell-expressed B7-H3 regulates the differentiation of tumor-associated macrophages in human colorectal carcinoma. Oncol. Lett. 2017, 14, 6177–6183. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, X.-H.; Jin, L. Macrophage Polarization in Physiological and Pathological Pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A. Tumour-associated macrophages as a prototypic type II polarised phagocyte population: Role in tumour progression. Eur. J. Cancer 2004, 40, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Kang, F.-B.; Wang, L.; Li, D.; Zhang, Y.-G.; Sun, D.-X. Hepatocellular carcinomas promote tumor-associated macrophage M2-polarization via increased B7-H3 expression. Oncol. Rep. 2015, 33, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Murakami, R.; Hamanishi, J.; Tanigaki, K.; Hosoe, Y.; Mise, N.; Takamatsu, S.; Mise, Y.; Ukita, M.; Taki, M.; et al. B7-H3 Suppresses Antitumor Immunity via the CCL2–CCR2–M2 Macrophage Axis and Contributes to Ovarian Cancer Progression. Cancer Immunol. Res. 2022, 10, 56–69. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, P.; Li, W.-J.; Zhang, J.; Wang, G.-P.; Jiang, D.-F.; Chen, F.-P. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol. Immunol. 2020, 117, 20–28. [Google Scholar] [CrossRef]
- Mager, L.F.; Wasmer, M.-H.; Rau, T.T.; Krebs, P. Cytokine-Induced Modulation of Colorectal Cancer. Front. Oncol. 2016, 6, 96. [Google Scholar] [CrossRef]
- Czajka-Francuz, P.; Cisoń-Jurek, S.; Czajka, A.; Kozaczka, M.; Wojnar, J.; Chudek, J.; Francuz, T. Systemic Interleukins’ Profile in Early and Advanced Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 124. [Google Scholar] [CrossRef]
- Razi, S.; Baradaran Noveiry, B.; Keshavarz-Fathi, M.; Rezaei, N. IL-17 and colorectal cancer: From carcinogenesis to treatment. Cytokine 2019, 116, 7–12. [Google Scholar] [CrossRef]
- Buhrmann, C.; Kunnumakkara, A.B.; Kumar, A.; Samec, M.; Kubatka, P.; Aggarwal, B.B.; Shakibaei, M. Multitargeting Effects of Calebin A on Malignancy of CRC Cells in Multicellular Tumor Microenvironment. Front. Oncol. 2021, 11, 650603. [Google Scholar] [CrossRef]
- Nalleweg, N.; Chiriac, M.T.; Podstawa, E.; Lehmann, C.; Rau, T.T.; Atreya, R.; Krauss, E.; Hundorfean, G.; Fichtner-Feigl, S.; Hartmann, A.; et al. IL-9 and its receptor are predominantly involved in the pathogenesis of UC. Gut 2015, 64, 743–755. [Google Scholar] [CrossRef]
- Wang, J.; Sun, M.; Zhao, H.; Huang, Y.; Li, D.; Mao, D.; Zhang, Z.; Zhu, X.; Dong, X.; Zhao, X. IL-9 Exerts Antitumor Effects in Colon Cancer and Transforms the Tumor Microenvironment In Vivo. Technol. Cancer Res. Treat. 2019, 18, 153303381985773. [Google Scholar] [CrossRef]
- Picarda, E.; Galbo, P.M.; Zong, H.; Rajan, M.R.; Wallenius, V.; Zheng, D.; Börgeson, E.; Singh, R.; Pessin, J.; Zang, X. The immune checkpoint B7-H3 (CD276) regulates adipocyte progenitor metabolism and obesity development. Sci. Adv. 2022, 8, eabm7012. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Marzese, D.M.; Wang, X.; Yang, Z.; Li, C.; Zhang, H.; Zhang, J.; Chen, C.C.; Kelly, D.F.; et al. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab. Investig. 2019, 99, 1117–1129. [Google Scholar] [CrossRef]


| Female | Male | All Cases | |
|---|---|---|---|
| 72 (45.57%) | 86 (54.43%) | 158 | |
| Age | 66.39 ± 9.32 | 63.83 ± 9.53 | 65.03 ± 9.49 |
| tumour localization | |||
| left-sided | 48 (69.57%) | 58 (71.60%) | 106 (70.67%) |
| right-sided | 21 (30.43%) | 23 (28.40%) | 44 (29.33%) |
| T parameter | |||
| T1 | 1 (1.43%) | 6 (7.41%) | 7 (4.64%) |
| T2 | 15 (21.43%) | 12 (14.81%) | 27 (17.88%) |
| T3 | 44 (62.86%) | 50 (61.73%) | 94 (62.25%) |
| T4 | 10 (14.29%) | 13 (16.05%) | 23 (15.23%) |
| N parameter | |||
| N0 | 29 (41.43%) | 34 (41.98%) | 63 (41.72%) |
| N1 | 28 (40.00%) | 33 (40.74%) | 61 (40.40%) |
| N2 | 13 (18.57%) | 14 (17.28%) | 27 (17.88%) |
| M parameter | |||
| M0 | 61 (87.14%) | 64 (79.01%) | 125 (82.78%) |
| M1 | 9 (12.86%) | 17 (20.99%) | 26 (17.22%) |
| TNM stage | |||
| I | 12 (17.14%) | 12 (14.81%) | 24 (15.89%) |
| II | 17 (24.29%) | 19 (23.46%) | 36 (23.84%) |
| III | 32 (45.71%) | 34 (41.98%) | 66 (43.71%) |
| IV | 9 (12.86%) | 16 (19.75%) | 25 (16.56%) |
| Grading | |||
| low | 60 (85.71%) | 68 (83.95%) | 128 (84.77%) |
| high | 10 (14.29%) | 13 (16.05%) | 23 (15.23%) |
| Adjuvant treatment | |||
| yes | 7 (9.86%) | 13 (15.48%) | 20 (12.66%) |
| no | 64 (90.14%) | 71 (84.52%) | 138 (87.34%) |
| Characteristics | B7H3 Tumour Expression | ||
|---|---|---|---|
| Positive | Negative | p | |
| All | 23 (29.87%) | 54 (70.13%) | - |
| Tumour localization | 0.932 | ||
| left-sided | 17 (28.81%) | 42 (71.19%) | |
| right-sided | 5 (27.78%) | 13 (72.22%) | |
| T parameter | 0.327 | ||
| T1 | 1 (25.00%) | 3 (75.00%) | |
| T2 | 5 (50.00%) | 5 (50.00%) | |
| T3 | 12 (23.08%) | 40 (76.92%) | |
| T4 | 4 (40.00%) | 6 (60.00%) | |
| N parameter | 0.587 | ||
| N0 | 11 (30.56%) | 25 (69.44%) | |
| N1 | 9 (32.14%) | 19 (67.86%) | |
| N2 | 2 (16.67%) | 10 (83.33%) | |
| M parameter | 0.156 | ||
| M0 | 21 (31.82%) | 45 (68.18%) | |
| M1 | 1 (10.00%) | 9 (90.00%) | |
| TNM stage | 0.313 | ||
| I | 5 (41.67%) | 7 (58.33%) | |
| II | 5 (22.73%) | 17 (77.27%) | |
| III | 11 (34.38%) | 21 (65.63%) | |
| IV | 1 (10.00%) | 9 (90.00%) | |
| Grading | 0.115 | ||
| low | 17 (25.76%) | 49 (74.24%) | |
| high | 5 (50.00%) | 5 (50.00%) | |
| MSS/MSI status | 0.52 | ||
| MSS tumours | 20 (29.85%) | 47 (70.15%) | |
| MSI tumours | 2 (20.00%) | 8 (80.00%) | |
| TILs | 0.86 | ||
| 0–5% | 11 (30.56%) | 25 (69.44%) | |
| 6–25% | 6 (28.57%) | 15 (71.43%) | |
| 26–50% | 5 (31.25% | 11 (68.75%) | |
| 51–75% | 0 (0.00%) | 3 (100.00%) | |
| 76–100% | 0 (0.00%) | 0 (0.00%) | |
| CD8 Lymphocytes | 0.128 | ||
| 0–5% | 5 (14.71%) | 29 (85.29%) | |
| 6–25% | 10 (47.62%) | 11 (52.38%) | |
| 26–50% | 4 (33.33%) | 8 (66.67%) | |
| 51–75% | 2 (33.33%) | 4 (66.67%) | |
| 76–100% | 1 (25.00%) | 3 (75.00%) | |
| Budding | 0.637 | ||
| 0–4 | 13 (32.5%) | 27 (67.5%) | |
| 5–9 | 5 (21.74%) | 18 (78.26%) | |
| >9 | 3 (25.00%) | 9 (75.00%) | |
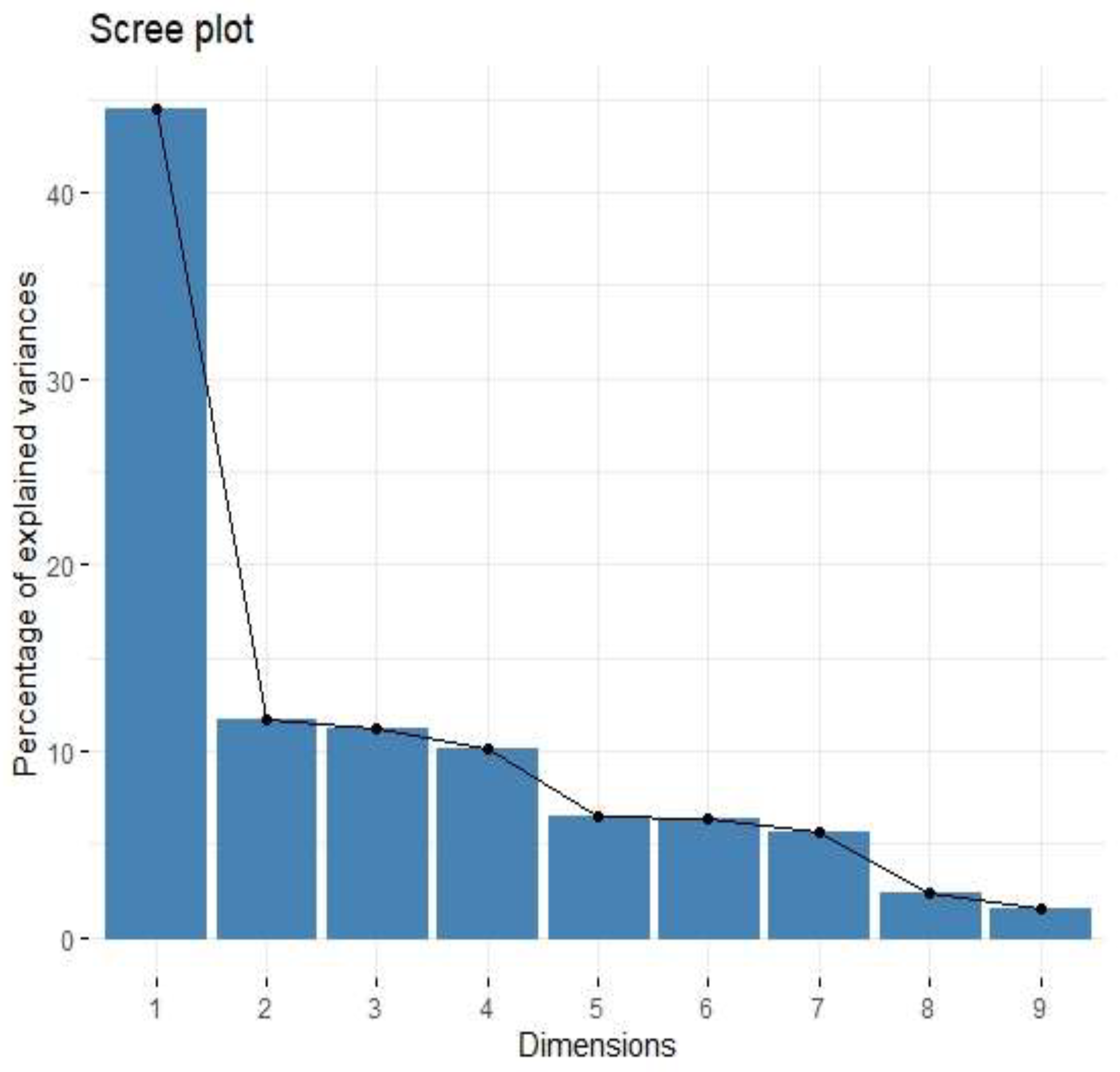
| Factor | Eigenvalue | Variance (%) | Cumulative Variance (%) |
|---|---|---|---|
| Factor 1 | 4.01 | 44.52 | 44.52 |
| Factor 2 | 1.05 | 11.68 | 56.20 |
| Factor 3 | 1.01 | 11.23 | 67.43 |
| Variable | Factor 1 | Factor 2 | Factor 3 |
|---|---|---|---|
| IL-1a | 0.64 | 0.28 | 0.11 |
| IL-1b | 0.49 | 0.05 | 0.71 |
| IL-1ra | 0.02 | 0.23 | 0.93 |
| IL-6 | 0.47 | 0.41 | 0.27 |
| IL-8 | 0.64 | 0.13 | 0.33 |
| IL-10 | 0.06 | 0.93 | 0.14 |
| TNF-alpha | 0.44 | 0.74 | 0.15 |
| VEGF-A | 0.85 | 0.07 | −0.02 |
| Basic FGF | 0.06 | 0.15 | 0.04 |
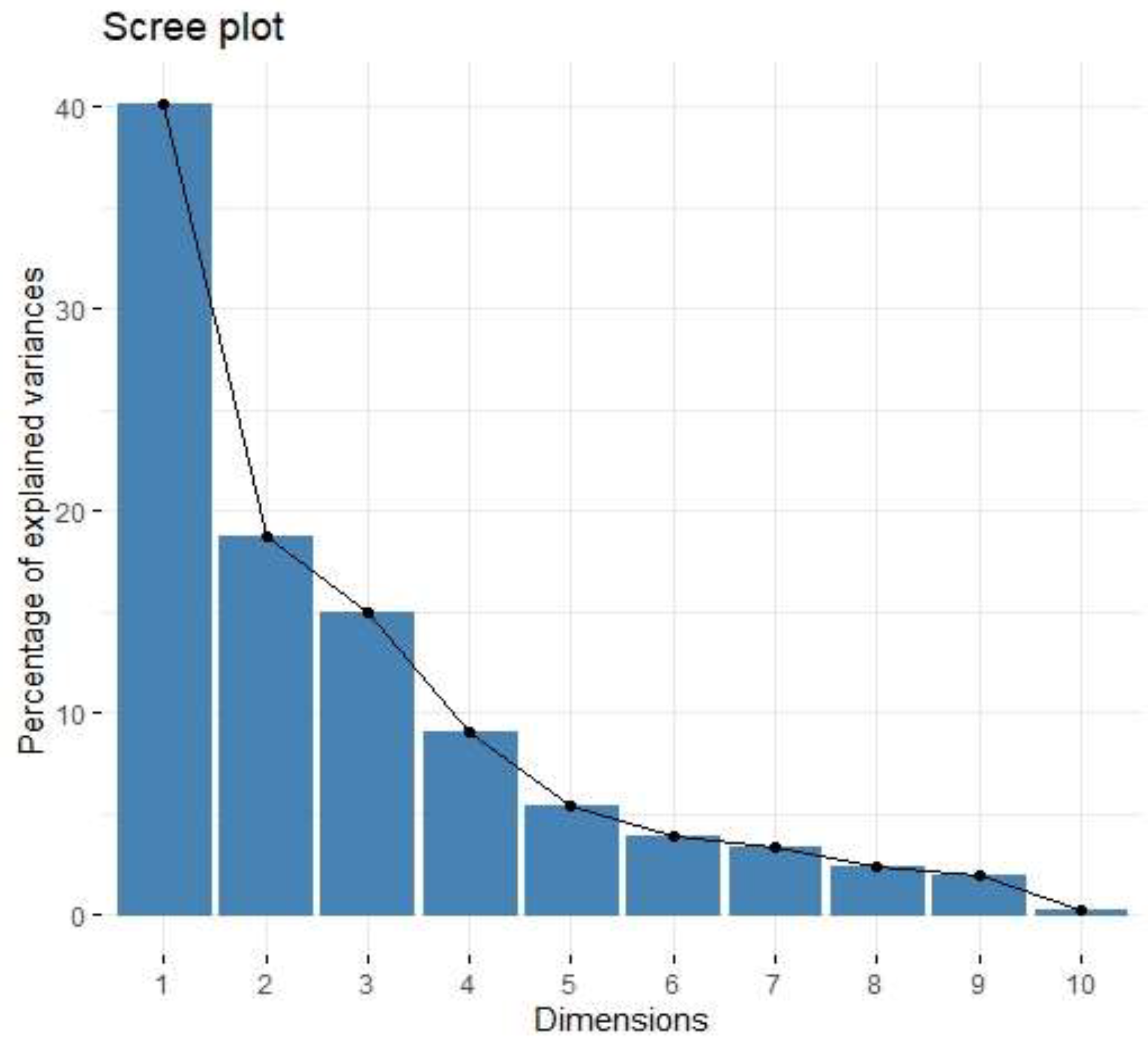
| Factor | Eigenvalue | Variance (%) | Cumulative Variance (%) |
|---|---|---|---|
| Factor 1 | 4.01 | 40.13 | 40.13 |
| Factor 2 | 1.87 | 18.72 | 58.85 |
| Factor 3 | 1.49 | 14.93 | 73.78 |
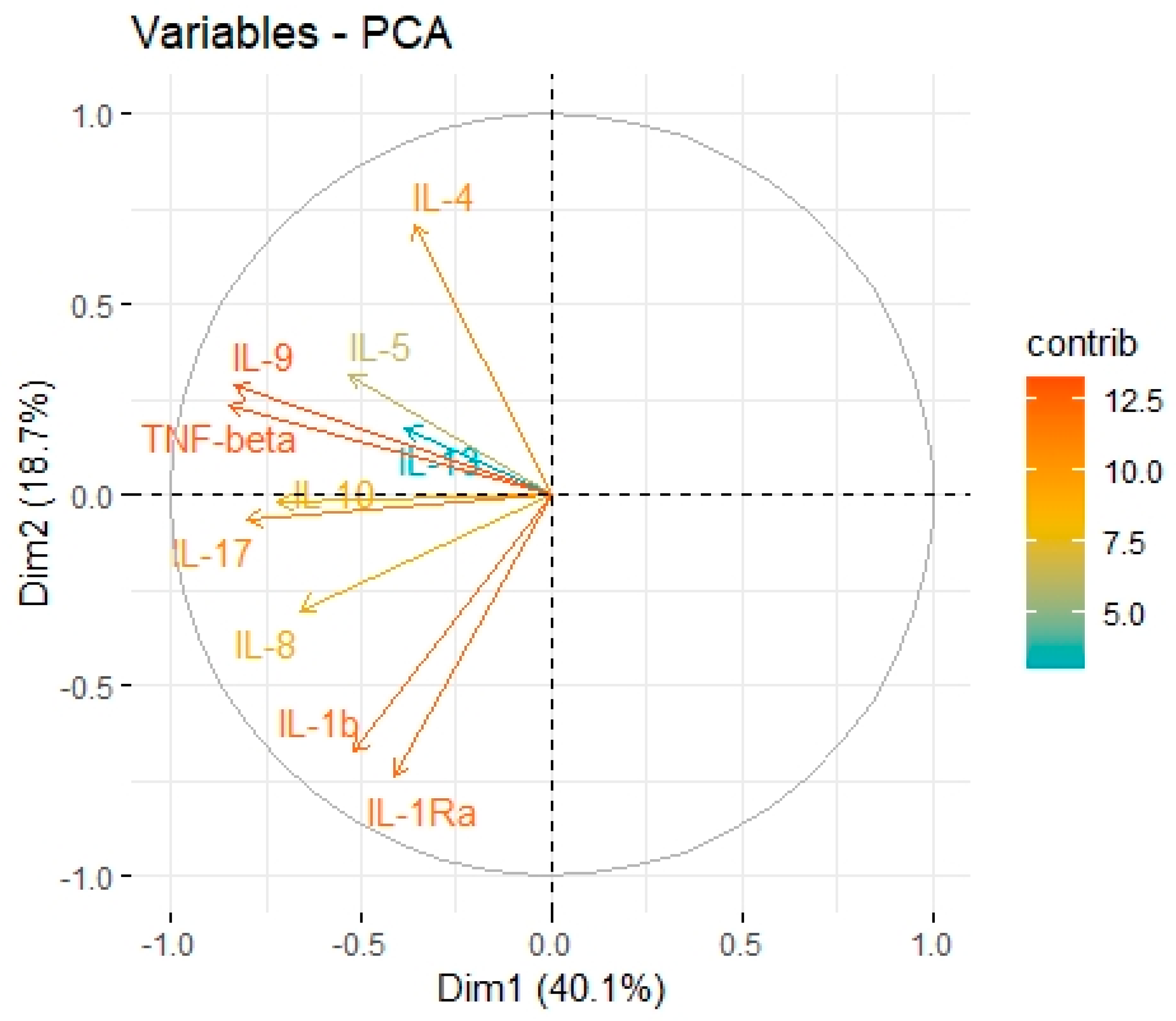
| Variable | Factor 1 | Factor 2 | Factor 3 |
|---|---|---|---|
| IL-1b | 0.05 | 0.92 | −0.01 |
| IL-1Ra | −0.03 | 0.70 | 0.41 |
| IL-4 | 0.46 | −0.23 | −0.18 |
| IL-5 | 0.05 | 0.17 | 0.18 |
| IL-8 | 0.45 | 0.70 | −0.14 |
| IL-9 | 0.90 | 0.11 | 0.32 |
| IL-10 | 0.18 | 0.27 | 0.78 |
| IL-13 | 0.34 | −0.23 | 0.78 |
| IL-17 | 0.38 | 0.46 | 0.44 |
| TNF-Beta | 0.91 | 0.18 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mielcarska, S.; Dawidowicz, M.; Kula, A.; Kiczmer, P.; Skiba, H.; Krygier, M.; Chrabańska, M.; Piecuch, J.; Szrot, M.; Ochman, B.; et al. B7H3 Role in Reshaping Immunosuppressive Landscape in MSI and MSS Colorectal Cancer Tumours. Cancers 2023, 15, 3136. https://doi.org/10.3390/cancers15123136
Mielcarska S, Dawidowicz M, Kula A, Kiczmer P, Skiba H, Krygier M, Chrabańska M, Piecuch J, Szrot M, Ochman B, et al. B7H3 Role in Reshaping Immunosuppressive Landscape in MSI and MSS Colorectal Cancer Tumours. Cancers. 2023; 15(12):3136. https://doi.org/10.3390/cancers15123136
Chicago/Turabian StyleMielcarska, Sylwia, Miriam Dawidowicz, Agnieszka Kula, Paweł Kiczmer, Hanna Skiba, Małgorzata Krygier, Magdalena Chrabańska, Jerzy Piecuch, Monika Szrot, Błażej Ochman, and et al. 2023. "B7H3 Role in Reshaping Immunosuppressive Landscape in MSI and MSS Colorectal Cancer Tumours" Cancers 15, no. 12: 3136. https://doi.org/10.3390/cancers15123136
APA StyleMielcarska, S., Dawidowicz, M., Kula, A., Kiczmer, P., Skiba, H., Krygier, M., Chrabańska, M., Piecuch, J., Szrot, M., Ochman, B., Robotycka, J., Strzałkowska, B., Czuba, Z., Waniczek, D., & Świętochowska, E. (2023). B7H3 Role in Reshaping Immunosuppressive Landscape in MSI and MSS Colorectal Cancer Tumours. Cancers, 15(12), 3136. https://doi.org/10.3390/cancers15123136






