Sex- and Co-Mutation-Dependent Prognosis in Patients with SMARCA4-Mutated Malignancies
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Population
2.2. StrataNGS
2.3. Treatment
2.4. Statistical Analysis
3. Results
3.1. Demographics
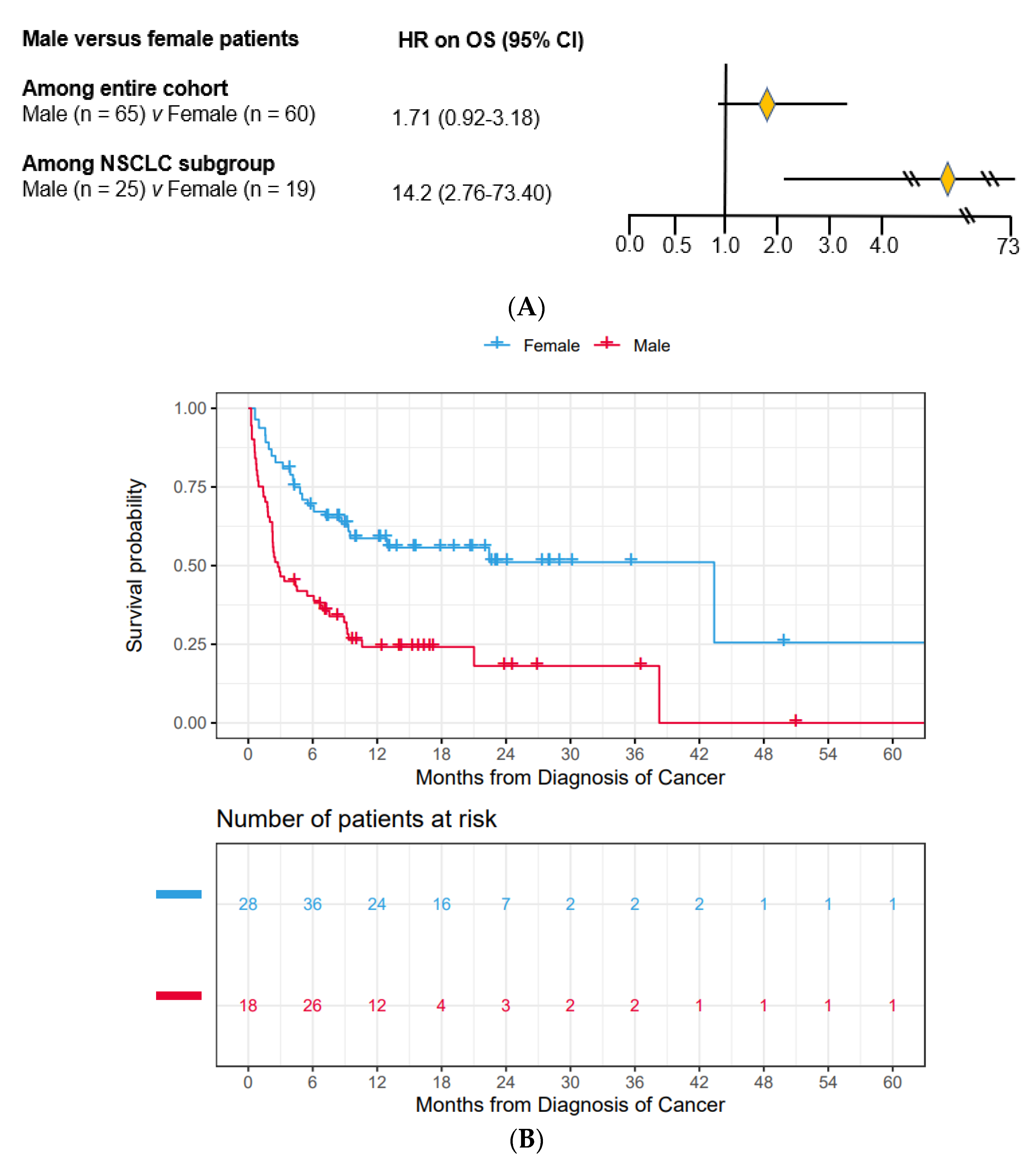
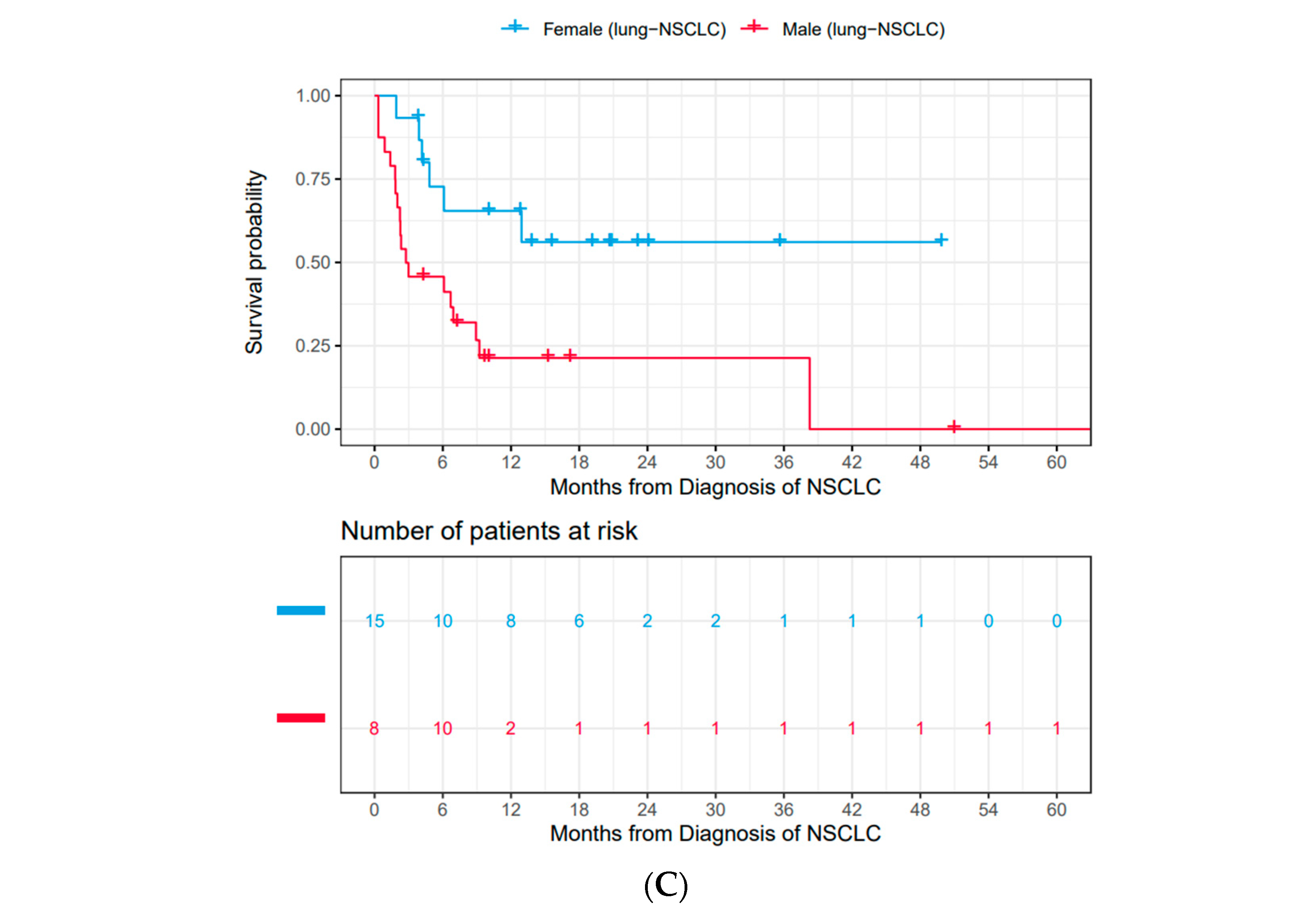
3.2. Male Patients Had Substantially Worse OS Compared to Female Patients
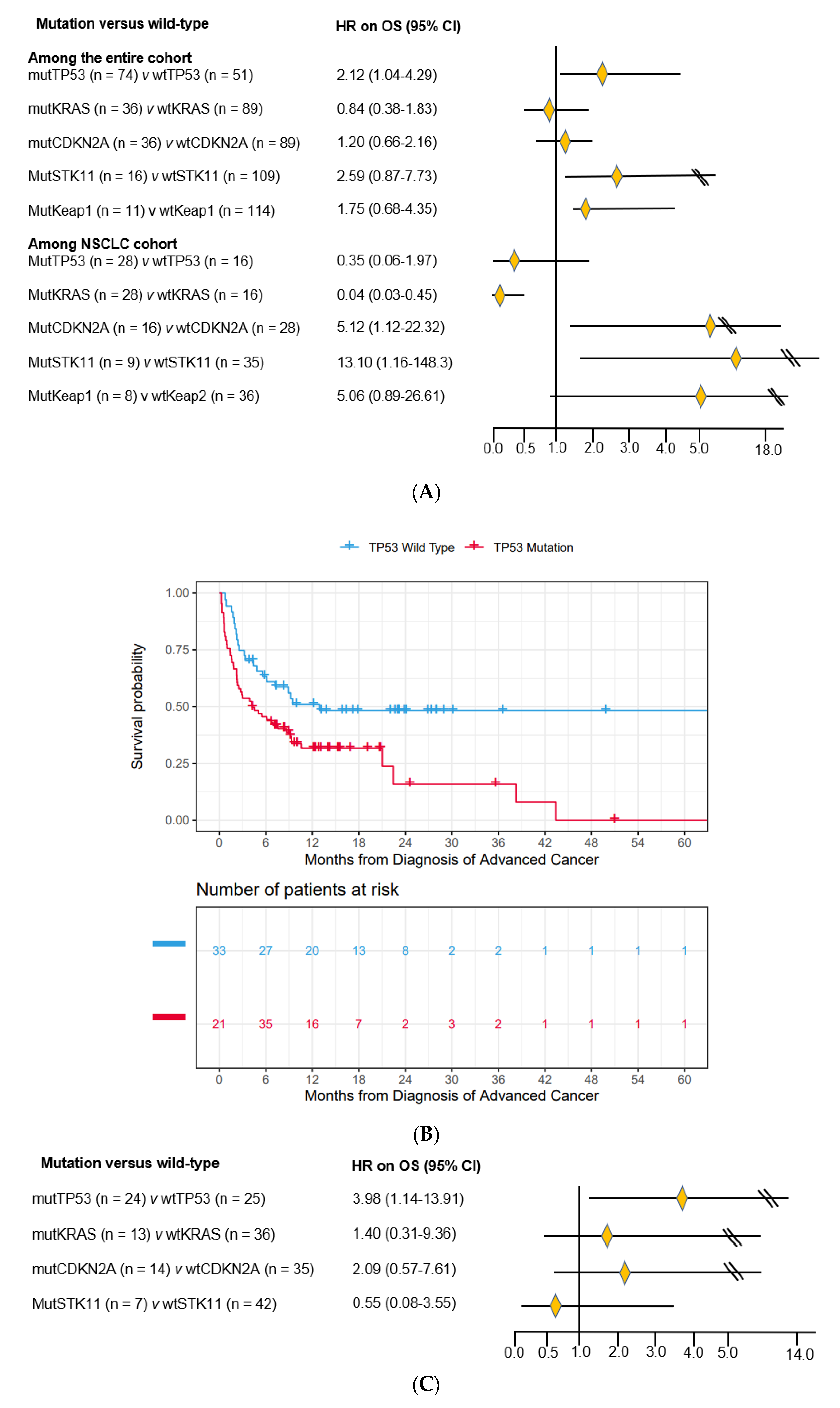
3.3. Impact of Co-Mutations on Prognosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiong, Y.; Shang, C.; Twu, K.Y.; Hang, C.T.; Yang, J.; Han, P.; Lin, C.Y.; Lin, C.J.; Tsai, F.C.; et al. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc. Natl. Acad. Sci. USA 2013, 110, 1738–1743. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Li, W.; Shang, C.; Chen, R.M.; Han, P.; Yang, J.; Stankunas, K.; Wu, B.; Pan, M.; Zhou, B.; et al. Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev. Cell 2013, 25, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Bultman, S.; Gebuhr, T.; Yee, D.; La Mantia, C.; Nicholson, J.; Gilliam, A.; Randazzo, F.; Metzger, D.; Chambon, P.; Crabtree, G.; et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 2000, 6, 1287–1295. [Google Scholar] [CrossRef]
- Pastorczak, A.; Krajewska, K.; Urbanska, Z.; Szmyd, B.; Salacinska-Los, E.; Kobos, J.; Mlynarski, W.; Trelinska, J. Ovarian carcinoma in children with constitutional mutation of SMARCA4: Single-family report and literature review. Fam. Cancer 2021, 20, 355–362. [Google Scholar] [CrossRef]
- Connor, Y.D.; Miao, D.; Lin, D.I.; Hayne, C.; Howitt, B.E.; Dalrymple, J.L.; DeLeonardis, K.R.; Hacker, M.R.; Esselen, K.M.; Shea, M. Germline mutations of SMARCA4 in small cell carcinoma of the ovary, hypercalcemic type and in SMARCA4-deficient undifferentiated uterine sarcoma: Clinical features of a single family and comparison of large cohorts. Gynecol. Oncol. 2020, 157, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Jelinic, P.; Mueller, J.J.; Olvera, N.; Dao, F.; Scott, S.N.; Shah, R.; Gao, J.; Schultz, N.; Gonen, M.; Soslow, R.A.; et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 2014, 46, 424–426. [Google Scholar] [CrossRef]
- Shain, A.H.; Pollack, J.R. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 2013, 8, e55119. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Crabtree, G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 2015, 1, e1500447. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Glaros, S.; Cirrincione, G.M.; Palanca, A.; Metzger, D.; Reisman, D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008, 68, 3689–3696. [Google Scholar] [CrossRef]
- Concepcion, C.P.; Ma, S.; LaFave, L.M.; Bhutkar, A.; Liu, M.; DeAngelo, L.P.; Kim, J.Y.; Del Priore, I.; Schoenfeld, A.J.; Miller, M.; et al. Smarca4 Inactivation Promotes Lineage-Specific Transformation and Early Metastatic Features in the Lung. Cancer Discov. 2022, 12, 562–585. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Crabtree, G.R. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013, 153, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Hornick, J.L.; Dal Cin, P.; Fletcher, C.D. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am. J. Surg. Pathol. 2009, 33, 542–550. [Google Scholar] [CrossRef]
- Fernando, T.M.; Piskol, R.; Bainer, R.; Sokol, E.S.; Trabucco, S.E.; Zhang, Q.; Trinh, H.; Maund, S.; Kschonsak, M.; Chaudhuri, S.; et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat. Commun. 2020, 11, 5551. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Schrock, A.B.; Kem, M.; Jessop, N.; Lee, J.; Ali, S.M.; Ross, J.S.; Lennerz, J.K.; Shaw, A.T.; Mino-Kenudson, M. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. J. Thorac. Oncol. 2020, 15, 766–776. [Google Scholar] [CrossRef]
- Liu, L.; Ahmed, T.; Petty, W.J.; Grant, S.; Ruiz, J.; Lycan, T.W.; Topaloglu, U.; Chou, P.C.; Miller, L.D.; Hawkins, G.A.; et al. SMARCA4 mutations in KRAS-mutant lung adenocarcinoma: A multi-cohort analysis. Mol. Oncol. 2021, 15, 462–472. [Google Scholar] [CrossRef]
- Miller, T.I.; Zoumberos, N.A.; Johnson, B.; Rhodes, D.R.; Tomlins, S.A.; Chan, M.P.; Andea, A.A.; Lucas, D.R.; McHugh, J.B.; Smith, N.; et al. A genomic survey of sarcomas on sun-exposed skin reveals distinctive candidate drivers and potentially targetable mutations. Hum. Pathol. 2020, 102, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Hovelson, D.H.; Harms, P.; Drewery, S.; Falkner, J.; Fischer, A.; Hipp, J.; Kwiatkowski, K.; Lazo de la Vega, L.; Mitchell, K.; et al. Development and Validation of StrataNGS, a Multiplex PCR, Semiconductor Sequencing-Based Comprehensive Genomic Profiling Test. J. Mol. Diagn. 2021, 23, 1515–1533. [Google Scholar] [CrossRef] [PubMed]
- Hernan, M.A.; Sauer, B.C.; Hernandez-Diaz, S.; Platt, R.; Shrier, I. Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J. Clin. Epidemiol. 2016, 79, 70–75. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Bandlamudi, C.; Lavery, J.A.; Montecalvo, J.; Namakydoust, A.; Rizvi, H.; Egger, J.; Concepcion, C.P.; Paul, S.; Arcila, M.E.; et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin. Cancer Res. 2020, 26, 5701–5708. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Cook, M.B.; McGlynn, K.A.; Devesa, S.S.; Freedman, N.D.; Anderson, W.F. Sex disparities in cancer mortality and survival. Cancer Epidemiol. Biomarkers Prev. 2011, 20, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- El-Tamer, M.B.; Komenaka, I.K.; Troxel, A.; Li, H.; Joseph, K.-A.; Ditkoff, B.-A.; Schnabel, F.R.; Kinne, D.W. Men With Breast Cancer Have Better Disease-Specific Survival Than Women. Arch. Surg. 2004, 139, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Micheli, A.; Ciampichini, R.; Oberaigner, W.; Ciccolallo, L.; de Vries, E.; Izarzugaza, I.; Zambon, P.; Gatta, G.; De Angelis, R.; Group, E.W. The advantage of women in cancer survival: An analysis of EUROCARE-4 data. Eur. J. Cancer 2009, 45, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Park, S.; Shin, A.; Oh, C.M.; Kong, H.J.; Jun, J.K.; Won, Y.J. Do female cancer patients display better survival rates compared with males? Analysis of the Korean National Registry data, 2005–2009. PLoS ONE 2012, 7, e52457. [Google Scholar] [CrossRef]
- Roy, N.; Malik, S.; Villanueva, K.E.; Urano, A.; Lu, X.; Von Figura, G.; Seeley, E.S.; Dawson, D.W.; Collisson, E.A.; Hebrok, M. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015, 29, 658–671. [Google Scholar] [CrossRef]
- Wilson, B.G.; Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 2011, 11, 481–492. [Google Scholar] [CrossRef] [PubMed]
- von Figura, G.; Fukuda, A.; Roy, N.; Liku, M.E.; Morris Iv, J.P.; Kim, G.E.; Russ, H.A.; Firpo, M.A.; Mulvihill, S.J.; Dawson, D.W.; et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2014, 16, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Holik, A.Z.; Young, M.; Krzystyniak, J.; Williams, G.T.; Metzger, D.; Shorning, B.Y.; Clarke, A.R. Brg1 loss attenuates aberrant wnt-signalling and prevents wnt-dependent tumourigenesis in the murine small intestine. PLoS Genet. 2014, 10, e1004453. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, N.; Dong, B.; Guo, W.; Wei, H.; Chen, Q.; Yuan, H.; Han, Y.; Chang, H.; Kan, S.; et al. Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. J. Clin. Investig. 2019, 129, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.H.; Chakraborty, A.R.; Mo, X.; Liu, Z.; Shilo, K.; Kirste, S.; Stegmaier, P.; McNulty, M.; Karachaliou, N.; Rosell, R.; et al. SMARCA4/BRG1 Is a Novel Prognostic Biomarker Predictive of Cisplatin-Based Chemotherapy Outcomes in Resected Non-Small Cell Lung Cancer. Clin. Cancer Res. 2016, 22, 2396–2404. [Google Scholar] [CrossRef]
- Hodges, C.; Kirkland, J.G.; Crabtree, G.R. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb. Perspect. Med. 2016, 6, a026930. [Google Scholar] [CrossRef]
- Hodges, H.C.; Stanton, B.Z.; Cermakova, K.; Chang, C.Y.; Miller, E.L.; Kirkland, J.G.; Ku, W.L.; Veverka, V.; Zhao, K.; Crabtree, G.R. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 2018, 25, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Molina-Vila, M.A.; Bertran-Alamillo, J.; Gasco, A.; Mayo-de-las-Casas, C.; Sanchez-Ronco, M.; Pujantell-Pastor, L.; Bonanno, L.; Favaretto, A.G.; Cardona, A.F.; Vergnenegre, A.; et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin. Cancer Res. 2014, 20, 4647–4659. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Hou, H.; Liang, Y.; Zhang, X. Prognostic value of TP53 concurrent mutations for EGFR- TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: A meta-analysis. BMC Cancer 2020, 20, 328. [Google Scholar] [CrossRef]
- VanderLaan, P.A.; Rangachari, D.; Mockus, S.M.; Spotlow, V.; Reddi, H.V.; Malcolm, J.; Huberman, M.S.; Joseph, L.J.; Kobayashi, S.S.; Costa, D.B. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017, 106, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, J.; Chen, X. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J. Biol. Chem. 2007, 282, 37429–37435. [Google Scholar] [CrossRef]
- Kurashima, K.; Kashiwagi, H.; Shimomura, I.; Suzuki, A.; Takeshita, F.; Mazevet, M.; Harata, M.; Yamashita, T.; Yamamoto, Y.; Kohno, T.; et al. SMARCA4 deficiency-associated heterochromatin induces intrinsic DNA replication stress and susceptibility to ATR inhibition in lung adenocarcinoma. NAR Cancer 2020, 2, zcaa005. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.J.; Stinchcombe, T.E.; Der, C.J.; Socinski, M.A. Personalized medicine in non-small-cell lung cancer: Is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J. Clin. Oncol. 2010, 28, 4769–4777. [Google Scholar] [CrossRef]
- Mascaux, C.; Iannino, N.; Martin, B.; Paesmans, M.; Berghmans, T.; Dusart, M.; Haller, A.; Lothaire, P.; Meert, A.P.; Noel, S.; et al. The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br. J. Cancer 2005, 92, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Man, J.; Lord, S.; Cooper, W.; Links, M.; Gebski, V.; Herbst, R.S.; Gralla, R.J.; Mok, T.; Yang, J.C. Clinical and Molecular Characteristics Associated With Survival Among Patients Treated With Checkpoint Inhibitors for Advanced Non-Small Cell Lung Carcinoma: A Systematic Review and Meta-analysis. JAMA Oncol. 2018, 4, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.; Lau, S.C.M.; Sharma, K.; Lee, J.; Ryan, M.I.; Schmid, S.; Bradbury, P.A.; Liu, G.; Shepherd, F.A.; Tsao, M.S.; et al. Impact of KRAS mutational variant on response to immunotherapy in metastatic NSCLC. J. Clin. Oncol. 2021, 39, e21127. [Google Scholar] [CrossRef]
- Bange, E.; Marmarelis, M.E.; Hwang, W.T.; Yang, Y.X.; Thompson, J.C.; Rosenbaum, J.; Bauml, J.M.; Ciunci, C.; Alley, E.W.; Cohen, R.B.; et al. Impact of KRAS and TP53 Co-Mutations on Outcomes After First-Line Systemic Therapy Among Patients With STK11-Mutated Advanced Non-Small-Cell Lung Cancer. JCO Precis. Oncol. 2019, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Zhong, W.Z.; Zhang, X.C.; Su, J.; Xie, Z.; Liu, S.Y.; Tu, H.Y.; Chen, H.J.; Sun, Y.L.; Zhou, Q.; et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef]
- Wang, P.; Wang, F.; He, H.; Chen, Y.; Lin, H.; Chen, P.; Chen, X.; Liu, S. TP53 and CDKN2A mutations in patients with early-stage lung squamous cell carcinoma: An analysis of the correlations and prognostic outcomes. Ann. Transl. Med. 2021, 9, 1330. [Google Scholar] [CrossRef]
- Gutiontov, S.I.; Turchan, W.T.; Spurr, L.F.; Rouhani, S.J.; Chervin, C.S.; Steinhardt, G.; Lager, A.M.; Wanjari, P.; Malik, R.; Connell, P.P.; et al. CDKN2A loss-of-function predicts immunotherapy resistance in non-small cell lung cancer. Sci. Rep. 2021, 11, 20059. [Google Scholar] [CrossRef]
- Ahn, E.R.; Mangat, P.K.; Garrett-Mayer, E.; Halabi, S.; Dib, E.G.; Haggstrom, D.E.; Alguire, K.B.; Calfa, C.J.; Cannon, T.L.; Crilley, P.A.; et al. Palbociclib in Patients With Non-Small-Cell Lung Cancer With CDKN2A Alterations: Results From the Targeted Agent and Profiling Utilization Registry Study. JCO Precis. Oncol. 2020, 4, 757–766. [Google Scholar] [CrossRef]
- Wang, Z.J.; Churchman, M.; Avizienyte, E.; McKeown, C.; Davies, S.; Evans, D.G.; Ferguson, A.; Ellis, I.; Xu, W.H.; Yan, Z.Y.; et al. Germline mutations of the LKB1 (STK11) gene in Peutz-Jeghers patients. J. Med. Genet. 1999, 36, 365–368. [Google Scholar]
- Sokol, E.; Danziger, N.; Pavlick, D.; Elvin, J.A.; Vergilio, J.-A.; Killian, J.K.; Lin, D.I.; Ramkissoon, S.H.; Severson, E.A.; Hemmerich, A.; et al. Clinically aggressive malignancies associated with STK11 germline mutations (STK11GCa): A comprehensive genomic profiling (CGP) study. J. Clin. Oncol. 2020, 38, 3558. [Google Scholar] [CrossRef]
- Krishnamurthy, N.; Goodman, A.M.; Barkauskas, D.A.; Kurzrock, R. STK11 alterations in the pan-cancer setting: Prognostic and therapeutic implications. Eur. J. Cancer 2021, 148, 215–229. [Google Scholar] [CrossRef]
- Naqash, A.R.; Floudas, C.S.; Maoz, A.; Xiu, J.; Baca, Y.; Zeng, J.; Kim, C.; Judd, J.; Raez, L.E.; Lopes, G.; et al. STK11/TP53 co-mutated non-small cell lung cancer (NSCLC) to display a unique tumor microenvironment (TME) and metabolic profile. J. Clin. Oncol. 2021, 39, 9087. [Google Scholar] [CrossRef]
- Xue, Y.; Meehan, B.; Fu, Z.; Wang, X.Q.D.; Fiset, P.O.; Rieker, R.; Levins, C.; Kong, T.; Zhu, X.; Morin, G.; et al. SMARCA4 loss is synthetic lethal with CDK4/6 inhibition in non-small cell lung cancer. Nat. Commun. 2019, 10, 557. [Google Scholar] [CrossRef]
| SMARCA4 (n = 125) | |
|---|---|
| Ampullary cancer (%) | 2 (1.6) |
| Bladder cancer (%) | 3 (2.4) |
| Breast cancer (%) | 5 (4.0) |
| Cancer of unknown primary (%) | 14 (11.2) |
| Colon cancer (%) | 12 (9.6) |
| Endometrial cancer (%) | 6 (4.8) |
| Esophageal/stomach cancer (%) | 16 (12.8) |
| Gallbladder/cholangiocarcinoma (%) | 3 (2.4) |
| Head and neck cancer (%) | 1 (0.8) |
| Kidney cancer (%) | 1 (0.8) |
| Non-small cell lung cancer (%) | 44 (35.2) |
| Melanoma (%) | 3 (2.4) |
| Ovarian cancer (%) | 3 (2.4) |
| Pancreatic cancer (%) | 5 (4.0) |
| Prostate cancer (%) | 2 (1.6) |
| Skin squamous cell carcinoma (%) | 2 (1.6) |
| Sarcoma (%) | 2 (1.6) |
| Small cell lung cancer (%) | 1 (0.8) |
| Females (n = 60) | Males (n = 65) | p-Value | ||
|---|---|---|---|---|
| Age | 68 (62–92) | 67 (65–75) | 0.56 | |
| Race | White | 34 (58.6) | 44 (68.8) | 0.60 |
| Black | 6 (10.3) | 4 (6.2) | ||
| Hispanic | 4 (6.9) | 5 (7.8) | ||
| Asian | 14 (24.1) | 11 (17.2) | ||
| Unknown | 2 (3.3) | 1 (1.5) | ||
| PS | 0–1 | 30 (50.0) | 26 (40.0) | 0.12 |
| 2–4 | 20 (33.3) | 33 (50.8) | ||
| Unknown | 10 (16.7) | 6 (9.2) | ||
| CCI | 0 (0–10) | 1 (0–9) | 0.004 | |
| Smoking status | Current smoker | 4 (6.7) | 8 (12.3) | 0.07 |
| Past smoker | 29 (48.3) | 40 (61.5) | ||
| Never smoker | 27 (45.0) | 17 (26.2) | ||
| TMB > 10 | 23 (38.3) | 25 (38.5) | 0.98 | |
| Treatment | Yes | 46 (76.7) | 40 (61.5) | 0.07 |
| No | 14 (23.3) | 25 (38.5) | ||
| MutTP53 | 28 (46.7) | 46 (70.8) | 0.006 | |
| MutKRAS | 17 (28.3) | 19 (29.2) | 0.91 | |
| MutCDKN2A | 16 (26.7) | 20 (30.8) | 0.61 | |
| MutSTK11 | 9 (15.0) | 7 (10.8) | 0.48 | |
| Keap1 | 6 (10) | 5 (7.7) | 0.65 | |
| Females (n = 19) | Males (n = 25) | p-Value | ||
|---|---|---|---|---|
| Age | 65 (46–96) | 70 (54–83) | 0.93 | |
| Race | White | 13 (68.4) | 16 (64.0) | 0.72 |
| Black | 3 (15.8) | 2 (8.0) | ||
| Hispanic | 1 (5.3) | 2 (8.0) | ||
| Asian | 2 (10.5) | 5 (20) | ||
| PS | 0–1 | 8 (42.1) | 11 (44.0) | 0.08 |
| 2–4 | 6 (31.6) | 13 (52.0) | ||
| Unknown | 5 (26.3) | 1 (4.0) | ||
| CCI | 0 (0–10) | 0 (0–9) | 0.32 | |
| Smoking status | Current smoker | 3 (15.8) | 6 (24.0) | 0.54 |
| Past smoker | 15 (78.9) | 16 (64.0) | ||
| Never smoker | 1 (5.3) | 3 (12.0) | ||
| TMB > 10 | 9 (47.4) | 14 (56.0) | 0.58 | |
| Treatment | Yes | 13 (68.3) | 15 (60.0) | 0.56 |
| No | 6 (31.6%) | 10 (40.0) | ||
| MutTP53 | 10 (52.6) | 18 (72.0) | 0.18 | |
| MutKRAS | 11 (57.9) | 17 (68.0) | 0.49 | |
| MutCDKN2A | 7 (36.8) | 9 (36.0) | 0.95 | |
| MutSTK11 | 5 (26.3) | 4 (16.0) | 0.40 | |
| Among All Patients | Among NSCLC Only | |||
|---|---|---|---|---|
| mutTP53 | wtTP53 | mutTP53 | wtTP53 | |
| mutSTK11 n = 16 (%) | 2 (12.5) | 14 (87.5) | 1 (6.25) | 15 (93.75) |
| p value | <0.001 | <0.001 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, M.; Jiang, C.; Zhang, Z.; Achacoso, N.; Solorzano-Pinto, A.V.; Tse, P.; Chung, E.; Suga, J.M.; Thomas, S.; Habel, L.A. Sex- and Co-Mutation-Dependent Prognosis in Patients with SMARCA4-Mutated Malignancies. Cancers 2023, 15, 2665. https://doi.org/10.3390/cancers15102665
Pan M, Jiang C, Zhang Z, Achacoso N, Solorzano-Pinto AV, Tse P, Chung E, Suga JM, Thomas S, Habel LA. Sex- and Co-Mutation-Dependent Prognosis in Patients with SMARCA4-Mutated Malignancies. Cancers. 2023; 15(10):2665. https://doi.org/10.3390/cancers15102665
Chicago/Turabian StylePan, Minggui, Chen Jiang, Zheyang Zhang, Ninah Achacoso, Aleyda V. Solorzano-Pinto, Pam Tse, Elaine Chung, Jennifer Marie Suga, Sachdev Thomas, and Laurel A. Habel. 2023. "Sex- and Co-Mutation-Dependent Prognosis in Patients with SMARCA4-Mutated Malignancies" Cancers 15, no. 10: 2665. https://doi.org/10.3390/cancers15102665
APA StylePan, M., Jiang, C., Zhang, Z., Achacoso, N., Solorzano-Pinto, A. V., Tse, P., Chung, E., Suga, J. M., Thomas, S., & Habel, L. A. (2023). Sex- and Co-Mutation-Dependent Prognosis in Patients with SMARCA4-Mutated Malignancies. Cancers, 15(10), 2665. https://doi.org/10.3390/cancers15102665





