K-Ras Binds Calmodulin-Related Centrin1 with Potential Implications for K-Ras Driven Cancer Cell Stemness
Abstract
Simple Summary
Abstract
1. Introduction
2. Experimental Procedures
2.1. Plasmids, siRNAs and Inhibitors
2.2. Protein Sequence Analyses
2.3. Protein Purification
2.4. Fluorescence Polarisation Binding Assay
2.5. Co-Immunoprecipitation Experiments
2.6. BRET Donor Saturation Titration Assays
2.7. Dose Response Analysis of Inhibitors and siRNA Knockdown in BRET Assays
2.8. siRNA-Mediated Knockdown and Western Blotting
2.9. Three-Dimensional Spheroid Assay
2.10. Confocal Microscopy
2.11. Data and Statistical Analysis
3. Results and Discussion
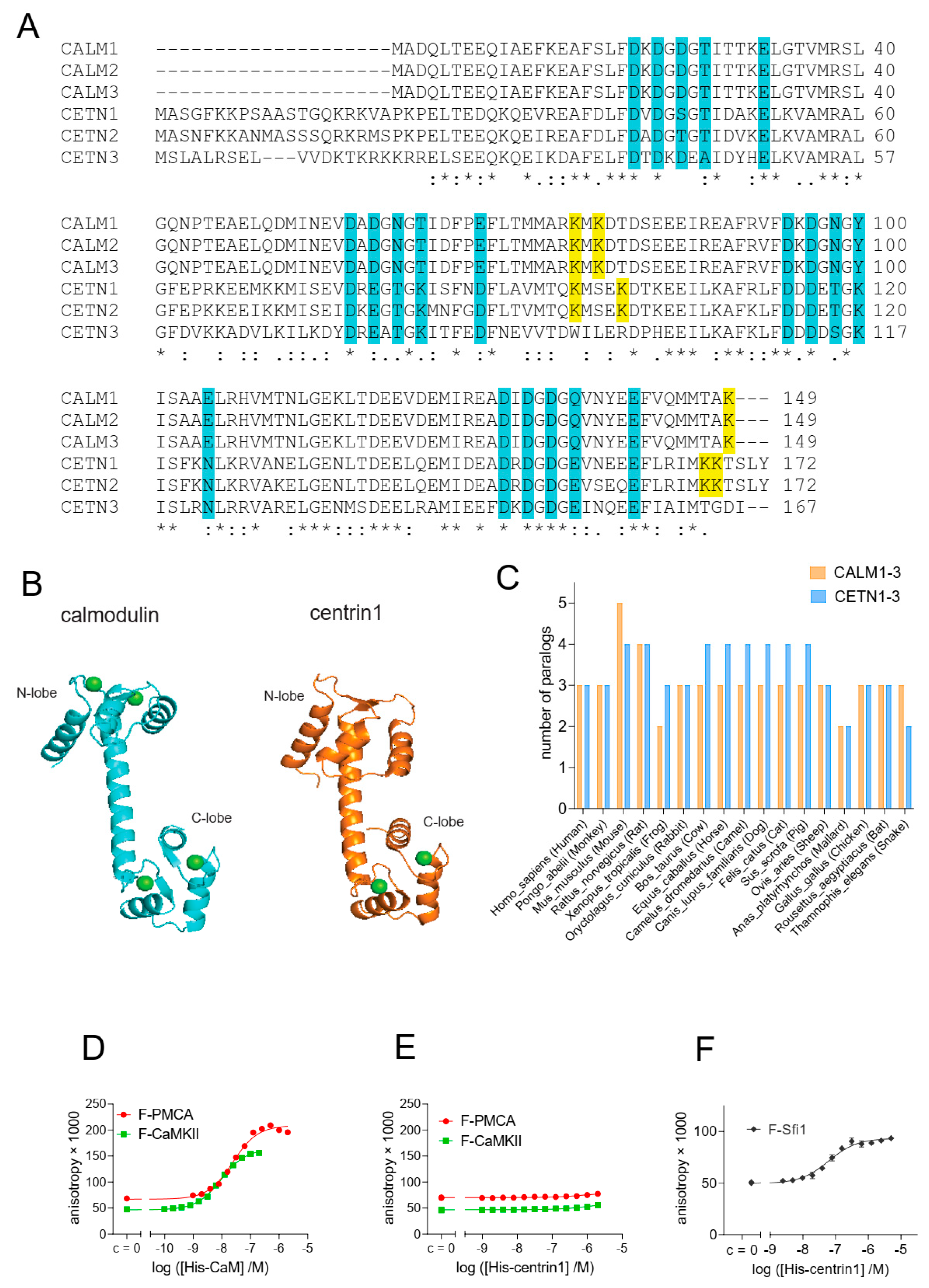
3.1. Binding Studies Support Specific Canonical Target Peptides for CaM or Centrin1
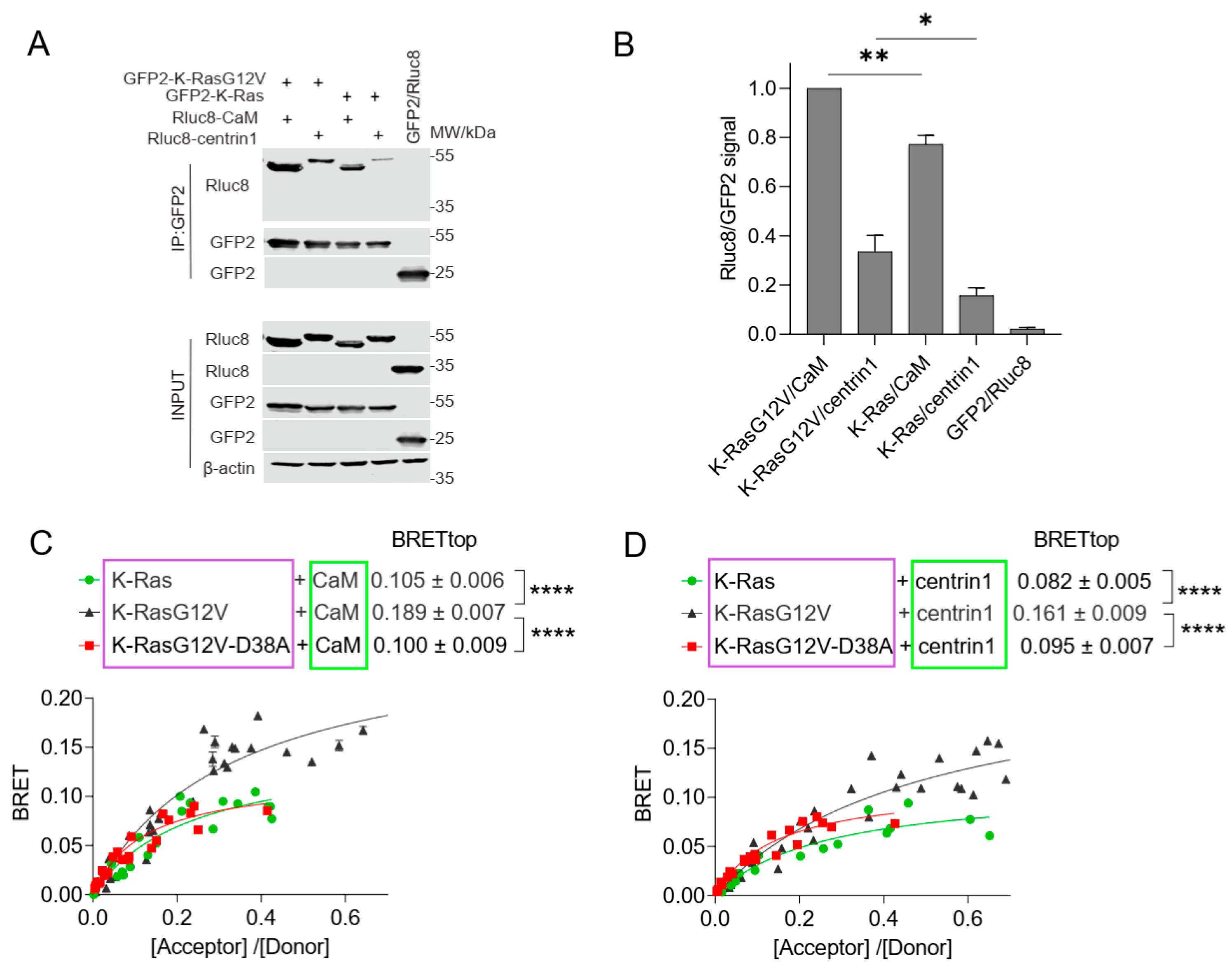
3.2. Cellular BRET Data Suggest That the K-Ras G-Domain Participates in Complexes with Either CaM or Centrin1
3.3. CaM Inhibitors Bind to Centrin
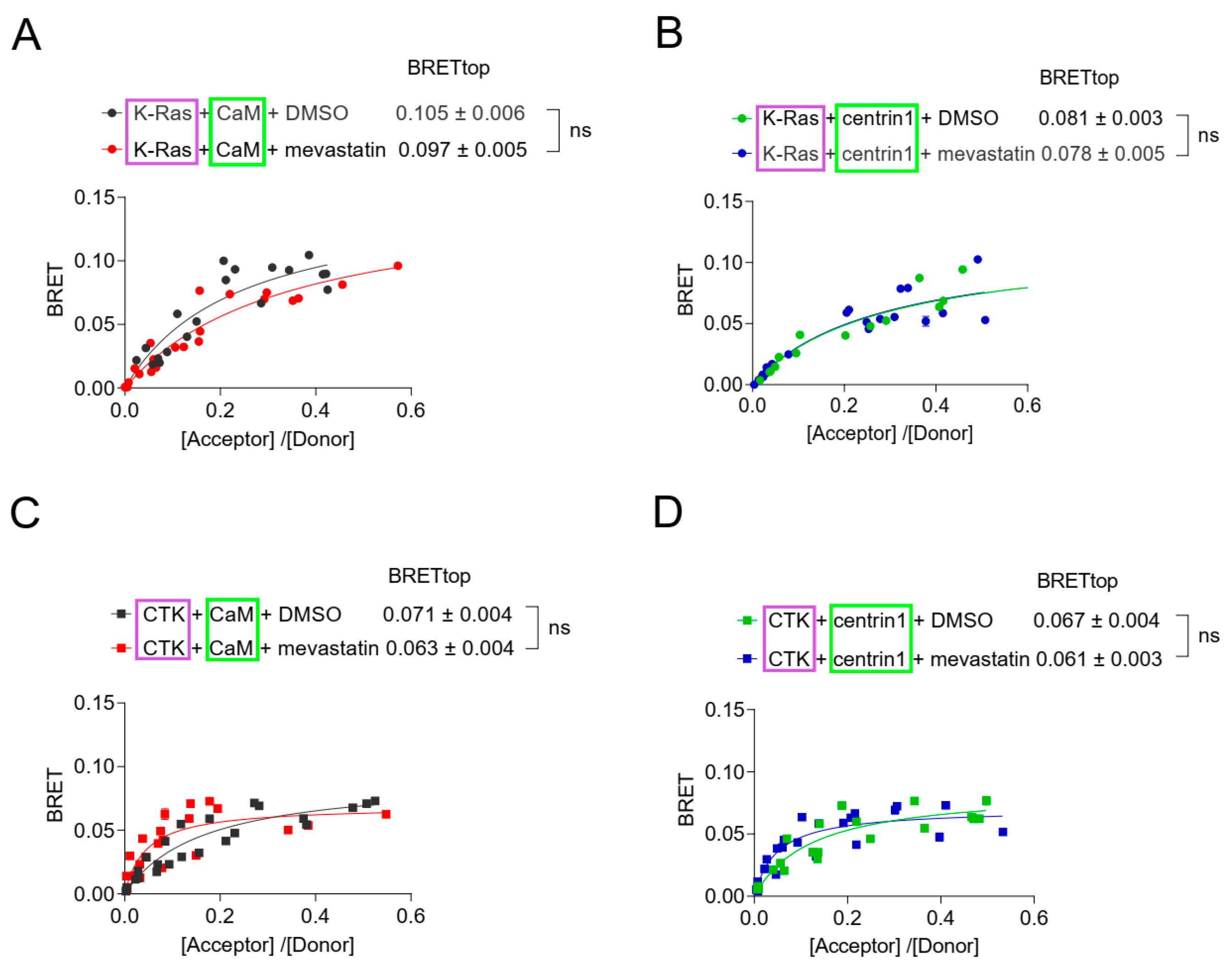
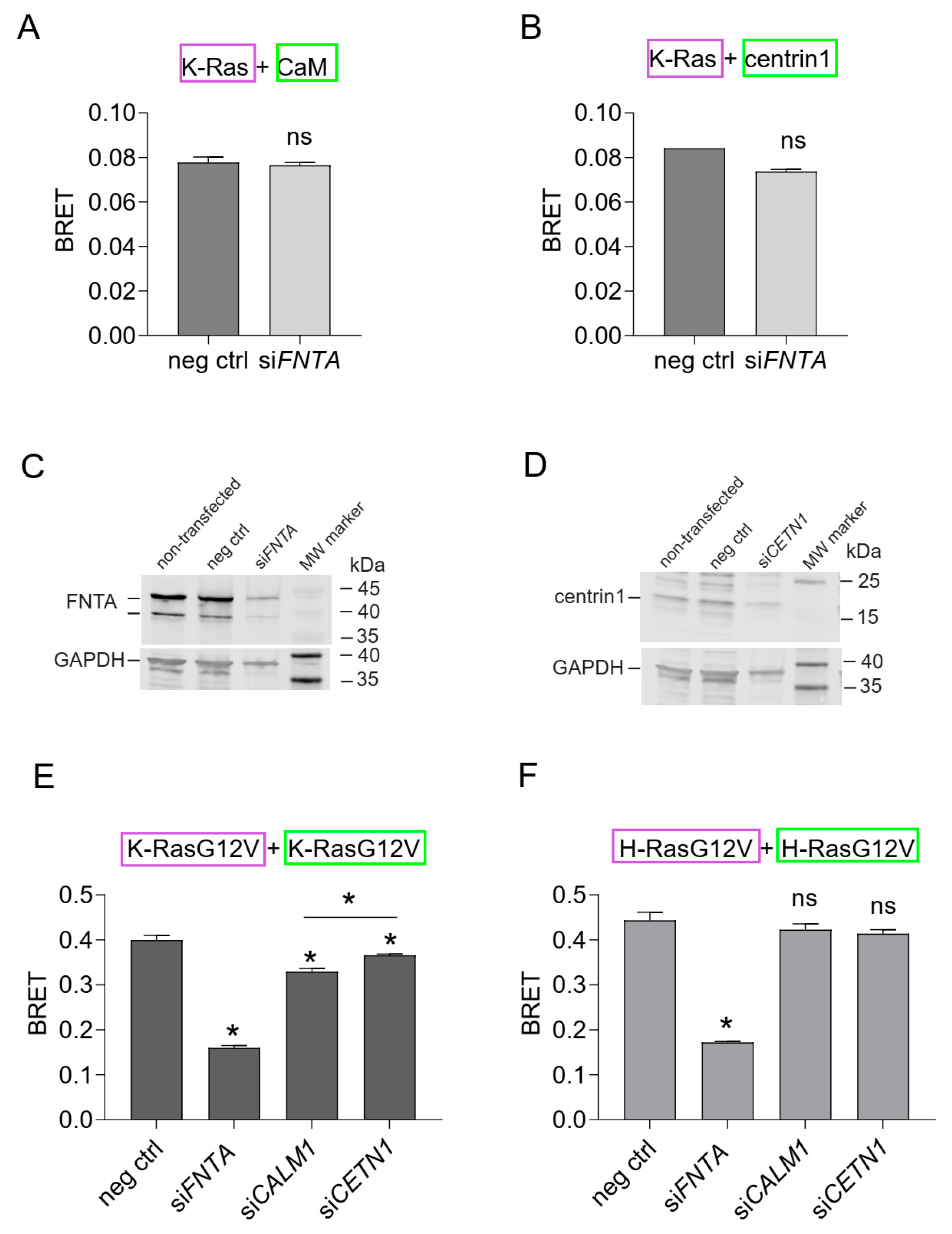
3.4. Inhibition of Prenylation Does Not Disrupt the BRET-Interaction of K-Ras with CaM or Centrin1 in Cells
3.5. Membrane Targeting and Anchorage of K-Ras Depends More on CaM Than on Centrin1
3.6. Centrin1 Co-Distributes with CaM during the Cell Cycle
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Castel, P.; Rauen, K.A.; McCormick, F. The duality of human oncoproteins: Drivers of cancer and congenital disorders. Nat. Rev. Cancer 2020, 20, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Abankwa, D.; Gorfe, A.A. Mechanisms of Ras Membrane Organization and Signaling: Ras Rocks Again. Biomolecules 2020, 10, 1522. [Google Scholar] [CrossRef] [PubMed]
- Schmick, M.; Kraemer, A.; Bastiaens, P.I. Ras moves to stay in place. Trends Cell Biol. 2015, 25, 190–197. [Google Scholar] [CrossRef]
- Newlaczyl, A.U.; Coulson, J.M.; Prior, I.A. Quantification of spatiotemporal patterns of Ras isoform expression during development. Sci. Rep. 2017, 7, 41297. [Google Scholar] [CrossRef]
- Wang, M.T.; Holderfield, M.; Galeas, J.; Delrosario, R.; To, M.D.; Balmain, A.; McCormick, F. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell 2015, 163, 1237–1251. [Google Scholar] [CrossRef]
- Quinlan, M.P.; Quatela, S.E.; Philips, M.R.; Settleman, J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol. Cell. Biol. 2008, 28, 2659–2674. [Google Scholar] [CrossRef]
- Najumudeen, A.K.; Jaiswal, A.; Lectez, B.; Oetken-Lindholm, C.; Guzman, C.; Siljamaki, E.; Posada, I.M.; Lacey, E.; Aittokallio, T.; Abankwa, D. Cancer stem cell drugs target K-ras signaling in a stemness context. Oncogene 2016, 35, 5248–5262. [Google Scholar] [CrossRef]
- Okutachi, S.; Manoharan, G.B.; Kiriazis, A.; Laurini, C.; Catillon, M.; McCormick, F.; Yli-Kauhaluoma, J.; Abankwa, D. A Covalent Calmodulin Inhibitor as a Tool to Study Cellular Mechanisms of K-Ras-Driven Stemness. Front. Cell Dev. Biol. 2021, 9, 665673. [Google Scholar] [CrossRef]
- Alvarez-Moya, B.; Lopez-Alcala, C.; Drosten, M.; Bachs, O.; Agell, N. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene 2010, 29, 5911–5922. [Google Scholar] [CrossRef]
- Dharmaiah, S.; Bindu, L.; Tran, T.H.; Gillette, W.K.; Frank, P.H.; Ghirlando, R.; Nissley, D.V.; Esposito, D.; McCormick, F.; Stephen, A.G.; et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEdelta. Proc. Natl. Acad. Sci. USA 2016, 113, E6766–E6775. [Google Scholar] [CrossRef]
- Chippalkatti, R.; Abankwa, D. Promotion of cancer cell stemness by Ras. Biochem. Soc. Trans. 2021, 49, 467–476. [Google Scholar] [CrossRef]
- Tidow, H.; Nissen, P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013, 280, 5551–5565. [Google Scholar] [CrossRef]
- Grant, B.M.M.; Enomoto, M.; Ikura, M.; Marshall, C.B. A Non-Canonical Calmodulin Target Motif Comprising a Polybasic Region and Lipidated Terminal Residue Regulates Localization. Int. J. Mol. Sci. 2020, 21, 2751. [Google Scholar] [CrossRef]
- Grant, B.M.M.; Enomoto, M.; Back, S.I.; Lee, K.Y.; Gebregiworgis, T.; Ishiyama, N.; Ikura, M.; Marshall, C.B. Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety. Sci. Signal. 2020, 13, eaaz0344. [Google Scholar] [CrossRef]
- Ismail, S.A.; Chen, Y.X.; Rusinova, A.; Chandra, A.; Bierbaum, M.; Gremer, L.; Triola, G.; Waldmann, H.; Bastiaens, P.I.; Wittinghofer, A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 2011, 7, 942–949. [Google Scholar] [CrossRef]
- Villalonga, P.; Lopez-Alcala, C.; Bosch, M.; Chiloeches, A.; Rocamora, N.; Gil, J.; Marais, R.; Marshall, C.J.; Bachs, O.; Agell, N. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 2001, 21, 7345–7354. [Google Scholar] [CrossRef]
- Chandra, A.; Grecco, H.E.; Pisupati, V.; Perera, D.; Cassidy, L.; Skoulidis, F.; Ismail, S.A.; Hedberg, C.; Hanzal-Bayer, M.; Venkitaraman, A.R.; et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011, 14, 148–158. [Google Scholar] [CrossRef]
- Li, C.J.; Heim, R.; Lu, P.; Pu, Y.; Tsien, R.Y.; Chang, D.C. Dynamic redistribution of calmodulin in HeLa cells during cell division as revealed by a GFP-calmodulin fusion protein technique. J. Cell Sci. 1999, 112, 1567–1577. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Chen, Y.; Dai, G.; Chen, J.; Sun, X.M.; Wen, C.J.; Zhao, D.H.; Chang, D.C.; Li, C.J. The association of calmodulin with central spindle regulates the initiation of cytokinesis in HeLa cells. Int. J. Biochem. Cell Biol. 2004, 36, 1562–1572. [Google Scholar] [CrossRef]
- Abraham, S.J.; Nolet, R.P.; Calvert, R.J.; Anderson, L.M.; Gaponenko, V. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry 2009, 48, 7575–7583. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Planchon, S.M.; Wolfman, J.C.; Wolfman, A. Growth factor-dependent AKT activation and cell migration requires the function of c-K(B)-Ras versus other cellular ras isoforms. J. Biol. Chem. 2006, 281, 29730–29738. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Julsgart, J.; Berchtold, M.W. High affinity calmodulin target sequence in the signalling molecule PI 3-kinase. FEBS Lett. 1998, 425, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.J.; Xu, L.R.; Liao, J.M.; Chen, J.; Liang, Y. Both the C-terminal polylysine region and the farnesylation of K-RasB are important for its specific interaction with calmodulin. PLoS ONE 2011, 6, e21929. [Google Scholar] [CrossRef] [PubMed]
- Agamasu, C.; Ghirlando, R.; Taylor, T.; Messing, S.; Tran, T.H.; Bindu, L.; Tonelli, M.; Nissley, D.V.; McCormick, F.; Stephen, A.G. KRAS Prenylation Is Required for Bivalent Binding with Calmodulin in a Nucleotide-Independent Manner. Biophys. J. 2019, 116, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, G.B.; Kopra, K.; Eskonen, V.; Härmä, H.; Abankwa, D. High-throughput amenable fluorescence-assays to screen for calmodulin-inhibitors. Anal. Biochem. 2019, 572, 25–32. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Villalobo, A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta 2014, 1843, 398–435. [Google Scholar] [CrossRef]
- Faust, F.M.; Slisz, M.; Jarrett, H.W. Calmodulin is labeled at lysine 148 by a chemically reactive phenothiazine. J. Biol. Chem. 1987, 262, 1938–1941. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Bunick, C.G.; Chazin, W.J. Target selectivity in EF-hand calcium binding proteins. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2004, 1742, 69–79. [Google Scholar] [CrossRef]
- Yang, A.; Miron, S.; Mouawad, L.; Duchambon, P.; Blouquit, Y.; Craescu, C.T. Flexibility and plasticity of human centrin 2 binding to the xeroderma pigmentosum group C protein (XPC) from nuclear excision repair. Biochemistry 2006, 45, 3653–3663. [Google Scholar] [CrossRef]
- Friedberg, F. Centrin isoforms in mammals. Relation to calmodulin. Mol. Biol. Rep. 2006, 33, 243–252. [Google Scholar] [CrossRef]
- Dantas, T.J.; Daly, O.M.; Morrison, C.G. Such small hands: The roles of centrins/caltractins in the centriole and in genome maintenance. Cell. Mol. Life Sci. 2012, 69, 2979–2997. [Google Scholar] [CrossRef]
- Puumalainen, M.R.; Ruthemann, P.; Min, J.H.; Naegeli, H. Xeroderma pigmentosum group C sensor: Unprecedented recognition strategy and tight spatiotemporal regulation. Cell. Mol. Life Sci. 2016, 73, 547–566. [Google Scholar] [CrossRef]
- Sanz, J.M.; Grecu, D.; Assairi, L. Ca2+ signaling and Target Binding Regulations: Calmodulin and Centrin In Vitro and In Vivo. Bioenergetics 2016, 5, 1000144. [Google Scholar] [CrossRef]
- Klein, U.R.; Nigg, E.A. SUMO-dependent regulation of centrin-2. J. Cell Sci. 2009, 122, 3312–3321. [Google Scholar] [CrossRef]
- Paoletti, A.; Moudjou, M.; Paintrand, M.; Salisbury, J.L.; Bornens, M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996, 109, 3089–3102. [Google Scholar] [CrossRef]
- Hodges, M.E.; Scheumann, N.; Wickstead, B.; Langdale, J.A.; Gull, K. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 2010, 123, 1407–1413. [Google Scholar] [CrossRef]
- Graser, S.; Stierhof, Y.D.; Lavoie, S.B.; Gassner, O.S.; Lamla, S.; Le Clech, M.; Nigg, E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007, 179, 321–330. [Google Scholar] [CrossRef]
- Wall, V.E.; Garvey, L.A.; Mehalko, J.L.; Procter, L.V.; Esposito, D. Combinatorial assembly of clone libraries using site-specific recombination. Methods Mol. Biol. 2014, 1116, 193–208. [Google Scholar] [CrossRef]
- Zimmermann, G.; Papke, B.; Ismail, S.; Vartak, N.; Chandra, A.; Hoffmann, M.; Hahn, S.A.; Triola, G.; Wittinghofer, A.; Bastiaens, P.I.; et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature 2013, 497, 638–642. [Google Scholar] [CrossRef]
- Vaasa, A.; Viil, I.; Enkvist, E.; Viht, K.; Raidaru, G.; Lavogina, D.; Uri, A. High-affinity bisubstrate probe for fluorescence anisotropy binding/displacement assays with protein kinases PKA and ROCK. Anal. Biochem. 2009, 385, 85–93. [Google Scholar] [CrossRef]
- Sinijarv, H.; Wu, S.; Ivan, T.; Laasfeld, T.; Viht, K.; Uri, A. Binding assay for characterization of protein kinase inhibitors possessing sub-picomolar to sub-millimolar affinity. Anal. Biochem. 2017, 531, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, G.B.; Okutachi, S.; Abankwa, D. Potential of phenothiazines to synergistically block calmodulin and reactivate PP2A in cancer cells. PLoS ONE 2022, 17, e0268635. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, M.R.; Zaidi, A.; Johnson, C.K. Fluorescence polarization assay for calmodulin binding to plasma membrane Ca2+-ATPase: Dependence on enzyme and Ca2+ concentrations. Anal. Biochem. 2009, 385, 1–6. [Google Scholar] [CrossRef]
- Waxham, M.N.; Tsai, A.L.; Putkey, J.A. A mechanism for calmodulin (CaM) trapping by CaM-kinase II defined by a family of CaM-binding peptides. J. Biol. Chem. 1998, 273, 17579–17584. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Guo, X.; Yang, B. Calcium-induced human centrin 1 self-assembly and double-regulating the binding with peptide R18-Sfi1p. Int. J. Biol. Macromol. 2019, 128, 314–323. [Google Scholar] [CrossRef]
- Bodle, J.C.; Loboa, E.G. Concise Review: Primary Cilia: Control Centers for Stem Cell Lineage Specification and Potential Targets for Cell-Based Therapies. Stem Cells 2016, 34, 1445–1454. [Google Scholar] [CrossRef]
- Pfleger, K.D.; Eidne, K.A. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods 2006, 3, 165–174. [Google Scholar] [CrossRef]
- Halling, D.B.; Liebeskind, B.J.; Hall, A.W.; Aldrich, R.W. Conserved properties of individual Ca2+-binding sites in calmodulin. Proc. Natl. Acad. Sci. USA 2016, 113, E1216–E1225. [Google Scholar] [CrossRef]
- Herrmann, C.; Horn, G.; Spaargaren, M.; Wittinghofer, A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 1996, 271, 6794–6800. [Google Scholar] [CrossRef]
- Vetter, I.R.; Linnemann, T.; Wohlgemuth, S.; Geyer, M.; Kalbitzer, H.R.; Herrmann, C.; Wittinghofer, A. Structural and biochemical analysis of Ras-effector signaling via RalGDS. FEBS Lett. 1999, 451, 175–180. [Google Scholar] [CrossRef]
- Thurnher, M.; Gruenbacher, G.; Nussbaumer, O. Regulation of mevalonate metabolism in cancer and immune cells. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1831, 1009–1015. [Google Scholar] [CrossRef]
- Abdelkarim, H.; Leschinsky, N.; Jang, H.; Banerjee, A.; Nussinov, R.; Gaponenko, V. The dynamic nature of the K-Ras/calmodulin complex can be altered by oncogenic mutations. Curr. Opin. Struct. Biol. 2021, 71, 164–170. [Google Scholar] [CrossRef]
- Parkkola, H.; Siddiqui, F.A.; Oetken-Lindholm, C.; Abankwa, D. FLIM-FRET Analysis of Ras Nanoclustering and Membrane-Anchorage. Methods Mol. Biol. 2021, 2262, 233–250. [Google Scholar] [CrossRef]
- Siddiqui, F.A.; Alam, C.; Rosenqvist, P.; Ora, M.; Sabt, A.; Manoharan, G.B.; Bindu, L.; Okutachi, S.; Catillon, M.; Taylor, T.; et al. PDE6D Inhibitors with a New Design Principle Selectively Block K-Ras Activity. ACS Omega 2020, 5, 832–842. [Google Scholar] [CrossRef]
- Kohnke, M.; Schmitt, S.; Ariotti, N.; Piggott, A.M.; Parton, R.G.; Lacey, E.; Capon, R.J.; Alexandrov, K.; Abankwa, D. Design and application of in vivo FRET biosensors to identify protein prenylation and nanoclustering inhibitors. Chem. Biol. 2012, 19, 866–874. [Google Scholar] [CrossRef]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef]
- Santoro, A.; Vlachou, T.; Carminati, M.; Pelicci, P.G.; Mapelli, M. Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep. 2016, 17, 1700–1720. [Google Scholar] [CrossRef]
- Cicalese, A.; Bonizzi, G.; Pasi, C.E.; Faretta, M.; Ronzoni, S.; Giulini, B.; Brisken, C.; Minucci, S.; Di Fiore, P.P.; Pelicci, P.G. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 2009, 138, 1083–1095. [Google Scholar] [CrossRef]
- Chen, C.; Yamashita, Y.M. Centrosome-centric view of asymmetric stem cell division. Open Biol. 2021, 11, 200314. [Google Scholar] [CrossRef]
- Nussinov, R.; Zhang, M.; Tsai, C.J.; Jang, H. Calmodulin and IQGAP1 activation of PI3Kalpha and Akt in KRAS, HRAS and NRAS-driven cancers. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864, 2304–2314. [Google Scholar] [CrossRef] [PubMed]
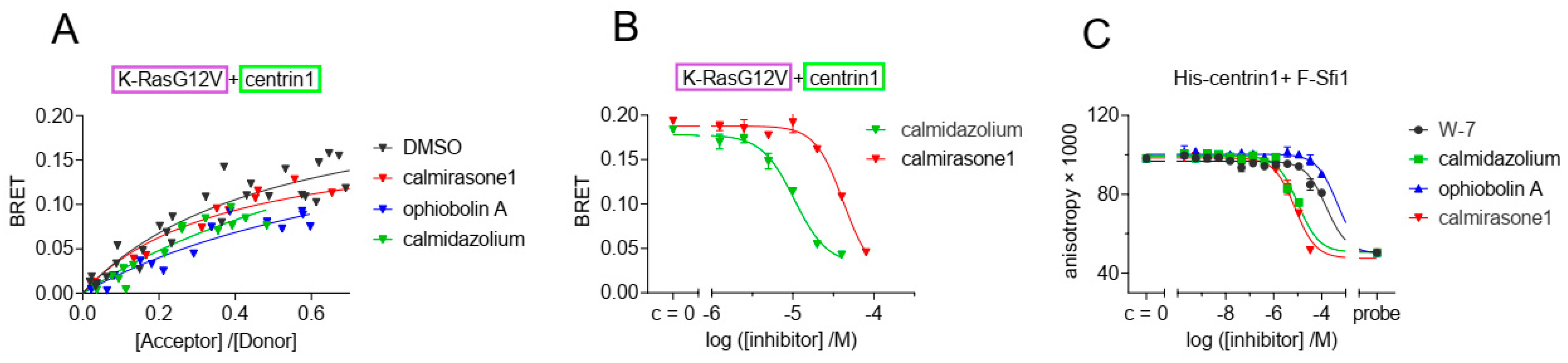

| Inhibitor | Centrin1 | CaM |
|---|---|---|
| Mean Kd (Repeat Values) | Kd (References) | |
| calmidazolium | 1.6 (1.4; 1.8) µM | 13.5 nM [26] |
| W-7 | 18.2 (17.8; 18.5) µM | 1.47 µM [26] |
| ophiobolin A | 49 (58; 39) µM | 3.5 µM [9] |
| calmirasone1 | 0.9 (1.0; 0.8) µM | 0.87 µM [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manoharan, G.b.; Laurini, C.; Bottone, S.; Ben Fredj, N.; Abankwa, D.K. K-Ras Binds Calmodulin-Related Centrin1 with Potential Implications for K-Ras Driven Cancer Cell Stemness. Cancers 2023, 15, 3087. https://doi.org/10.3390/cancers15123087
Manoharan Gb, Laurini C, Bottone S, Ben Fredj N, Abankwa DK. K-Ras Binds Calmodulin-Related Centrin1 with Potential Implications for K-Ras Driven Cancer Cell Stemness. Cancers. 2023; 15(12):3087. https://doi.org/10.3390/cancers15123087
Chicago/Turabian StyleManoharan, Ganesh babu, Christina Laurini, Sara Bottone, Nesrine Ben Fredj, and Daniel Kwaku Abankwa. 2023. "K-Ras Binds Calmodulin-Related Centrin1 with Potential Implications for K-Ras Driven Cancer Cell Stemness" Cancers 15, no. 12: 3087. https://doi.org/10.3390/cancers15123087
APA StyleManoharan, G. b., Laurini, C., Bottone, S., Ben Fredj, N., & Abankwa, D. K. (2023). K-Ras Binds Calmodulin-Related Centrin1 with Potential Implications for K-Ras Driven Cancer Cell Stemness. Cancers, 15(12), 3087. https://doi.org/10.3390/cancers15123087






