The Impact of Tumor Cell-Intrinsic Expression of Cyclic GMP-AMP Synthase (cGAS)-Stimulator of Interferon Genes (STING) on the Infiltration of CD8+ T Cells and Clinical Outcomes in Mismatch Repair Proficient/Microsatellite Stable Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. IHC
2.3. Data Analyses of TCGA and GEO Database
2.4. Statistical Analysis
3. Results
3.1. The Expression Patterns of Tumor Cell-Intrinsic cGAS-STING in pMMR/MSS CRC
3.2. Down-Regulation of the Expression of cGAS-STING in Tumor Cells in the Advanced Stages of pMMR CRC
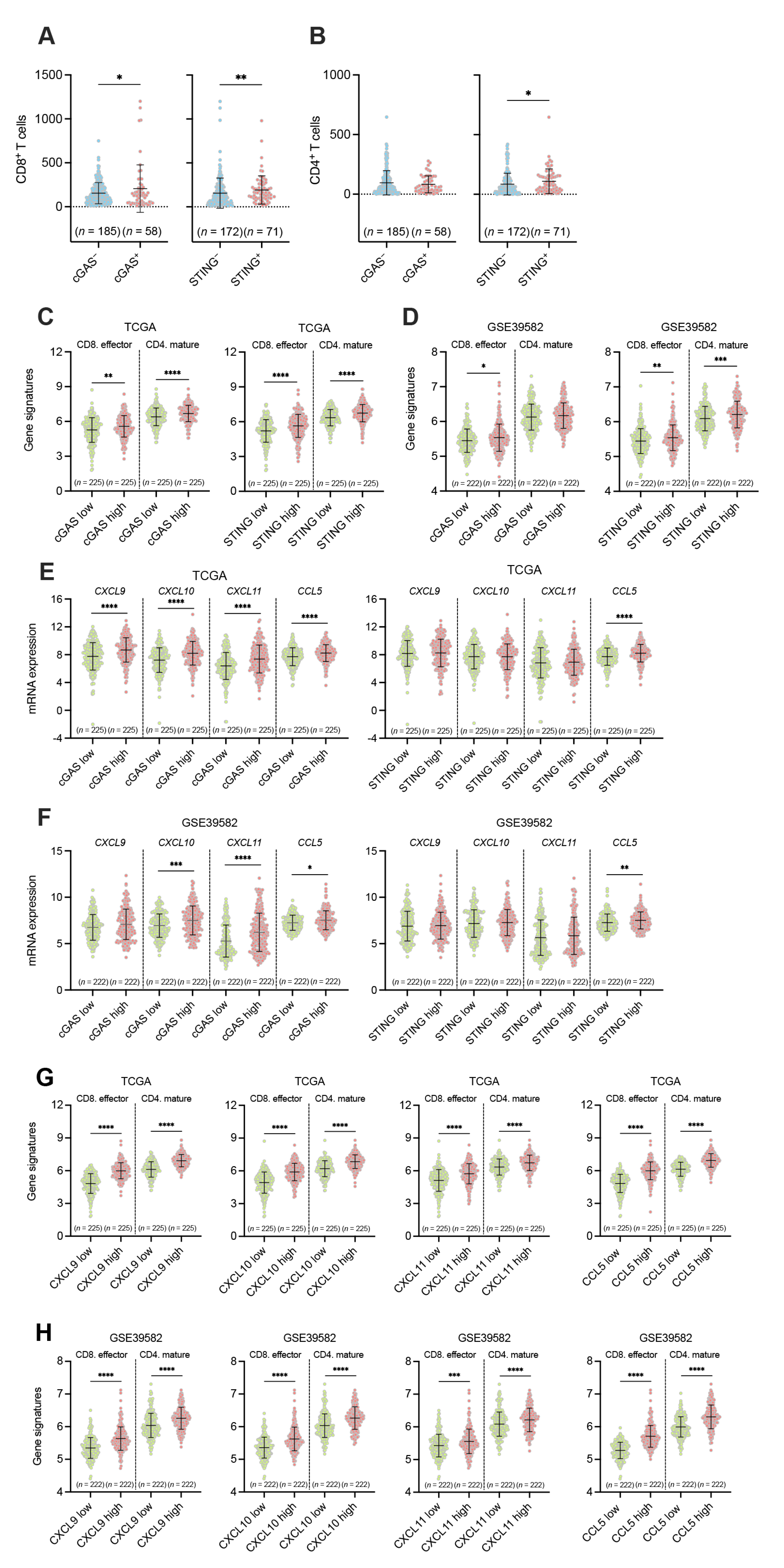
3.3. Associations between the Tumor Cell-Intrinsic Expression of cGAS-STING and the Infiltration of CD8+ and CD4+ T Cells in pMMR/MSS CRC
3.4. Associations between the Tumor Cell-Intrinsic Expression of cGAS-STING and Clinical Outcomes in Patients with pMMR/MSS CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
- de la Chapelle, A.; Hampel, H. Clinical relevance of microsatellite instability in colorectal cancer. J. Clin. Oncol. 2010, 28, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Bronner, C.E.; Baker, S.M.; Morrison, P.T.; Warren, G.; Smith, L.G.; Lescoe, M.K.; Kane, M.; Earabino, C.; Lipford, J.; Lindblom, A.; et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 1994, 368, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Fishel, R.; Lescoe, M.K.; Rao, M.R.; Copeland, N.G.; Jenkins, N.A.; Garber, J.; Kane, M.; Kolodner, R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993, 75, 1027–1038. [Google Scholar] [CrossRef]
- Miyaki, M.; Konishi, M.; Tanaka, K.; Kikuchi-Yanoshita, R.; Muraoka, M.; Yasuno, M.; Igari, T.; Koike, M.; Chiba, M.; Mori, T. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat. Genet. 1997, 17, 271–272. [Google Scholar] [CrossRef]
- Nicolaides, N.C.; Papadopoulos, N.; Liu, B.; Wei, Y.F.; Carter, K.C.; Ruben, S.M.; Rosen, C.A.; Haseltine, W.A.; Fleischmann, R.D.; Fraser, C.M.; et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 1994, 371, 75–80. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Nicolaides, N.C.; Wei, Y.F.; Ruben, S.M.; Carter, K.C.; Rosen, C.A.; Haseltine, W.A.; Fleischmann, R.D.; Fraser, C.M.; Adams, M.D.; et al. Mutation of a mutL homolog in hereditary colon cancer. Science 1994, 263, 1625–1629. [Google Scholar] [CrossRef]
- Peltomäki, P.; Vasen, H.F. Mutations predisposing to hereditary nonpolyposis colorectal cancer: Database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 1997, 113, 1146–1158. [Google Scholar] [CrossRef]
- Kane, M.F.; Loda, M.; Gaida, G.M.; Lipman, J.; Mishra, R.; Goldman, H.; Jessup, J.M.; Kolodner, R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997, 57, 808–811. [Google Scholar]
- Cunningham, J.M.; Christensen, E.R.; Tester, D.J.; Kim, C.Y.; Roche, P.C.; Burgart, L.J.; Thibodeau, S.N. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998, 58, 3455–3460. [Google Scholar]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef]
- Kreiter, S.; Vormehr, M.; van de Roemer, N.; Diken, M.; Löwer, M.; Diekmann, J.; Boegel, S.; Schrörs, B.; Vascotto, F.; Castle, J.C.; et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015, 520, 692–696. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef]
- Vanpouille-Box, C.; Demaria, S.; Formenti, S.C.; Galluzzi, L. Cytosolic DNA Sensing in Organismal Tumor Control. Cancer Cell 2018, 34, 361–378. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef]
- Shen, Y.J.; Le Bert, N.; Chitre, A.A.; Koo, C.X.; Nga, X.H.; Ho, S.S.; Khatoo, M.; Tan, N.Y.; Ishii, K.J.; Gasser, S. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015, 11, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Konno, H.; Ahn, J.; Barber, G.N. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates with Tumorigenesis. Cell Rep. 2016, 14, 282–297. [Google Scholar] [CrossRef] [PubMed]
- Chon, H.J.; Kim, H.; Noh, J.H.; Yang, H.; Lee, W.S.; Kong, S.J.; Lee, S.J.; Lee, Y.S.; Kim, W.R.; Kim, J.H.; et al. STING signaling is a potential immunotherapeutic target in colorectal cancer. J. Cancer 2019, 10, 4932–4938. [Google Scholar] [CrossRef] [PubMed]
- Kaneta, A.; Nakajima, S.; Okayama, H.; Matsumoto, T.; Saito, K.; Kikuchi, T.; Endo, E.; Ito, M.; Mimura, K.; Kanke, Y.; et al. Role of the cGAS-STING pathway in regulating the tumor-immune microenvironment in dMMR/MSI colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 2765–2776. [Google Scholar] [CrossRef]
- Kikuchi, T.; Mimura, K.; Okayama, H.; Nakayama, Y.; Saito, K.; Yamada, L.; Endo, E.; Sakamoto, W.; Fujita, S.; Endo, H.; et al. A subset of patients with MSS/MSI-low-colorectal cancer showed increased CD8(+) TILs together with up-regulated IFN-γ. Oncol. Lett. 2019, 18, 5977–5985. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.Y.; Molina, J.; Kim, J.H.; Arvis, C.D.; Ahn, M.J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Noda, M.; Okayama, H.; Tachibana, K.; Sakamoto, W.; Saito, K.; Thar Min, A.K.; Ashizawa, M.; Nakajima, T.; Aoto, K.; Momma, T.; et al. Glycosyltransferase Gene Expression Identifies a Poor Prognostic Colorectal Cancer Subtype Associated with Mismatch Repair Deficiency and Incomplete Glycan Synthesis. Clin. Cancer Res. 2018, 24, 4468–4481. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef]
- Konno, H.; Yamauchi, S.; Berglund, A.; Putney, R.M.; Mulé, J.J.; Barber, G.N. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 2018, 37, 2037–2051. [Google Scholar] [CrossRef]
- Harlin, H.; Meng, Y.; Peterson, A.C.; Zha, Y.; Tretiakova, M.; Slingluff, C.; McKee, M.; Gajewski, T.F. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009, 69, 3077–3085. [Google Scholar] [CrossRef]
- Heide, T.; Househam, J.; Cresswell, G.D.; Spiteri, I.; Lynn, C.; Mossner, M.; Kimberley, C.; Fernandez-Mateos, J.; Chen, B.; Zapata, L.; et al. The co-evolution of the genome and epigenome in colorectal cancer. Nature 2022, 611, 733–743. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Nakamura, Y.; White, R.; Smits, A.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef]
- Baker, S.J.; Fearon, E.R.; Nigro, J.M.; Hamilton, S.R.; Preisinger, A.C.; Jessup, J.M.; vanTuinen, P.; Ledbetter, D.H.; Barker, D.F.; Nakamura, Y.; et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 1989, 244, 217–221. [Google Scholar] [CrossRef]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359, 235–237. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e2073. [Google Scholar] [CrossRef]
- Langner, C. Serrated and non-serrated precursor lesions of colorectal cancer. Dig. Dis. 2015, 33, 28–37. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef]
- Luo, N.; Nixon, M.J.; Gonzalez-Ericsson, P.I.; Sanchez, V.; Opalenik, S.R.; Li, H.; Zahnow, C.A.; Nickels, M.L.; Liu, F.; Tantawy, M.N.; et al. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat. Commun. 2018, 9, 248. [Google Scholar] [CrossRef]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.W.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Kuang, C.; Park, Y.; Augustin, R.C.; Lin, Y.; Hartman, D.J.; Seigh, L.; Pai, R.K.; Sun, W.; Bahary, N.; Ohr, J.; et al. Pembrolizumab plus azacitidine in patients with chemotherapy refractory metastatic colorectal cancer: A single-arm phase 2 trial and correlative biomarker analysis. Clin. Epigenetics 2022, 14, 3. [Google Scholar] [CrossRef] [PubMed]
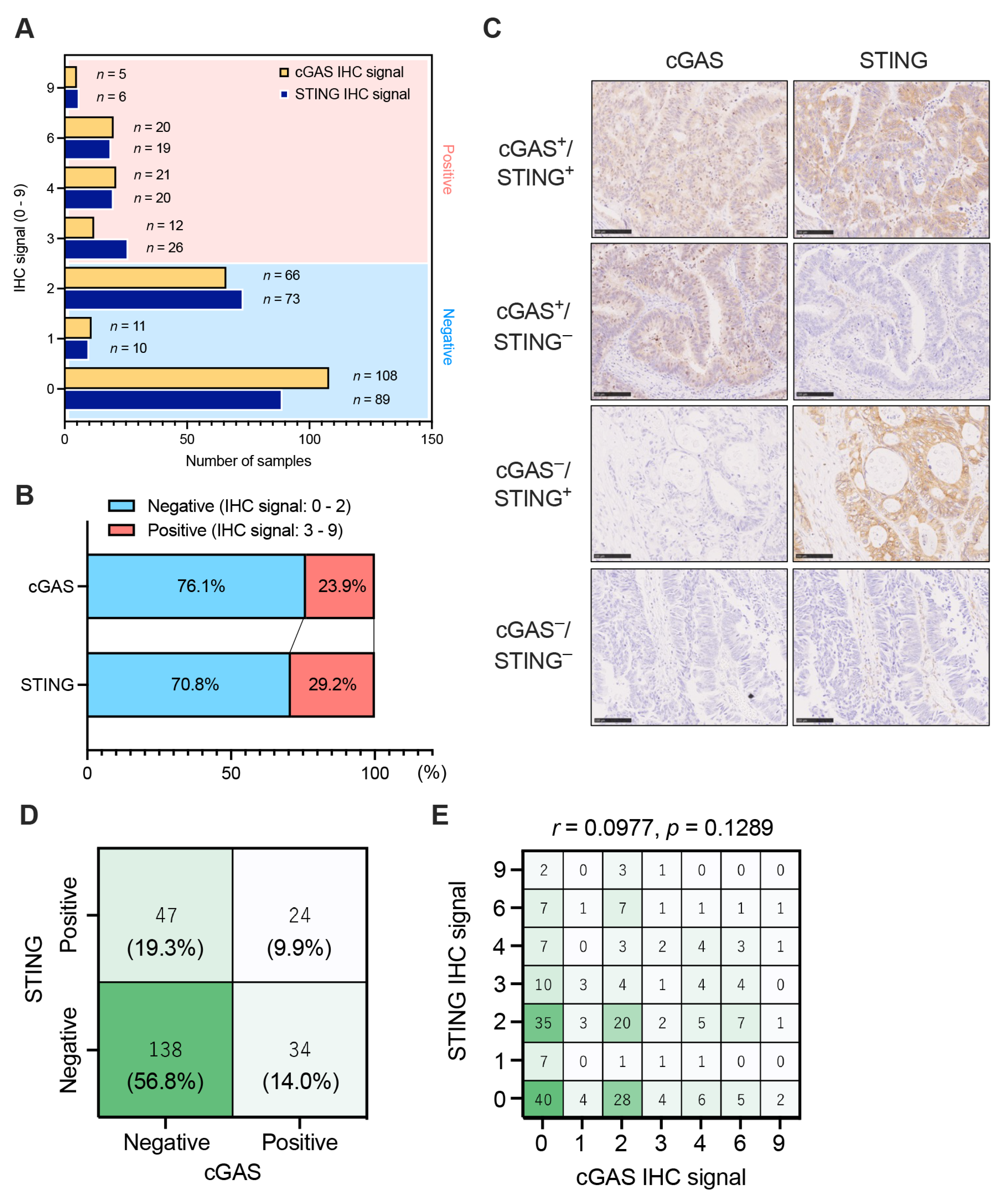


| pMMR CRC All | cGAS+/STING+ | cGAS+/STING− | cGAS−/STING+ | cGAS−/STING− | |||
|---|---|---|---|---|---|---|---|
| (n = 243) | (n = 24) | (n = 34) | (n = 47) | (n = 138) | p-Value | ||
| Age | <70 | 121 | 11 | 20 | 17 | 73 | 0.1523 |
| 70≤ | 122 | 13 | 14 | 30 | 65 | ||
| Gender | Male | 154 | 18 | 25 | 30 | 81 | 0.2393 |
| Female | 89 | 6 | 9 | 17 | 57 | ||
| Tumor location | Proximal | 78 | 12 | 16 | 16 | 34 | 0.0379 |
| Distal | 68 | 4 | 10 | 15 | 39 | ||
| Rectum | 97 | 8 | 8 | 16 | 65 | ||
| Tumor differentiation | Well/Moderate | 234 | 22 | 31 | 47 | 134 | 0.1116 |
| Poor | 9 | 2 | 3 | 0 | 4 | ||
| T stage | Tis | 13 | 3 | 0 | 6 | 4 | 0.0113 |
| T1 | 31 | 3 | 4 | 6 | 18 | ||
| T2 | 35 | 1 | 5 | 14 | 15 | ||
| T3 | 101 | 10 | 16 | 13 | 62 | ||
| T4 | 63 | 7 | 9 | 8 | 39 | ||
| N stage | N0 | 144 | 13 | 17 | 31 | 83 | 0.4967 |
| N1-3 | 99 | 11 | 17 | 16 | 55 | ||
| M stage | M0 | 213 | 20 | 30 | 44 | 119 | 0.5267 |
| M1 | 30 | 4 | 4 | 3 | 19 | ||
| TNM stage | 0 | 13 | 3 | 0 | 6 | 4 | 0.0658 |
| I | 52 | 4 | 7 | 15 | 26 | ||
| II | 72 | 6 | 9 | 9 | 48 | ||
| III | 76 | 7 | 14 | 14 | 41 | ||
| IV | 30 | 4 | 4 | 3 | 19 | ||
| PD-L1 | Positive | 12 | 2 | 3 | 2 | 5 | 0.5246 |
| Negative | 231 | 22 | 31 | 45 | 133 | ||
| Recurrence | Yes | 40 | 2 | 5 | 8 | 25 | 0.6752 |
| No | 180 | 19 | 26 | 37 | 98 | ||
| Not available | 23 | 3 | 3 | 2 | 15 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakajima, S.; Kaneta, A.; Okayama, H.; Saito, K.; Kikuchi, T.; Endo, E.; Matsumoto, T.; Fukai, S.; Sakuma, M.; Sato, T.; et al. The Impact of Tumor Cell-Intrinsic Expression of Cyclic GMP-AMP Synthase (cGAS)-Stimulator of Interferon Genes (STING) on the Infiltration of CD8+ T Cells and Clinical Outcomes in Mismatch Repair Proficient/Microsatellite Stable Colorectal Cancer. Cancers 2023, 15, 2826. https://doi.org/10.3390/cancers15102826
Nakajima S, Kaneta A, Okayama H, Saito K, Kikuchi T, Endo E, Matsumoto T, Fukai S, Sakuma M, Sato T, et al. The Impact of Tumor Cell-Intrinsic Expression of Cyclic GMP-AMP Synthase (cGAS)-Stimulator of Interferon Genes (STING) on the Infiltration of CD8+ T Cells and Clinical Outcomes in Mismatch Repair Proficient/Microsatellite Stable Colorectal Cancer. Cancers. 2023; 15(10):2826. https://doi.org/10.3390/cancers15102826
Chicago/Turabian StyleNakajima, Shotaro, Akinao Kaneta, Hirokazu Okayama, Katsuharu Saito, Tomohiro Kikuchi, Eisei Endo, Takuro Matsumoto, Satoshi Fukai, Mei Sakuma, Takahiro Sato, and et al. 2023. "The Impact of Tumor Cell-Intrinsic Expression of Cyclic GMP-AMP Synthase (cGAS)-Stimulator of Interferon Genes (STING) on the Infiltration of CD8+ T Cells and Clinical Outcomes in Mismatch Repair Proficient/Microsatellite Stable Colorectal Cancer" Cancers 15, no. 10: 2826. https://doi.org/10.3390/cancers15102826
APA StyleNakajima, S., Kaneta, A., Okayama, H., Saito, K., Kikuchi, T., Endo, E., Matsumoto, T., Fukai, S., Sakuma, M., Sato, T., Mimura, K., Saito, M., Saze, Z., Sakamoto, W., Onozawa, H., Momma, T., & Kono, K. (2023). The Impact of Tumor Cell-Intrinsic Expression of Cyclic GMP-AMP Synthase (cGAS)-Stimulator of Interferon Genes (STING) on the Infiltration of CD8+ T Cells and Clinical Outcomes in Mismatch Repair Proficient/Microsatellite Stable Colorectal Cancer. Cancers, 15(10), 2826. https://doi.org/10.3390/cancers15102826







