mRNA-Lipid Nanoparticle (LNP) Delivery of Humanized EpCAM-CD3 Bispecific Antibody Significantly Blocks Colorectal Cancer Tumor Growth
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Antibodies
2.3. Design and Cloning of Bispecific Antibody DNA Constructs
2.4. In Vitro Transcription
2.5. Embedding of mRNA into LNP and Transfection of mRNA-LNP into Cells
2.6. Transfection of HEK-293 Cells with DNA Encoding Bispecific Antibodies
2.7. Flow Cytometry (FACS)
2.8. Real-Time Cytotoxicity Assay (RTCA)
2.9. ELISA (Enzyme-Linked Immunoassay)
2.10. Mouse Xenograft Tumor Model
2.11. Statistical Analysis
3. Results
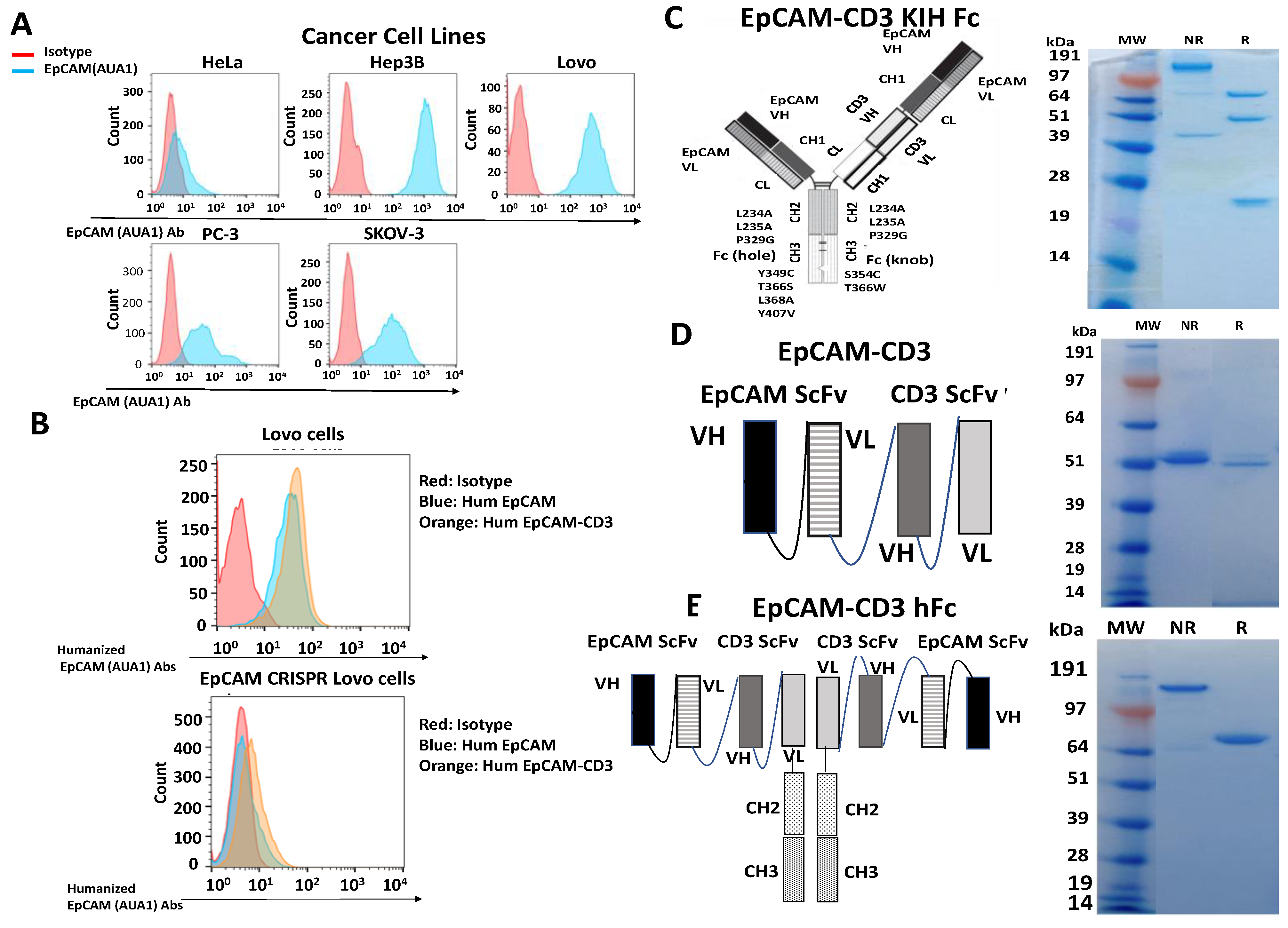
3.1. The AUA1 Mouse Antibody Was Humanized and Used to Engineer Three Designs of Humanized AUA1-CD3 Bispecific Antibodies
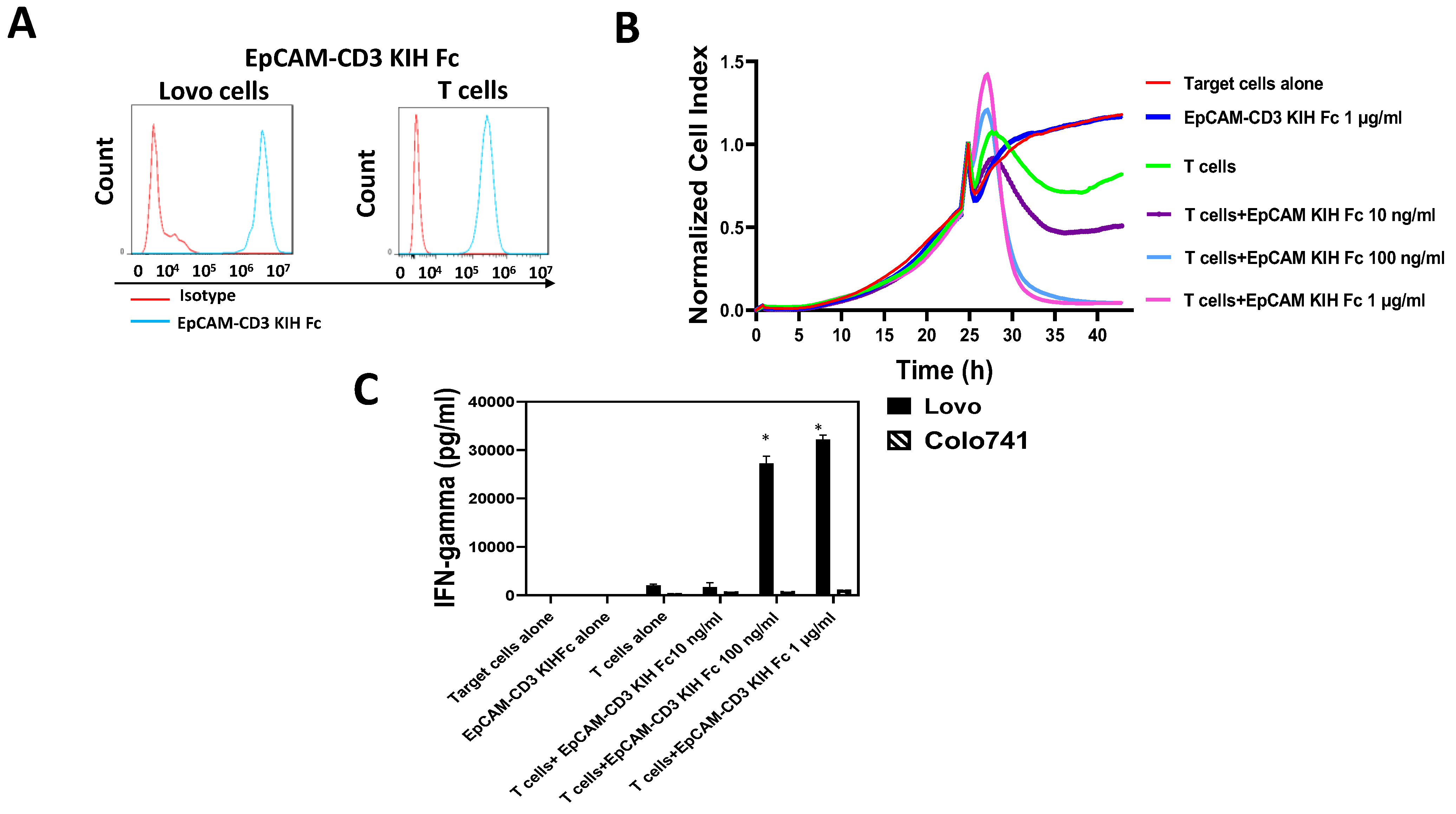
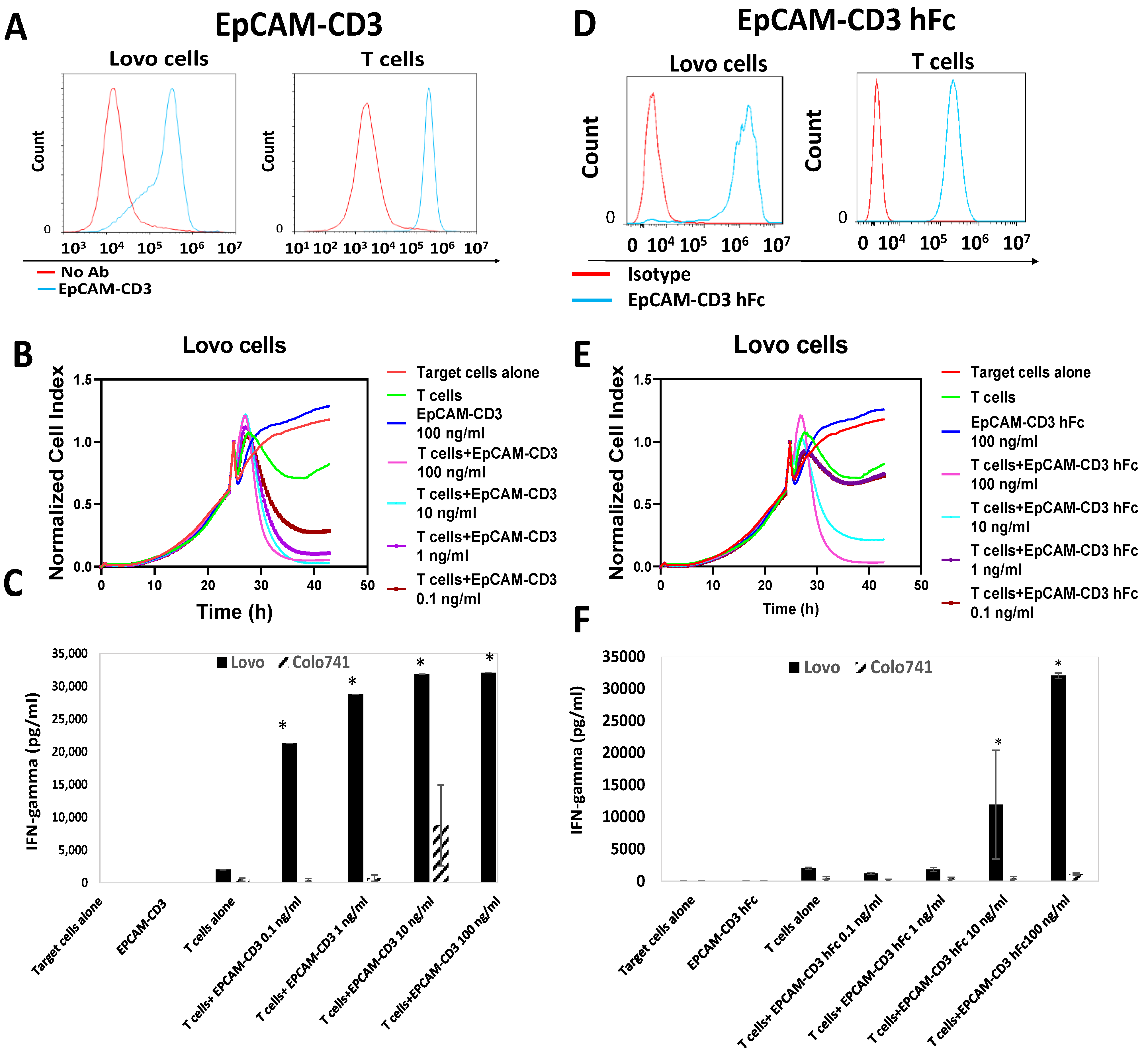
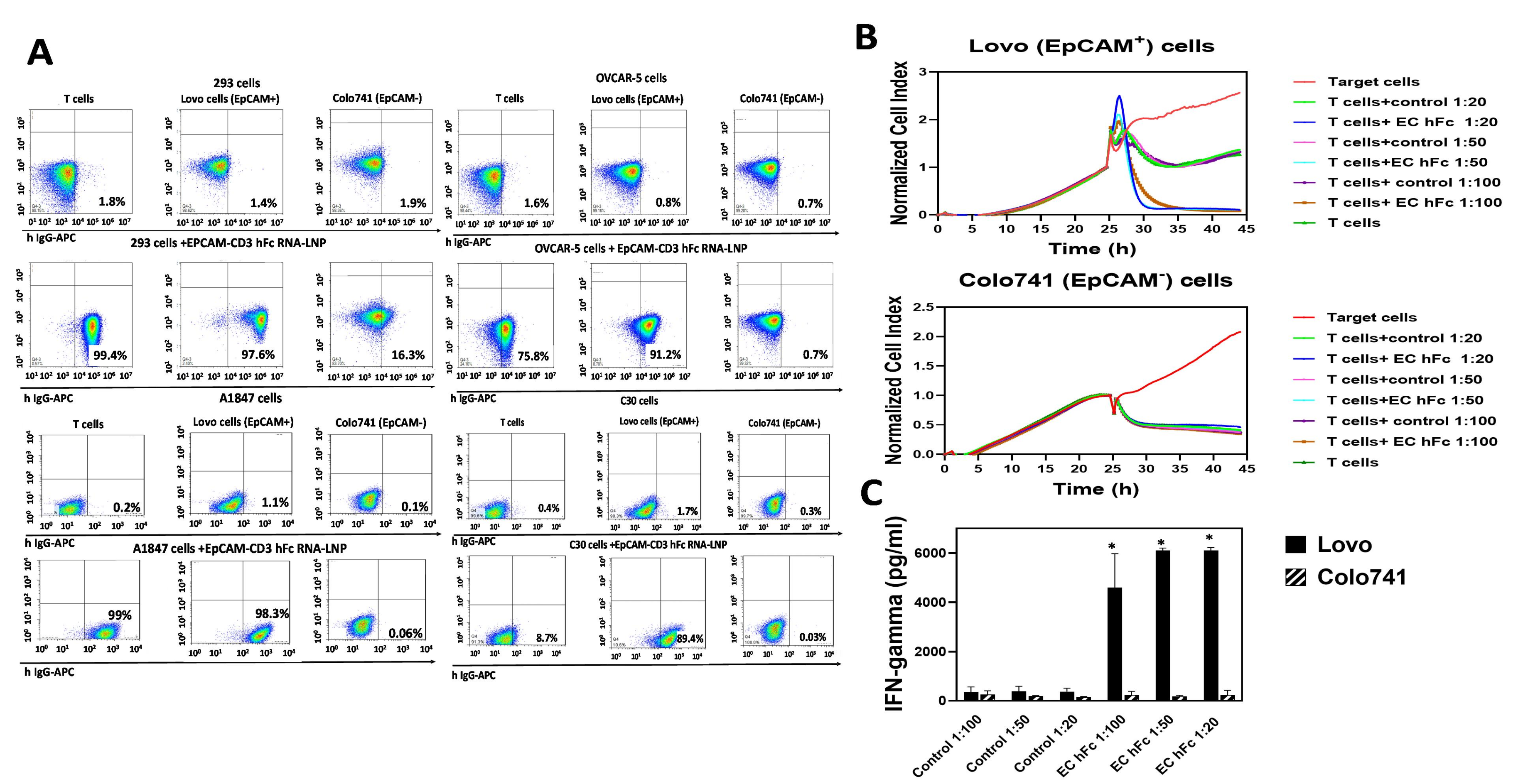
3.2. EpCAM-CD3 Bispecific Antibodies Caused High Binding, Killing, and IFN-Gamma Secretion with EpCAM-Positive Target Cancer Cells
3.3. EpCAM-CD3 hFc mRNA-LNPs Transfected in HEK293 Cells Produce EpCAM-CD3 hFc Antibody with Highly Specific Functional Activity against EpCAM-Positive Cells
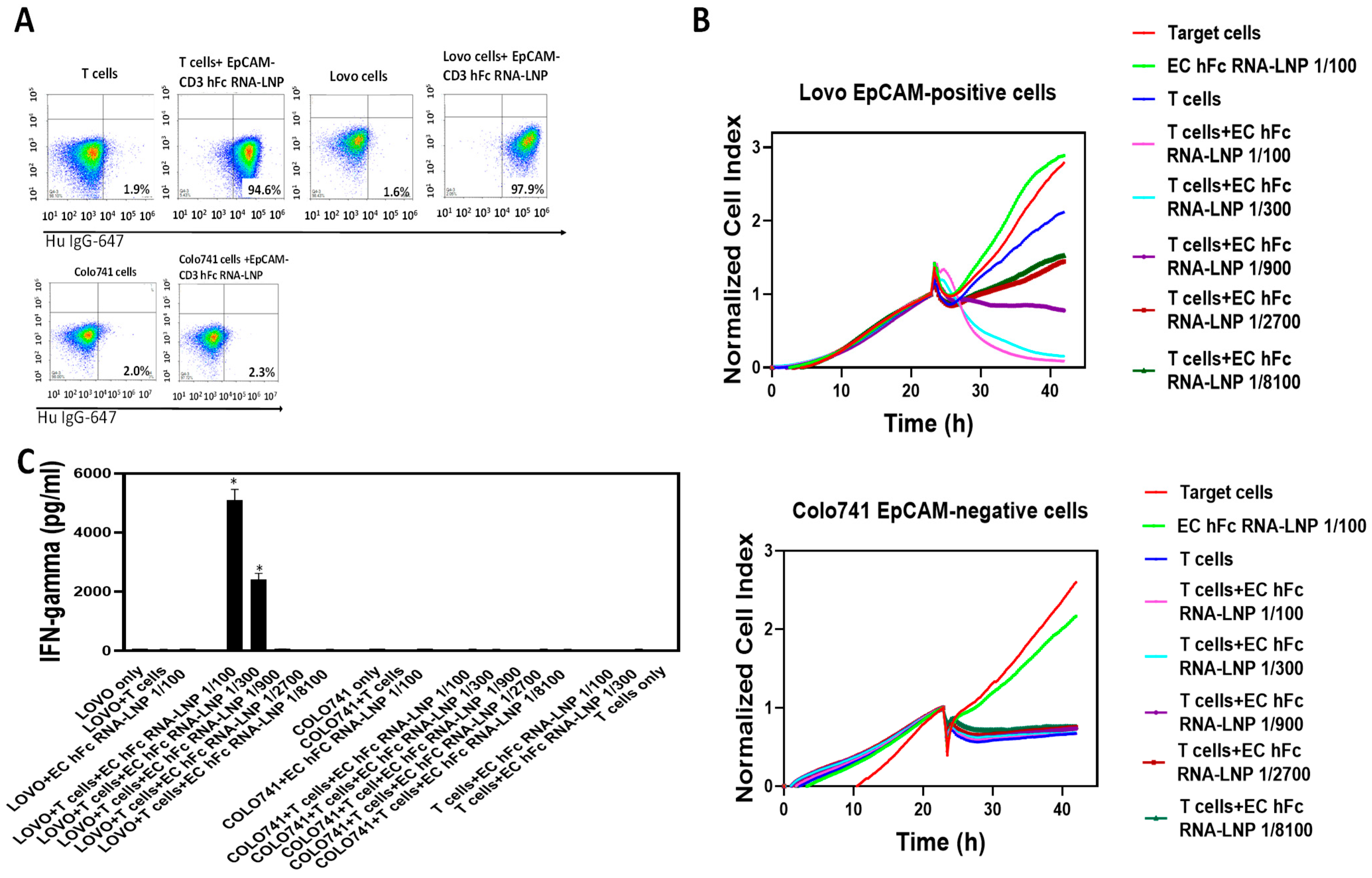
3.4. EpCAM-CD3 hFc mRNA-LNPs Transfected to Cancer Cells Produced Bispecific Antibody with Specific Functional Activity against EpCAM-Positive Cells
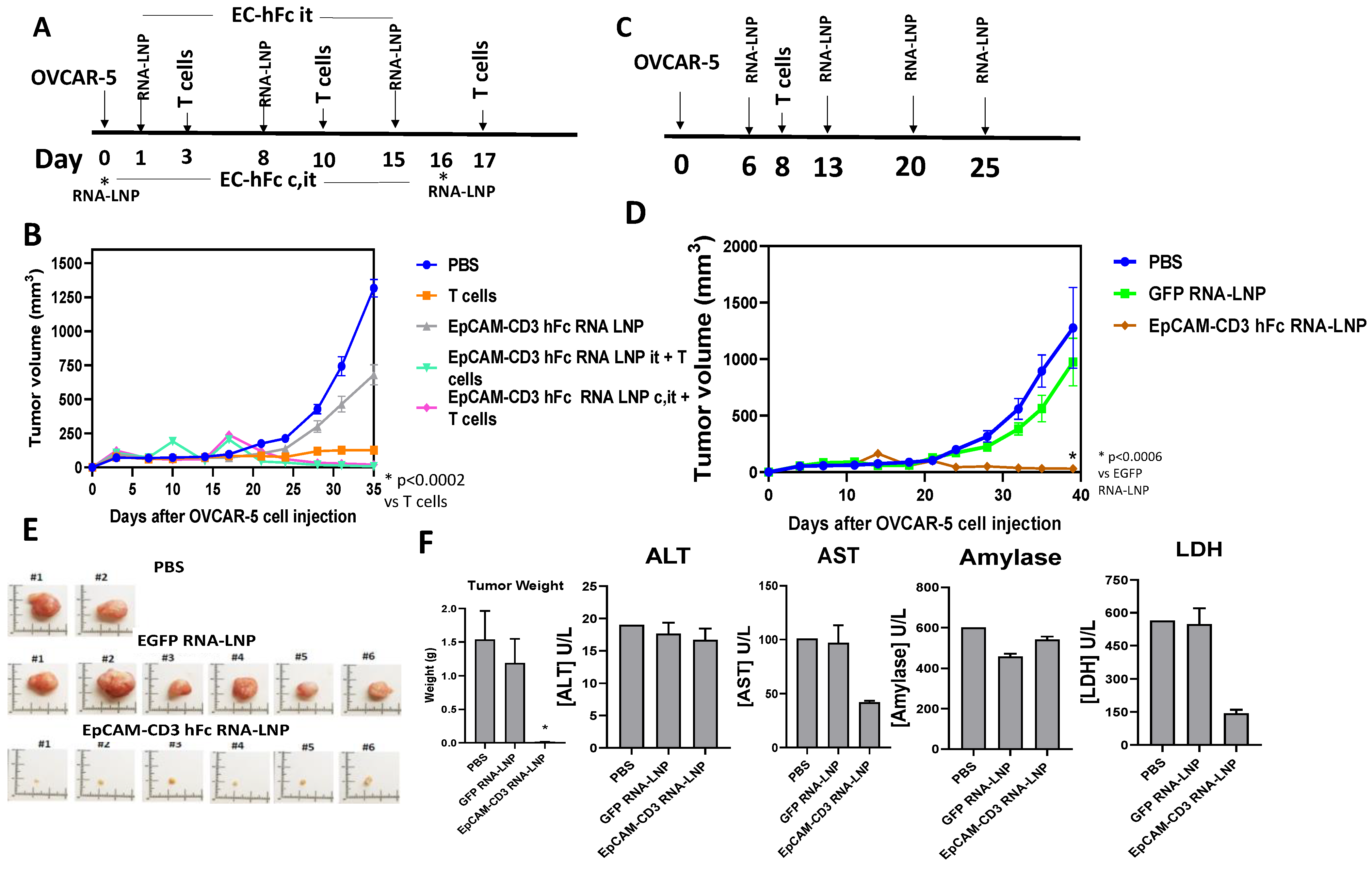
3.5. EpCAM-CD3 hFc mRNA-LNPs with T Cells Significantly Blocked OVCAR-5 Xenograft Tumor Growth
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bodmer, W.F. Cancer genetics: Colorectal cancer as a model. J. Hum. Genet. 2006, 51, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ruff, S.M.; Pawlik, T.M. A Review of Translational Research for Targeted Therapy for Metastatic Colorectal Cancer. Cancers 2023, 15, 1395. [Google Scholar] [CrossRef]
- Ciardiello, D.; Vitiello, P.P.; Cardone, C.; Martini, G.; Troiani, T.; Martinelli, E. Immunotherapy of colorectal cancer: Challenges for therapeutic efficacy. Cancer Treat. Rev. 2019, 76, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.; Eck, S.L. EpCAM: A new therapeutic target for an old cancer antigen. Cancer Biol. Ther. 2003, 2, 320–326. [Google Scholar] [CrossRef]
- Schnell, U.; Cirulli, V.; Giepmans, B.N. EpCAM: Structure and function in health and disease. Biochim. Biophys. Acta 2013, 1828, 1989–2001. [Google Scholar] [CrossRef]
- Imrich, S.; Hachmeister, M.; Gires, O. EpCAM and its potential role in tumor-initiating cells. Cell Adhes. Migr. 2012, 6, 30–38. [Google Scholar] [CrossRef]
- van der Gun, B.T.; Melchers, L.J.; Ruiters, M.H.; de Leij, L.F.; McLaughlin, P.M.; Rots, M.G. EpCAM in carcinogenesis: The good, the bad or the ugly. Carcinogenesis 2010, 31, 1913–1921. [Google Scholar] [CrossRef]
- Munz, M.; Baeuerle, P.A.; Gires, O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009, 69, 5627–5629. [Google Scholar] [CrossRef]
- Ntouroupi, T.G.; Ashraf, S.Q.; McGregor, S.B.; Turney, B.W.; Seppo, A.; Kim, Y.; Wang, X.; Kilpatrick, M.W.; Tsipouras, P.; Tafas, T.; et al. Detection of circulating tumour cells in peripheral blood with an automated scanning fluorescence microscope. Br. J. Cancer 2008, 99, 789–795. [Google Scholar] [CrossRef]
- Cetin, D.; Okan, M.; Bat, E.; Kulah, H. A comparative study on EpCAM antibody immobilization on gold surfaces and microfluidic channels for the detection of circulating tumor cells. Colloids Surf. B Biointerfaces 2020, 188, 110808. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Henri, J.; Roy, K.; Hays, E.; Bauer, M.; Veedu, R.N. EpCAM Immunotherapy versus Specific Targeted Delivery of Drugs. Cancers 2018, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Alhabbab, R.Y. Targeting Cancer Stem Cells by Genetically Engineered Chimeric Antigen Receptor T Cells. Front. Genet. 2020, 11, 312. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Gires, O. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 2007, 96, 417–423. [Google Scholar] [CrossRef]
- Bremer, E.; Helfrich, W. EpCAM-targeted induction of apoptosis. Front. Biosci.-Landmark 2008, 13, 5042–5049. [Google Scholar] [CrossRef]
- Carpenter, G.; Red Brewer, M. EpCAM: Another surface-to-nucleus missile. Cancer Cell 2009, 15, 165–166. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.T. The Critical Contribution of Pseudouridine to mRNA COVID-19 Vaccines. Front. Cell Dev. Biol. 2021, 9, 789427. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Dong, Y. Lipid Nanoparticle-mRNA Formulations for Therapeutic Applications. Acc. Chem. Res. 2021, 54, 4283–4293. [Google Scholar] [CrossRef]
- Jung, H.N.; Lee, S.Y.; Lee, S.; Youn, H.; Im, H.J. Lipid nanoparticles for delivery of RNA therapeutics: Current status and the role of in vivo imaging. Theranostics 2022, 12, 7509–7531. [Google Scholar] [CrossRef]
- Zhou, L.; Zou, M.; Xu, Y.; Lin, P.; Lei, C.; Xia, X. Nano Drug Delivery System for Tumor Immunotherapy: Next-Generation Therapeutics. Front. Oncol. 2022, 12, 864301. [Google Scholar] [CrossRef]
- Bacac, M.; Fauti, T.; Sam, J.; Colombetti, S.; Weinzierl, T.; Ouaret, D.; Bodmer, W.; Lehmann, S.; Hofer, T.; Hosse, R.J.; et al. A Novel Carcinoembryonic Antigen T-Cell Bispecific Antibody (CEA TCB) for the Treatment of Solid Tumors. Clin. Cancer Res. 2016, 22, 3286–3297. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Huang, Y.; Sienkiewicz, J.; Sun, J.; Guiang, L.; Li, F.; Yang, L.; Golubovskaya, V. Bispecific BCMA-CD3 Antibodies Block Multiple Myeloma Tumor Growth. Cancers 2022, 14, 2518. [Google Scholar] [CrossRef] [PubMed]
- Berahovich, R.; Zhou, H.; Xu, S.; Wei, Y.; Guan, J.; Guan, J.; Harto, H.; Fu, S.; Yang, K.; Zhu, S.; et al. Golubovskaya. CAR-T Cells Based on Novel BCMA Monoclonal Antibody Block Multiple Myeloma Cell Growth. Cancers 2018, 10, 323. [Google Scholar] [CrossRef] [PubMed]
- Berahovich, R.; Xu, S.; Zhou, H.; Harto, H.; Xu, Q.; Garcia, A.; Liu, F.; Golubovskaya, V.M.; Wu, L. FLAG-tagged CD19-specific CAR-T cells eliminate CD19-bearing solid tumor cells in vitro and in vivo. Front. Biosci.-Landmark 2017, 22, 1644–1654. [Google Scholar]
- Berahovich, R.; Liu, X.; Zhou, H.; Tsadik, E.; Xu, S.; Golubovskaya, V. Hypoxia Selectively Impairs CAR-T Cells In Vitro. Cancers 2019, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Spurr, N.K.; Durbin, H.; Sheer, D.; Parkar, M.; Bobrow, L.; Bodmer, W.F. Characterization and chromosomal assignment of a human cell surface antigen defined by the monoclonal antibody AUAI. Int. J. Cancer 1986, 38, 631–636. [Google Scholar] [CrossRef]
- Wong, N.A.; Warren, B.F.; Piris, J.; Maynard, N.; Marshall, R.; Bodmer, W.F. EpCAM and gpA33 are markers of Barrett’s metaplasia. J. Clin. Pathol. 2006, 59, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Almagro, J.C.; Fransson, J. Humanization of antibodies. Front. Biosci. 2008, 13, 1619–1633. [Google Scholar]
- Golubovskaya, V.; Berahovich, R.; Zhou, H.; Xu, S.; Harto, H.; Li, L.; Chao, C.C.; Mao, M.M.; Wu, L. CD47-CAR-T Cells Effectively Kill Target Cancer Cells and Block Pancreatic Tumor Growth. Cancers 2017, 9, 139. [Google Scholar] [CrossRef]
- Golubovskaya, V.; Zhou, H.; Li, F.; Valentine, M.; Sun, J.; Berahovich, R.; Xu, S.; Quintanilla, M.; Ma, M.C.; Sienkiewicz, J.; et al. Novel CD37, Humanized CD37 and Bi-Specific Humanized CD37-CD19 CAR-T Cells Specifically Target Lymphoma. Cancers 2021, 13, 981. [Google Scholar] [CrossRef]
- Reinhard, K.; Rengstl, B.; Oehm, P.; Michel, K.; Billmeier, A.; Hayduk, N.; Klein, O.; Kuna, K.; Ouchan, Y.; Woll, S.; et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science 2020, 367, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wang, X.; Xing, M.; Yang, X.; Wu, M.; Shi, H. Intratumoral delivery of a novel oncolytic adenovirus encoding human antibody against PD-1 elicits enhanced antitumor efficacy. Mol. Ther.-Oncolytics 2022, 25, 236–248. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golubovskaya, V.; Sienkiewicz, J.; Sun, J.; Huang, Y.; Hu, L.; Zhou, H.; Harto, H.; Xu, S.; Berahovich, R.; Bodmer, W.; et al. mRNA-Lipid Nanoparticle (LNP) Delivery of Humanized EpCAM-CD3 Bispecific Antibody Significantly Blocks Colorectal Cancer Tumor Growth. Cancers 2023, 15, 2860. https://doi.org/10.3390/cancers15102860
Golubovskaya V, Sienkiewicz J, Sun J, Huang Y, Hu L, Zhou H, Harto H, Xu S, Berahovich R, Bodmer W, et al. mRNA-Lipid Nanoparticle (LNP) Delivery of Humanized EpCAM-CD3 Bispecific Antibody Significantly Blocks Colorectal Cancer Tumor Growth. Cancers. 2023; 15(10):2860. https://doi.org/10.3390/cancers15102860
Chicago/Turabian StyleGolubovskaya, Vita, John Sienkiewicz, Jinying Sun, Yanwei Huang, Liang Hu, Hua Zhou, Hizkia Harto, Shirley Xu, Robert Berahovich, Walter Bodmer, and et al. 2023. "mRNA-Lipid Nanoparticle (LNP) Delivery of Humanized EpCAM-CD3 Bispecific Antibody Significantly Blocks Colorectal Cancer Tumor Growth" Cancers 15, no. 10: 2860. https://doi.org/10.3390/cancers15102860
APA StyleGolubovskaya, V., Sienkiewicz, J., Sun, J., Huang, Y., Hu, L., Zhou, H., Harto, H., Xu, S., Berahovich, R., Bodmer, W., & Wu, L. (2023). mRNA-Lipid Nanoparticle (LNP) Delivery of Humanized EpCAM-CD3 Bispecific Antibody Significantly Blocks Colorectal Cancer Tumor Growth. Cancers, 15(10), 2860. https://doi.org/10.3390/cancers15102860






