The Sweet Side of HIPK2
Abstract
Simple Summary
Abstract
1. Introduction
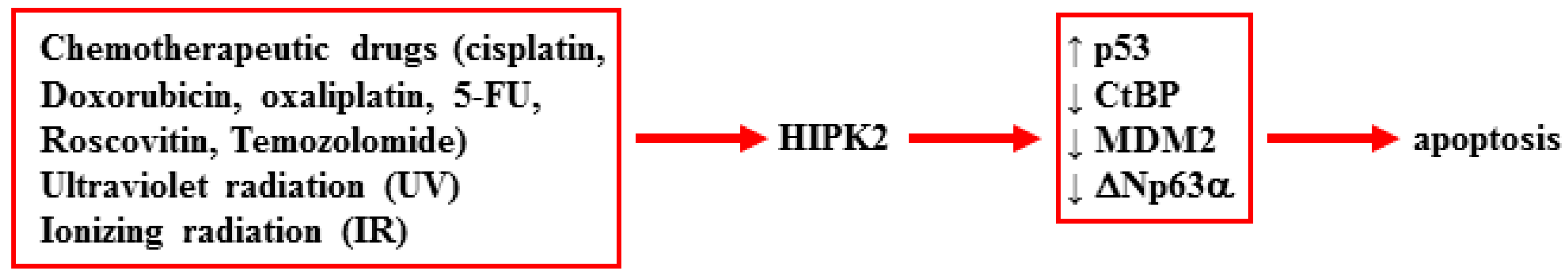
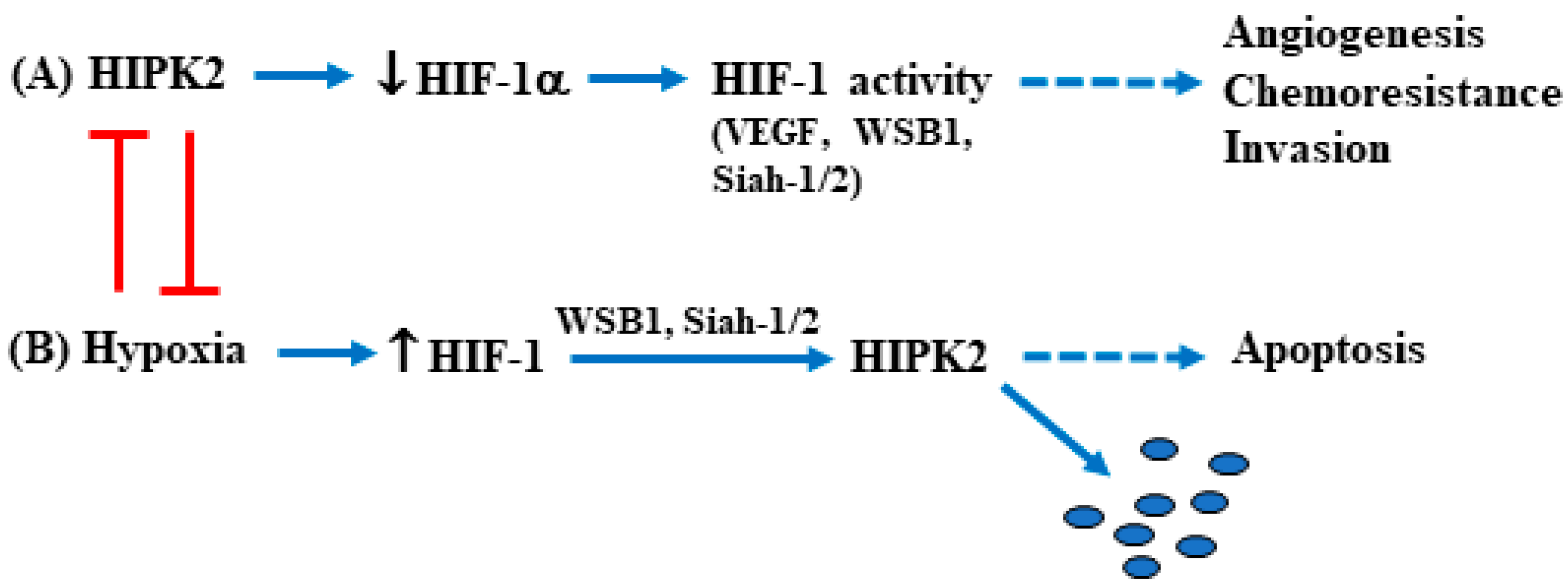
2. HIPK2 Function and Dysfunction
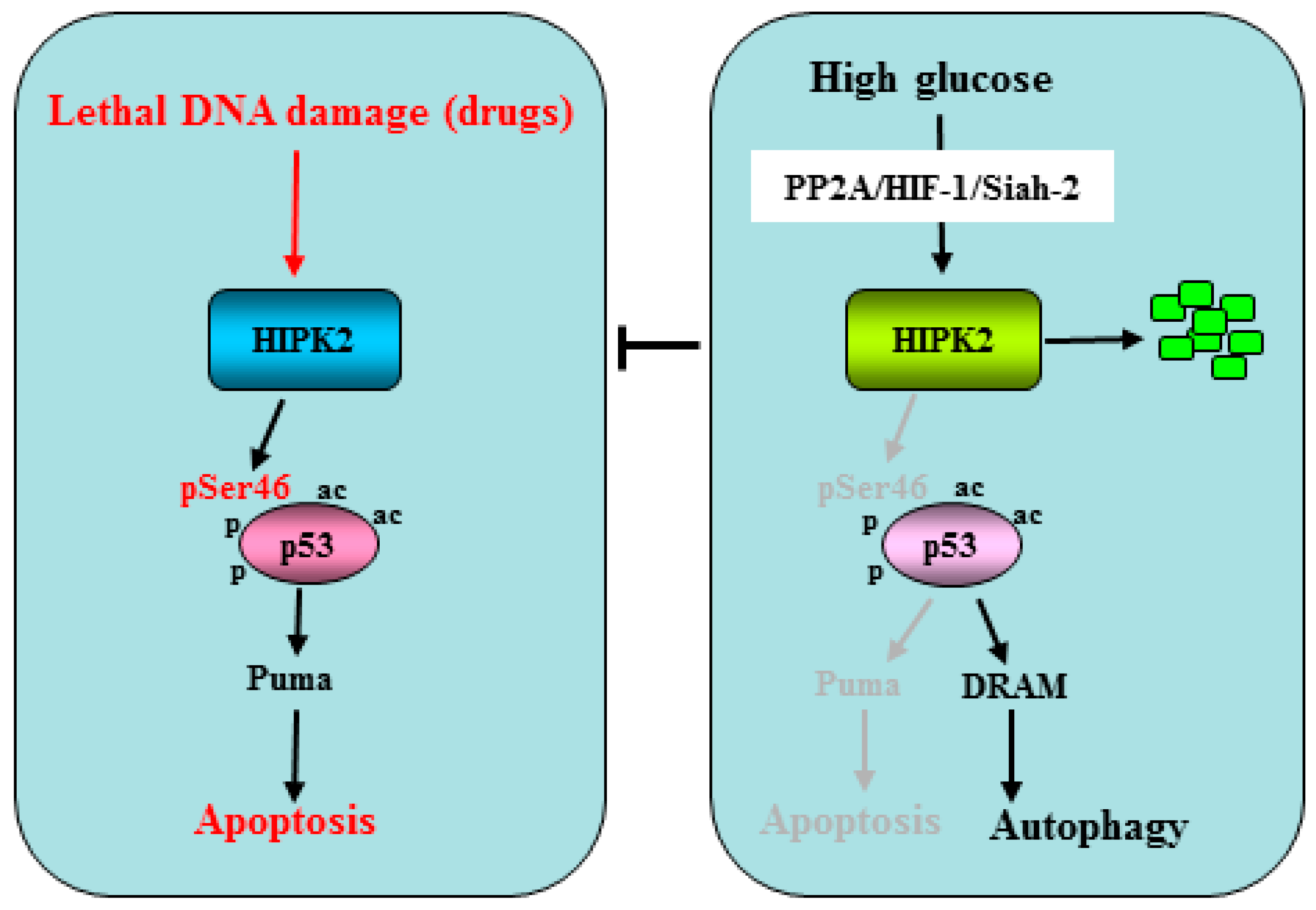
2.1. HIPK2 Regulation by Hyperglycemia in Tumors
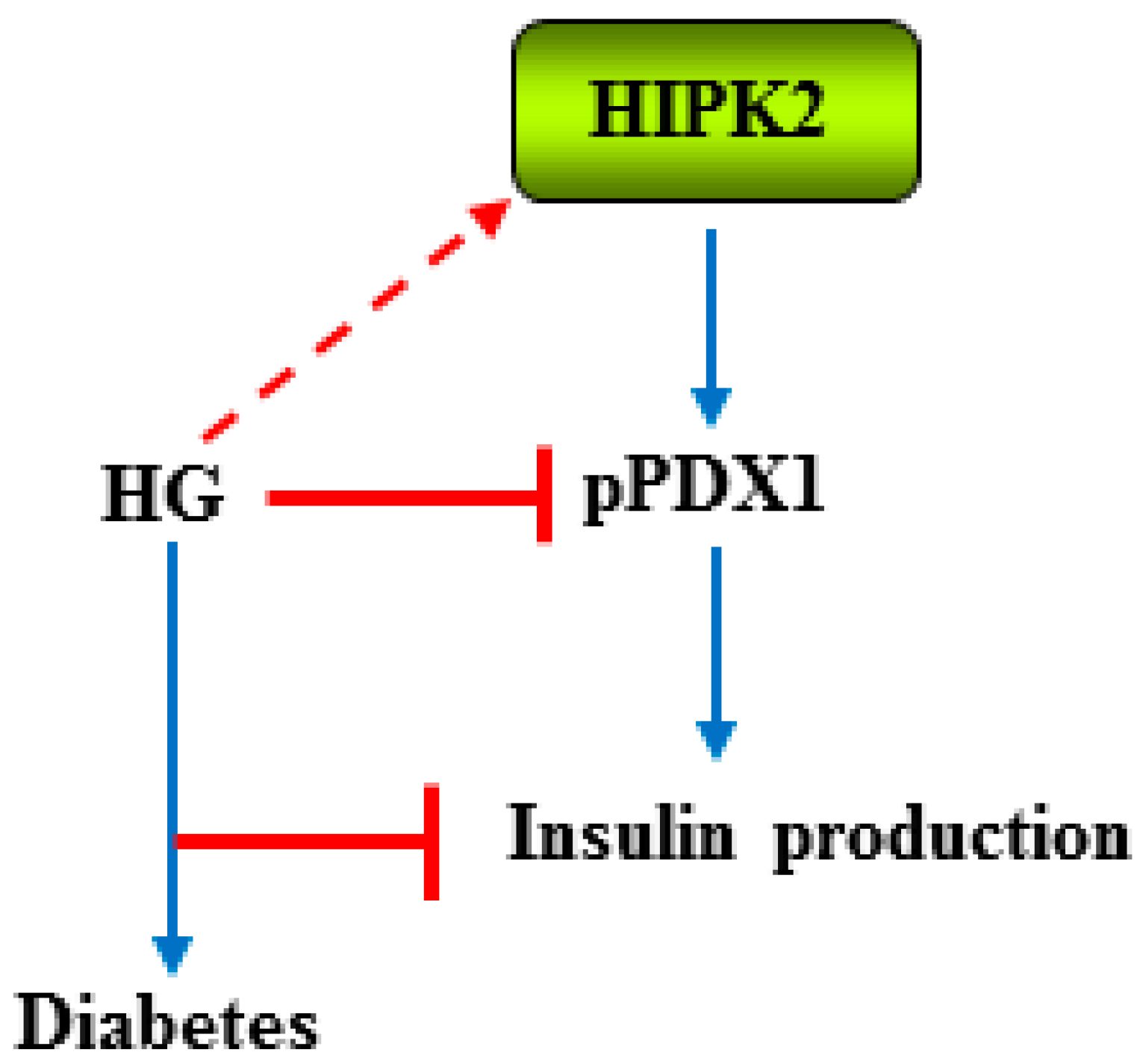
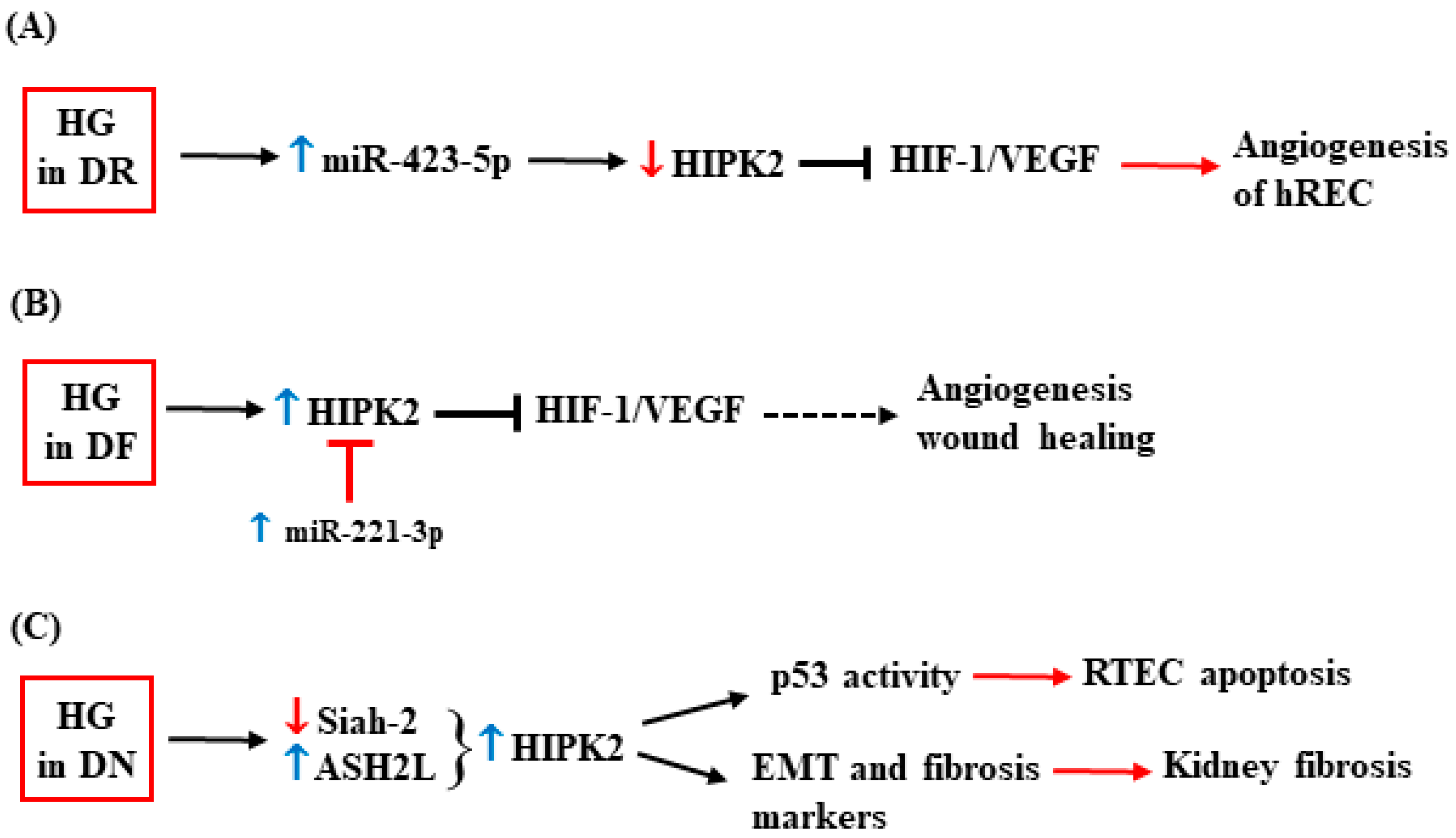
2.2. HIPK2 and Diabetes Complications
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schmidt, A.M. Highlighting diabetes mellitus: The epidemic continues. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e1–e8. [Google Scholar] [CrossRef] [PubMed]
- Engerman, R.L.; Kern, T.S. Hyperglycemia as a cause of diabetic retinopathy. Metabolism 1986, 35, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.T. Review of diabetes: Identification of markers for early detection, glycemic control, and monitoring clinical implications. J. Clin. Lab. Anal. 1993, 7, 293–300. [Google Scholar] [CrossRef]
- Ansari, P.; Akther, S.; Khan, J.; Islam, S.S.; Masud, S.R.; Rahman, A.; Seidel, V.; Abdel-Wahab, Y.H.A. Hyperglycaemia-linked diabetic foot complications and their management using conventional and alternative therapies. Appl. Sci. 2022, 12, 11777. [Google Scholar] [CrossRef]
- Martyn, J.A.J.; Kaneki, M.; Yasuhara, S. Obesity-induced insulin resistance and hyperglycemia: Etiologic factors and molecular mechanisms. Anesthesiology 2008, 109, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Mechanick, J.I. Metabolic mechanisms of stress hyperglycemia. J. Parent. Enter. Nutr. 2006, 30, 157–163. [Google Scholar] [CrossRef]
- Ryu, T.Y.; Park, J.; Scherer, P.E. Hyperglycemia as a risk factor for cancer progression. Diabetes Metab. J. 2014, 38, 330–336. [Google Scholar] [CrossRef]
- Li, W.; Zhang, X.; Sang, H.; Zhou, Y.; Shang, C.; Wang, Y.; Zhu, H. Effects of hyperglycemia on the progression of tumor diseases. J. Exp. Clin. Cancer Res. 2019, 38, 327. [Google Scholar] [CrossRef]
- Vishvakarma, N.K.; Kumar, A.; Singh, V.; Singh, S.M. Hyperglicaemia of tumor microenvironment modulates stage-dependent tumor progression and multidrug resistance: Implication of cell survival regulatory molecules and altered glucose transport. Mol. Carcinog. 2013, 52, 932–945. [Google Scholar] [CrossRef]
- Rahman, I.; Athar, T.; Islam, M. Type 2 diabetes, obesity, and cancer share some common and critical pathways. Front. Oncol. 2021, 10, 2020. [Google Scholar] [CrossRef]
- Gilardini Montani, M.S.; Granato, M.; Cuomo, L.; Valia, S.; Di Renzo, L.; D’Orazi, G.; Faggioni, A.; Cirone, M. High glucose and hyperglycemic sera from type 2 diabetic patients impair DC differentiation by inducing ROS and activating Wnt/beta-catenin and p38 MAPK. Biochem. Biophys. Acta-DIS 2016, 1862, 805–813. [Google Scholar] [CrossRef]
- Kim, Y.H.; Choi, C.Y.; Lee, S.J.; Conti, M.A.; Kim, Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 1998, 273, 25875–25879. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.G.; Mincheva, A.; Lichter, P.; Droge, W.; Schimtz, M.L. Human homeodomain-interacting protein kinase-2 (HIPK2) is a member of the DYRK family of protein kinases abd maps to chromosome 7q32-q34. Biochimie 2000, 82, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, C.; Prodosmo, A.; Mancini, F.; Iacovelli, S.; Sacchi, A.; Moretti, F.; Soddu, S. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell 2007, 25, 739–750. [Google Scholar] [CrossRef]
- He, Q.; Shi, J.; Sun, H.; An, J.; Huang, Y.; Sheikh, M.S. Characterization of human homeodomain-interacting protein kinase 4 (HIPK4) as a unique member of the HIPK family. Mol. Cell. Pharmacol. 2010, 2, 61–68. [Google Scholar]
- Feng, Y.; Zhou, L.; Sun, X.; Li, Q. Homeodomain-interacting protein kinase 2 (HIPK2): A promising target for anti-cancer therapies. Oncotarget 2017, 8, 20452–20461. [Google Scholar] [CrossRef]
- Schmitz, M.L.; Rodriguez-Gil, A.; Hornung, J. Integration of stress signals by homeodomain interacting protein kinases. Biol. Chem. 2014, 395, 375–386. [Google Scholar] [CrossRef]
- Calzado, M.A.; Renner, F.; Roscic, A.; Schitz, M.L. HIPK2: A versatile switchboard regulating the transcription machinery and cell death. Cell Cycle 2014, 6, 139–143. [Google Scholar] [CrossRef]
- Haas, J.; Bloesel, D.; Bacher, S.; Kracht, M.; Schmitz, M.L. Chromatin targeting of HIPK2 leads to acetylation-dependent chromatin decondensation. Front. Cell Dev. Biol. 2020, 8, 852. [Google Scholar] [CrossRef]
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain interacting protein kinase-2 phosphorylates p53 at Ser46 and mediates apoptosis. Nat. Cell Biol. 2002, 4, 11–19. [Google Scholar] [CrossRef]
- Iacovelli, S.; Ciuffini, L.; Lazzari, C.; Bracaglia, G.; Rinaldo, C.; Prodosmo, A.; Bartolazzi, A.; Sacchi, A.; Soddu, S. HIPK2 is involved in cell proliferation and its suppression promotes growth arrest independently of DNA damage. Cell Prolif. 2009, 42, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.G.; Glas, C.; Bitomsky, N. HIPK2: A tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. BioEssays 2013, 35, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Valente, D.; Bossi, G.; Moncada, A.; Tornincasa, M.; Indelicato, S.; Piscuoglio, S.; Karamitopoulou, E.D.; Bartolazzi, A.; Pierantoni, G.M.; Fusco, A.; et al. HIPK2 deficiency causes chromosomal instability by cytokinesis failure and increases tumorigenicity. Oncotarget 2015, 6, 10320–10334. [Google Scholar] [CrossRef]
- Blaquiere, J.A.; Verheyen, E.M. Homeodomain-interacting protein kinases: Diverse and complex roles in development and diseases. Curr. Opin. Dev. Biol. 2017, 123, 73–103. [Google Scholar] [CrossRef]
- Garufi, A.; Pistritto, G.; D’Orazi, G. HIPK2 as a novel regulator of fibrosis. Cancers 2023, 15, 1059. [Google Scholar] [CrossRef] [PubMed]
- Garufi, A.; D’Orazi, V.; Pistritto, G.; D’Orazi, G. HIPK2 in angiogenesis: A promising biomarker in cancer progression and in angiogenic diseases. Cancers 2023, 14, 1566. [Google Scholar] [CrossRef]
- Stanga, S.; Lanni, C.; Govono, S.; Uberti, D.; D’Orazi, G.; Racchi, M. Unfolded p53 in the pathogenesis of Alzheimer’s disease: Is HIPK2 the link? Aging 2010, 2, 545–554. [Google Scholar] [CrossRef]
- Sardina, F.; Conte, A.; Paladino, S.; Pierantoni, G.M.; Rinaldo, C. HIPK2 in the physiology of nervous system and its implications in neurological disorders. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119465. [Google Scholar] [CrossRef]
- Hofmann, T.G.; Moller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Dröge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoshimatsu, Y.; Hildebrand, J.; Frisch, S.M.; Goodman, R.H. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 2003, 115, 177–186. [Google Scholar] [CrossRef]
- Di Stefano, V.; Blandino, G.; Sacchi, A.; Soddu, S.; D’Orazi, G. HIPK2 neutralizes MDM2 inhibition by rescuing p53 transcriptional activity and apoptotic function. Oncogene 2004, 23, 5185–5192. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, C.; Prodosmo, A.; Siepi, F.; Rinaldo, C.; Galli, F.; Gentileschi, M.; Bartolazzi, A.; Costanzo, A.; Sacchi, A.; Guerrini, L.; et al. HIPK2 phosphorylates DNp63α and promotes its degradation in response to DNA damage. Oncogene 2011, 30, 4802–4813. [Google Scholar] [CrossRef] [PubMed]
- Puca, R.; Nardinocchi, L.; Gal, H.; Rechavi, G.; Amariglio, N.; Domany, E.; Notterman, D.A.; Scarsella, M.; Leonetti, C.; Sacchi, A.; et al. Reversible dysfunction of wild-type p53 following homeodomain interacting protein kinase-2 knockdown. Cancer Res. 2008, 15, 3707–3714. [Google Scholar] [CrossRef]
- Puca, R.; Nardinocchi, L.; Givol, D.; D’Orazi, G. Regulation of p53 activity by HIPK2: Molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 2010, 29, 4378–4387. [Google Scholar] [CrossRef]
- de la Vega, L.; Frobius, K.; Moreno, R.; Calzado, M.A.; Geng, H.; Schmitz, M.L. Control of nuclear HIPK2 localization and function by a SUMO interaction motif. Biochim. Biophys. Acta 2011, 1813, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Saul, V.V.; Schmitz, M.L. Posttranslational modifications regulate HIPK2, a driver of proliferative diseases. J. Mol. Med. 2013, 91, 1051–1058. [Google Scholar] [CrossRef]
- Choi, D.W.; Choi, C.Y. HIPK2 modification code for cell death and survival. Mol. Cell Oncol. 2014, 1, e955999. [Google Scholar] [CrossRef]
- de la Vega, L.; Grishina, I.; Moreno, R.; Krüger, M.; Braun, T.; Schmitz, M.L. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell 2012, 46, 472–483. [Google Scholar] [CrossRef]
- Wong, K.K.L.; Liu, T.W.; Parker, J.M.; Sinclair, D.A.R.; Chen, Y.Y.; Khoo, K.H.; Vocadlo, D.J.; Verheyen, E.M. The nutrient sensor OGT regulates Hipk stability and tumorigenic-like activities in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 2004–2013. [Google Scholar] [CrossRef]
- Chen, J.; Verheyen, E.M. Homeodomain-interacting protein kinase regulates Yorkie activity to promote tissue growth. Curr. Biol. 2012, 22, 1582–1586. [Google Scholar] [CrossRef]
- Oh, H.; Kato, M.; Deshpande, S.; Zhang, E.; Das, S.; Lanting, L.; Wang, M.; Natarajan, R. Inhibition of the processing of miR-25 by HIPK2-phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy. Sci. Rep. 2016, 6, 38789. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.; Sombroek, D.; Dauth, I.; Moehlenbrink, J.; Scheuermann, K.; Crone, J.; Hofmann, T.G. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint ATM and ATR. Nat. Cell Biol. 2008, 10, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Choi, D.W.; Kim, E.A.; Choi, C.Y. Stabilization of HIPK2 by escape from proteasomal degradation by the E3 ubiquitin ligase Siah1. Cancer Lett. 2009, 279, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.P.; Sabatini, D.M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, C.M.; Sodi, V.L.; Reginato, M.J. O-GlcNAcylation in cancer biology: Linking metabolism and signaling. J. Mol. Biol. 2016, 428, 3282–3294. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, H.; Yeh, T.H.; Yu, J.; Sharma, M.K.; Perry, A.; Leonard, J.R.; Watson, M.A.; Gutmann, D.H.; Nagarajan, R. High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene 2008, 27, 4745–4751. [Google Scholar] [CrossRef] [PubMed]
- Al-Beiti, M.A.; Lu, X. Expression of HIPK2 in cervical cancer: Correlation with clinicopathology and prognosis. Aust. N. Z. J. Obstet. Gynaecol. 2008, 48, 329–336. [Google Scholar] [CrossRef]
- Bon, G.; Di Carlo, S.E.; Folgiero, V.; Avetrani, P.; Lazzari, C.; D’Orazi, G.; Brizzi, M.F.; Sacchi, A.; Soddu, S.; Blandino, G.; et al. Negative regulation of beta(β) integrin transcription by homeodomain-interacting protein kinase e and p53 impairs tumor progression. Cancer Res. 2009, 69, 5978–5986. [Google Scholar] [CrossRef]
- Polonio-Vallon, T.; Kirkpatrick, J.; Krijgsveld, J.; Hofmann, T.G. Src kinase modulates the apoptotic p53 pathway by altering HIPK2 localization. Cell Cycle 2014, 13, 115–125. [Google Scholar] [CrossRef]
- Bon, G.; Folgiero, V.; Di Carlo, S.; Sacchi, A.; Falcioni, R. The involvement of alpha(6)beta(4) integrin in the mechanisms that regulate breast cancer progression. Breast Cancer Res. 2007, 9, 203. [Google Scholar] [CrossRef]
- Nardinocchi, L.; Puca, R.; Guidolin, D.; Belloni, A.S.; Bossi, G.; Michiels, C.; Sacchi, A.; Onisto, M.; D’Orazi, G. Transcriptional regulation of hypoxia-inducible factor 1alpha by HIPK2 suggests a novel mechanism to restrain tumor growth. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2009, 1793, 368–377. [Google Scholar] [CrossRef] [PubMed]
- D’Orazi, G.; Sciulli, M.G.; Di Stefano, V.; Riccioni, S.; Frattini, M.; Falcioni, R.; Bertario, L.; Sacchi, A.; Patrignani, P. Homeodomaininteracting protein kinase-2 restrains cytosolic phospholipase A2-dependent prostaglandin E2 generation in human colorectal cancer cells. Clin. Cancer Res. 2006, 12, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Wee, H.J.; Voon, D.C.C.; Bae, S.C.; Ito, Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: Implications for leukemogenesis. Blood 2008, 112, 3777–3787. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.; Bhadauriya, P.; Ganesh, S. Heat shock modulates the subcellular localization, stability and activity of HIPK2. Biochem. Biophys. Res. Commun. 2016, 472, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Arai, Y.; Harada, H.; Shima, Y.; Yoshida, H.; Rokudai, S.; Aikawa, Y.; Kimura, A.; Kitabayashi, I. Mutations of the HIPK2 gene in acute myeloid leukemia and myelodisplatic syndrome impair AML-1 and p53-mediated transcription. Oncogene 2007, 26, 7231–7239. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.S.; Kim, S.J.; Cho, S.W.; Park, Y.J.; Tae, K.; Choi, C.Y. Functional impairment of the HIPK2 small ubiquitin-like modifier (SUMO)-interacting motif in acute myeloid leukemia. Am. J. Cancer Res. 2019, 9, 94–107. [Google Scholar]
- Ricci, A.; Cherubini, E.; Ulivieri, A.; Lavra, L.; Sciacchitano, S.; Scozzi, D.; Mancini, R.; Ciliberto, G.; Bartolazzi, A.; Bruno, P.; et al. Homeodomain-interacting protein kinase2 in human idiopathic pulmonary fibrosis. J. Cell Physiol. 2013, 228, 235–241. [Google Scholar] [CrossRef]
- Muschik, D.; Braspenning-Wesch, I.; Stockgleth, E.; Rosl, F.; Hofmann, T.G.; Nindl, I. Cutaneous HPV23 E6 prevents p53 phosphorylation through interaction with HIPK2. PLoS ONE 2011, 6, e27655. [Google Scholar] [CrossRef]
- Orth, G.; Jablonska, S.; Favre, M.; Croissant, O.; Jarzabek-Chorzelska, M.; Rzesa, G. Characterization of two types of human papillomaviruses in lesions of epidermodysplasia verruciformis. Proc. Natl. Acad. Sci. USA 1978, 75, 1537–1541. [Google Scholar] [CrossRef]
- Di Stefano, V.; Mattiussi, M.; Sacchi, A.; D’Orazi, G. HIPK2 inhibits both MDM2 gene and protein by, respectively, p53-dependent and independent regulations. FEBS Lett. 2005, 579, 5473–5480. [Google Scholar] [CrossRef]
- Lee, I.; Kim, C.E.; Cho, H.; Im, H.; Shin, K.S.; Kang, S.J. TRAF2 regulates the protein stability of HIPK2. Biochem. Biophys. Res. Commun. 2022, 627, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Rego, E.M.; Wang, Z.G.; Peruzzi, D.; He, L.Z.; Cordon-Cardo, C.; Pandolfi, P.P. Role of promyelocytic leukemia (Pml) protein in tumor suppression. J. Exp. Med. 2001, 193, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Shima, Y.; Shima, T.; Chiba, T.; Irimura, T.; Pandolfi, P.P.; Kitabayashi, I. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol. Cell. Biol. 2008, 28, 7126–7138. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W.; Seo, Y.M.; Kim, E.A.; Sung, K.S.; Ahn, J.W.; Park, S.J.; Lee, S.R.; Choi, C.Y. Ubiquitination and degradation of homeodomain- interacting protein kinase-2 by WD40 repeat/SOCS box protein WSB-1. J. Biol. Chem. 2008, 283, 4682–4689. [Google Scholar] [CrossRef] [PubMed]
- Benita, Y.; Kikuchi, H.; Smith, A.D.; Zhang, M.Q.; Chung, D.C.; Xavier, R.J. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucl. Acid Res. 2009, 37, 4587–4602. [Google Scholar] [CrossRef] [PubMed]
- Calzado, M.A.; de la Vega, L.; Möller, A.; Bowtell, D.D.; Schmitz, M.L. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat. Cell Biol. 2009, 11, 85–91. [Google Scholar] [CrossRef]
- Hernandez, C.; Huebener, P.; Pradere, J.P.; Antoine, D.J.; Friedman, R.A.; Schwabe, R.F. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J. Clin. Investig. 2018, 128, 2436–2451. [Google Scholar] [CrossRef]
- Zhu, W.; Li, J.; Zhang, Y.; Zhu, Z.; Liu, H.; Lin, Y.; Hu, A.; Zhou, J.; Ren, H.; Shi, X. Inhibition of HMGB1 suppresses hepatocellular carcinoma progression via HIPK2-mediated autophagic degradation of ZEB1. Front. Oncol. 2021, 11, 599124. [Google Scholar] [CrossRef]
- Jiang, B.H.; Semenza, G.L.; Bauer, H.; Marti, H. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am. J. Physiol. 1996, 271, C1172–C1180. [Google Scholar] [CrossRef]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Nardinocchi, L.; Puca, R.; Sacchi, A.; Rechavi, G.; Givol, D.; D’Orazi, G. Targeting hypoxia in cancer cells by restoring homeodomain interacting protein kinase 2 and p53 activity and suppressing HIF-1alpha. PLoS ONE 2009, 4, e6819. [Google Scholar] [CrossRef] [PubMed]
- Nardinocchi, L.; Puca, R.; Givol, D.; D’Orazi, G. HIPK2-a therapeutical target to be (re)activated for tumor suppression: Role in p53 activation and HIF-1 α inhibition. Cell Cycle 2010, 9, 1270–1275. [Google Scholar] [CrossRef] [PubMed]
- Nardinocchi, L.; Pantisano, V.; Puca, R.; Porru, M.; Aiello, A.; Grasselli, A.; Leonetti, C.; Safran, M.; Rechavi, G.; Givol, D.; et al. Zinc downregulates HIF-1alpha and inhibits its activity in tumor cells in vitro and in vivo. PLoS ONE 2010, 5, e15048. [Google Scholar] [CrossRef] [PubMed]
- Conte, A.; Pierantoni, G.M. Update on the regulation of KIPK1, HIPK2 and HIPK3 protein kinases by microRNAs. Microrna 2018, 7, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Y.; Yu, C.H.; Zhang, H.H.; Zhang, S.Z.; Yu, W.Y.; Yang, Y.; Chen, Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int. J. Biol. Macromol. 2019, 132, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, H.; Choi, Y.J.; Kang, M.H. Exosomal miR-1260b derived from non-small cell lung cancer promotes tumor metastasis through the inhibition of HIPK2. Cell Death Dis. 2021, 12, 747. [Google Scholar] [CrossRef]
- Garufi, A.; Trisciuoglio, D.; Cirone, M.; D’Orazi, G. ZnCl2 sustains the Adriamycin-induced cell death inhibited by high glucose. Cell Death Dis. 2016, 7, e2280. [Google Scholar] [CrossRef]
- Isoe, T.; Makino, Y.; Mizumoto, K.; Sakagami, H.; Fujita, Y.; Honjo, J.; Takiyama, Y.; Itoh, H.; Haneda, M. High glucose activates HIF-1-mediated signal transduction in glomerular mesangial cells through a carbohydrate response element binding protein. Kidney Int. 2010, 78, 48–59. [Google Scholar] [CrossRef]
- Baldari, S.; Garufi, A.; Granato, M.; Cuomo, L.; Pistritto, G.; Cirone, M.; D’Orazi, G. Hyperglycemia triggers HIPK2 protein degradation. Oncotarget 2017, 8, 1190–1203. [Google Scholar] [CrossRef]
- Garufi, A.; D’Orazi, G. High glucose dephosphorylates serine 46 and inhibits p53 apoptotic activity. J. Exp. Clin. Cancer Res. 2014, 33, 79. [Google Scholar] [CrossRef]
- Mi, J.; Bolesta, E.; Brautigan, D.L.; Larner, J.M. PP2A regulates ionizing radiation-induced apoptosis through Ser46 phosphorylation of p53. Mol. Cancer Ther. 2009, 8, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Garufi, A.; Pistritto, G.; Baldari, S.; Toietta, G.; Cirone, M.; D’Orazi, G. p53-dependent PUMA to DRAM antagonistic interplay as a key molecular switch in cell-fate decision in normal/high glucose conditions. J. Exp. Clin. Cancer Res. 2017, 36, 126. [Google Scholar] [CrossRef] [PubMed]
- Cirone, M.; Gilardini Montani, M.S.; Granato, M.; Garufi, A.; D’Orazi, G. Autophagy manipulation as a strategy for efficient anticancer therapies: Possible consequences. J. Exp. Clin. Cancer Res. 2019, 38, 262. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.; Gueven, N. The complexity of p53 stabilization and activation. Cell Death Diff. 2006, 13, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, J.; Gan, S.; Guan, H.; Zhou, Y.; Ouyang, Q.; Shi, J. DNA damage strength modulates a bimodal switch of p53 dynamics for cell-fate control. BMC Biology 2013, 11, 73. [Google Scholar] [CrossRef]
- Gerards, M.C.; van der Velde, D.L.; Baars, J.W.; Brandjes, D.P.; Hoekstra, J.B.; Vriesendorp, T.M.; Gerdes, V.E. Impact of hyperglycemia on the efficacy of chemotherapy—A systematic review of preclinical studies. Crit. Rev. Oncol. Hematol. 2017, 113, 235–341. [Google Scholar] [CrossRef]
- Garufi, A.; Traversi, G.; Gilardini Montani, M.S.; D’Orazi, V.; Pistritto, G.; Cirone, M.; D’Orazi, G. Reduced chemotherapeutic sensitivity in high glucose condition: Implication of antioxidant response. Oncotarget 2019, 10, 4691–4702. [Google Scholar] [CrossRef]
- Ma, Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the hallmarks of cancer. Cancer Cell 2018, 34, 20–43. [Google Scholar] [CrossRef]
- Garufi, A.; Pistritto, G.; D’Orazi, V.; Cirone, M.; D’Orazi, G. The impact of NRF2 inhibition on drug-induced colon cancer cell death and p53 activity: A pilot study. Biomolecules 2022, 12, 461. [Google Scholar] [CrossRef]
- Torrente, L.; Sanchez, C.; Moreno, R.; Chowdhry, S.; Cabello, P.; Isono, K.; Koseki, H.; Honda, T.; Hayes, J.D.; Dinkova-Kostova, A.T.; et al. Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene 2017, 36, 6204–6212. [Google Scholar] [CrossRef] [PubMed]
- Toth, R.K.; Warfel, N.A. Strange bedfellows: Nuclear factor, Erythroid 2-like 2 (Nrf2) and hypoxia-inducible factor 1 (HIF-1) in tumor hypoxia. Antioxidants 2017, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Kong, F.; Wu, X.; Ren, X.; Wu, S.; Wu, K.; Iang, Z.; Zhang, W. High glucose promotes tumor invasion and increases metastasis-associated protein expression in human lung epithelial cells by upregulating heme oxygenase-1 via reactive oxygen species or the TGF-β1/PI3K/Akt signaling pathway. Cell. Physiol. Biochem. 2015, 35, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, J.; Xi, Y.; Wang, F.; Sha, H.; Luo, L.; Zhu, Y.; Hong, X.; Bu, S. High glucose induces epithelial-mesenchymal transition and results in the migration and invasion of colorectal cancer cells. Exp. Ther. Med. 2018, 16, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Nodale, C.; Sheffer, M.; Jacob-Hirsch, J.; Folgiero, V.; Falcioni, R.; Aiello, A.; Garufi, A.; Rechavi, G.; Givol, D.; D’Orazi, D. HIPK2 downregulates vimentin and inhibits breast cancer cell invasion. Cancer Biol. Ther. 2012, 13, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S.; Beck, F.W.J.; Snell, D.C.; Kucuk, O. Zinc in cancer prevention. Nutr. Cancer 2009, 61, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Sheffer, S.; Simon, A.J.; Jacob-Hirsch, J.; Rechavi, G.; Domany, E.; Givol, D.; D’Orazi, G. Genome-wide analysis discloses reversal of the hypoxia-induced changes of gene expression in colon cancer cells by zinc supplementation. Oncotarget 2011, 2, 1191–1202. [Google Scholar] [CrossRef]
- Cirone, M.; Garufi, A.; Di Renzo, L.; Granato, M.; Faggioni, A.; D’Orazi, G. Zinc supplementation is required for the cytotoxic and immunogenic effects of chemotherapy in chemoresistant p53-functionally deficient cells. Oncoimmunology 2013, 2, e26198. [Google Scholar] [CrossRef]
- Barman, S.; Srinivasan, K. Zinc supplementation alleviates hyperglycemia and associated metabolic abnormalities in streptozotocin-induced diabetic rats. Can. J. Physiol. Pharmacol. 2016, 94, 1356–1365. [Google Scholar] [CrossRef]
- Wang, X.; Wu, W.; Zheng, W.; Fang, X.; Chen, L.; Rink, L.; Min, J.; Wang, F. Zinc supplementation improves glycemic control for diabetes prevention and management: A systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 2019, 110, 76–90. [Google Scholar] [CrossRef]
- Boucher, M.J.; Simoneau, M.; Edlund, H. The homeodomain-interacting protein kinase 2 regulates insulin promoter factor-1 pancreatic duodenal homeobox-1 transcriptional activity. Endocrinology 2009, 150, 87–97. [Google Scholar] [CrossRef] [PubMed]
- An, R.; da Silva, X.G.; Semplici, F.; Vakhshouri, S.; Hao, H.X.; Rutter, J.; Pagano, M.A.; Meggio, F.; Pinna, L.A.; Rutter, G.A. Pancreatic and duodenal homeobox 1 (PDX1) phosphorylation at serine-269 is HIPK2-dependent and affects PDX1 subnuclear localization. Biochem. Biophys. Res. Commun. 2010, 399, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, R.K.; Yu, S.M.; Flores, L.E.; Jhala, U.S. Glucose regulates steady-state levels of PDX1 via the reciprocal actions of GSK3 and Akt kinases. J. Biol. Chem. 2009, 285, 3406–3416. [Google Scholar] [CrossRef] [PubMed]
- Ahlgren, U.; Jonsson, J.; Jonsson, L.; Simu, K.; Edlund, H. Beta-cell-specific inactivation of the mouse Ipf1/PDX1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998, 12, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Hani, E.H.; Stoffers, D.A.; Chevre, J.C.; Durand, E.; Stanojevic, V.; Dina, C.; Habener, J.F.; Froguel, P. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J. Clin. Investig. 1999, 104, R41–R48. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lo, A.C.-Y. Diabetic retinopathy: Pathophysiology and treatments. Int. J. Mol. Sci. 2018, 19, 1816. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, S.; Tramontano, M.; Costa, V.; Aprile, M.; Ciccodicola, A. Diabetic retinopathy: Are lncRNAs new molecular players and targets? Antioxidant 2022, 11, 2021. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Zhao, Y.; Xu, J.; Li, W.J.; Chen, Y.; Sun, H.J. NFE2/miR-423-5p/TFF1 axis regulates high glucose-induced apoptosis in retinal pigment epithelial cells. BMC Mol. Cell Biol. 2019, 20, 39. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhao, Y.; Sun, H.; Hu, J.; Li, W.; Gao, L. MiR-423-5p activated by E2F1 promotes neovascularization in diabetic retinopathy by targeting HIPK2. Diabetol. Metab. Syndr. 2021, 13, 152. [Google Scholar] [CrossRef]
- Fui, L.W.; Lok, M.P.W.; Govindasamy, V.; Yong, T.K.; Lek, T.K.; Das, A.K. Understanding the multifaceted mechanisms of diabetic wound healing and therapeutic applications of stem cells conditioned medium in the healing process. J. Tissue Eng. Regen. Med. 2019, 13, 2218–2233. [Google Scholar] [CrossRef]
- Zhang, P.; Lu, J.; Jing, Y.; Tang, S.; Zhu, D.; Bi, Y. Global epidemiology of diabetic foot ulceration: A systematic review and meta-analysis. Ann. Med. 2017, 49, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Panunzi, A.; Madotto, F.; Sangalli, E.; Riccio, F.; Sganzaroli, A.B.; Galenda, P.; Bertulessi, A.; Barmina, M.F.; Ludovico, O.; Fortunato, O.; et al. Results of a prospective observational study of autologous peripheral blood mononuclear cell therapy for no-option critical limb-threatening ischemia and severe diabetic foot ulcers. Cardiovasc. Diabetol. 2022, 21, 196. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Bai, S.; Cao, Y.; Liu, L.; Fang, Y.; Du, J.; Luo, L.; Chen, M.; Shen, B.; Zhang, Q. miRNA-221-3p in endothelial progenitor cell-derived exosomes accelerates skin wound healing in diabetic mice. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Liu, L.; Zhang, X.; Chang, H.; Ma, S.; Xie, Z.; Tang, S.; Ju, X.; Zhu, H.; Shen, B.; et al. MiR-221-3p targets HIPK2 to promote diabetic wound healing. Microvasc. Res. 2022, 140, 104306. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C.; Brownlee, M.; Susztak, K.; Sharma, K.; Jandeleit-Dahm, K.A.M.; Zoungas, S.; Rossing, P.; Groop, P.-H.; Cooper, M.E. Diabetic kidney disease. Nat. Rev. Dis. Primers 2015, 1, 15018. [Google Scholar] [CrossRef]
- Jin, Y.; Ratnam, K.; Chuang, P.Y.; Fan, Y.; Zhong, Y.; Dai, Y.; Mazloom, A.R.; Chen, E.Y.; D’Agati, V.; Xiong, H.; et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat. Med. 2012, 18, 580–588. [Google Scholar] [CrossRef]
- Chen, W.; Mook, R.A., Jr.; Premont, R.T.; Wang, J. Niclosamide: Beyond an antihelminthic drug. Cell. Signal. 2018, 41, 89–96. [Google Scholar] [CrossRef]
- Chang, X.; Zhen, X.; Liu, J.; Ren, X.; Hu, Z.; Zhou, Z.; Zhu, F.; Ding, K.; Nie, J. The antihelmenthic phosphate niclosamide impedes renal fibrosis by inhibiting homeodomain-interacting protein kinase 2 expression. Kidney Int. 2017, 92, 612–624. [Google Scholar] [CrossRef]
- Zhong, W.; Hong, C.; Dong, Y.; Li, Y.; Xiao, C.; Liu, X. ASH2L aggravates fibrosis and inflammation through HIPK2 in high glucose-induced glomerular mesangial cells. Genes 2022, 13, 2244. [Google Scholar] [CrossRef]
| Molecules and Stimuli Involved | Mechanisms of HIPK2 Inactivation | Tissue | Cells | Ref. |
|---|---|---|---|---|
| Integrin alpha(6)beta(4) | Cytoplasmic localization | + | + | [48] |
| Src | Cytoplasmic localization | + | [49] | |
| CBF-β-SMMHC | Cytoplasmic localization | + | [53] | |
| HPV23 E6 | Impairment of PML-NB localization | + | + | [58] |
| Sub-lethal heat shock | Cytoplasmic localization | + | [54] | |
| Gene mutation (in AML) | Downregulation | + | + | [55,56] |
| LOH (in IPF) | Downregulation | + | [57] | |
| Overexpressed ExomiR-1229 | Downregulation | + | + | [75] |
| Overexpressed ExomiR-1260b | Downregulation | + | + | [76] |
| MDM2 | Protein degradation | + | [14] | |
| Siah-1 | Protein degradation | + | [42,43] | |
| Siah-2 | Protein degradation | + | [66,68] | |
| PML-RARα | Protein degradation | [63] | ||
| TRAF-2 | Protein degradation | + | [61] | |
| WSB-1 | Protein degradation | + | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garufi, A.; D’Orazi, V.; Pistritto, G.; Cirone, M.; D’Orazi, G. The Sweet Side of HIPK2. Cancers 2023, 15, 2678. https://doi.org/10.3390/cancers15102678
Garufi A, D’Orazi V, Pistritto G, Cirone M, D’Orazi G. The Sweet Side of HIPK2. Cancers. 2023; 15(10):2678. https://doi.org/10.3390/cancers15102678
Chicago/Turabian StyleGarufi, Alessia, Valerio D’Orazi, Giuseppa Pistritto, Mara Cirone, and Gabriella D’Orazi. 2023. "The Sweet Side of HIPK2" Cancers 15, no. 10: 2678. https://doi.org/10.3390/cancers15102678
APA StyleGarufi, A., D’Orazi, V., Pistritto, G., Cirone, M., & D’Orazi, G. (2023). The Sweet Side of HIPK2. Cancers, 15(10), 2678. https://doi.org/10.3390/cancers15102678







