Circulating Let-7 Family Members as Non-Invasive Biomarkers for Predicting Hepatocellular Carcinoma Risk after Antiviral Treatment among Chronic Hepatitis C Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. MicroRNA Extraction from Plasma
2.3. Quantification of Circulating miRNAs
2.4. Statistical Analysis
3. Results
3.1. Characteristics of Patients with CHC
3.2. Comparison of Expression among Circulating Let-7 Family between Patients with and Those without HCC Development
3.3. Correlation Analysis between Let-7 Family Members and Laboratory Parameters
3.4. Let-7 Family Performance for Prediction of HCC Development in CHC Patients after Anti-Viral Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ioannou, G.N. Hcc surveillance after SVR in patients with f3/f4 fibrosis. J. Hepatol. 2021, 74, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Donato, F.; Tagger, A.; Gelatti, U.; Parrinello, G.; Boffetta, P.; Albertini, A.; Decarli, A.; Trevisi, P.; Ribero, M.L.; Martelli, C.; et al. Alcohol and hepatocellular carcinoma: The effect of lifetime intake and hepatitis virus infections in men and women. Am. J. Epidemiol. 2002, 155, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, W.F.; Tsai, P.C.; Chen, C.Y.; Tseng, K.C.; Lai, H.C.; Kuo, H.T.; Hung, C.H.; Tung, S.Y.; Wang, J.H.; Chen, J.J.; et al. Hepatitis c virus eradication decreases the risks of liver cirrhosis and cirrhosis-related complications (taiwanese chronic hepatitis c cohort). J. Gastroenterol. Hepatol. 2021, 36, 2884–2892. [Google Scholar] [CrossRef] [PubMed]
- Besheer, T.; Elalfy, H.; Abd El-Maksoud, M.; Abd El-Razek, A.; Taman, S.; Zalata, K.; Elkashef, W.; Zaghloul, H.; Elshahawy, H.; Raafat, D.; et al. Diffusion-weighted magnetic resonance imaging and micro-RNA in the diagnosis of hepatic fibrosis in chronic hepatitis c virus. World J. Gastroenterol. 2019, 25, 1366–1377. [Google Scholar] [CrossRef]
- Biselli, M.; Conti, F.; Gramenzi, A.; Frigerio, M.; Cucchetti, A.; Fatti, G.; D’Angelo, M.; Dall’Agata, M.; Giannini, E.G.; Farinati, F.; et al. A new approach to the use of alpha-fetoprotein as surveillance test for hepatocellular carcinoma in patients with cirrhosis. Br. J. Cancer 2015, 112, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Lowey, B.; Sodroski, C.; Krishnamurthy, S.; Alao, H.; Cha, H.; Chiu, S.; El-Diwany, R.; Ghany, M.G.; Liang, T.J. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat. Commun. 2017, 8, 1789. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y.; et al. Identification of mirnomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar] [CrossRef] [Green Version]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natil. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [Green Version]
- Mayr, C.; Hemann, M.T.; Bartel, D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007, 315, 1576–1579. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.S.; Yeh, M.L.; Tsai, P.C.; Huang, C.I.; Huang, C.F.; Hsieh, M.H.; Liu, T.W.; Lin, Y.H.; Liang, P.C.; Lin, Z.Y.; et al. Clusters of Circulating let-7 Family Tumor Suppressors Are Associated with Clinical Characteristics of Chronic Hepatitis C. Int. J. Mol. Sci. 2020, 21, 4945. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; De Giorgi, V.; Schechterly, C.; Wang, R.Y.; Farci, P.; Tanaka, Y.; Alter, H.J. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology 2016, 64, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Liu, S.M.; Ma, H.; Yang, Y.; Zhang, X.; Sun, H.; Zhang, X.; Xu, J.; Wang, J. Systematic review and meta-analysis: Circulating mirnas for diagnosis of hepatocellular carcinoma. J. Cell. Physiol. 2016, 231, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [Green Version]
- Omata, M.; Lesmana, L.A.; Tateishi, R.; Chen, P.J.; Lin, S.M.; Yoshida, H.; Kudo, M.; Lee, J.M.; Choi, B.I.; Poon, R.T.; et al. Asian Pacific Association for the Study of the Liver consensus recommendations on hepatocellular carcinoma. Hepatol. Int. 2010, 4, 439–474. [Google Scholar] [CrossRef] [Green Version]
- Chou, W.W.; Huang, C.F.; Yeh, M.L.; Tsai, Y.S.; Hsieh, M.Y.; Huang, C.I.; Huang, J.F.; Tsai, P.C.; Hsi, E.; Juo, S.H.; et al. MicroRNA let-7g cooperates with interferon/ribavirin to repress hepatitis C virus replication. J. Mol. Med. 2016, 94, 311–320. [Google Scholar] [CrossRef]
- Matsuura, K.; Aizawa, N.; Enomoto, H.; Nishiguchi, S.; Toyoda, H.; Kumada, T.; Iio, E.; Ito, K.; Ogawa, S.; Isogawa, M.; et al. Circulating let-7 Levels in Serum Correlate with the Severity of Hepatic Fibrosis in Chronic Hepatitis C. Open Forum Infect Dis. 2018, 5, ofy268. [Google Scholar] [CrossRef]
- Ali, N.; Allam, H.; May, R.; Sureban, S.M.; Bronze, M.S.; Bader, T.; Umar, S.; Anant, S.; Houchen, C.W. Hepatitis C virus-induced cancer stem cell-like signatures in cell culture and murine tumor xenografts. J. Virol. 2011, 85, 12292–12303. [Google Scholar] [CrossRef] [Green Version]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Moss, E.G.; Lee, R.C.; Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. Elegans and is regulated by the lin-4 RNA. Cell 1997, 88, 637–646. [Google Scholar] [CrossRef] [Green Version]
- Moss, E.G.; Tang, L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003, 258, 432–442. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Powers, J.T.; Einhorn, W.; Hoshida, Y.; Ng, T.L.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.A.; Lockhart, V.L.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Chen, Y.; Ito, H.; Watanabe, A.; Ge, X.; Kodama, T.; Aburatani, H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006, 384, 51–61. [Google Scholar] [CrossRef]
- Cheng, J.C.; Yeh, Y.J.; Tseng, C.P.; Hsu, S.D.; Chang, Y.L.; Sakamoto, N.; Huang, H.D. Let-7b is a novel regulator of hepatitis C virus replication. Cell. Mol. Life Sci. CMLS 2012, 69, 2621–2633. [Google Scholar] [CrossRef]
- Cheng, M.; Si, Y.; Niu, Y.; Liu, X.; Li, X.; Zhao, J.; Jin, Q.; Yang, W. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGFBP1. J. Virol. 2013, 87, 9707–9718. [Google Scholar] [CrossRef] [Green Version]
- Lan, F.F.; Wang, H.; Chen, Y.C.; Chan, C.Y.; Ng, S.S.; Li, K.; Xie, D.; He, M.L.; Lin, M.C.; Kung, H.F. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int. J. Cancer 2011, 128, 319–331. [Google Scholar] [CrossRef]
- Bussing, I.; Slack, F.J.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Vijayalekshmi, R.V.; Sung, B. Targeting inflammatory pathways for prevention and therapy of cancer: Short-term friend, long-term foe. Clin. Cancer Res. 2009, 15, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Vespasiani-Gentilucci, U.; Carotti, S.; Onetti-Muda, A.; Perrone, G.; Ginanni-Corradini, S.; Latasa, M.U.; Avila, M.A.; Carpino, G.; Picardi, A.; Morini, S. Toll-like receptor-4 expression by hepatic progenitor cells and biliary epithelial cells in HCV-related chronic liver disease. Mod. Pathol. 2012, 25, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.L.; Chen, P.J.; Dai, C.Y.; Hu, T.H.; Huang, C.F.; Huang, Y.H.; Hung, C.H.; Lin, C.Y.; Liu, C.H.; Liu, C.J.; et al. 2020 taiwan consensus statement on the management of hepatitis C: Part (II) special populations. J. Formos. Med. Assoc. 2020, 119, 1135–1157. [Google Scholar] [CrossRef]
- Conti, F.; Buonfiglioli, F.; Scuteri, A.; Crespi, C.; Bolondi, L.; Caraceni, P.; Foschi, F.G.; Lenzi, M.; Mazzella, G.; Verucchi, G.; et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J. Hepatol. 2016, 65, 727–733. [Google Scholar] [CrossRef]
- Wang, B.; Jha, J.C.; Hagiwara, S.; McClelland, A.D.; Jandeleit-Dahm, K.; Thomas, M.C.; Cooper, M.E.; Kantharidis, P. Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014, 85, 352–361. [Google Scholar] [CrossRef] [Green Version]
| Parameters | Total (n = 227) | Without HCC Development (n = 173) | De Novo HCC (n = 54) | p |
|---|---|---|---|---|
| Age (yr, mean ± SD) | 55.15 ± 8.75 | 54.31 ± 8.73 | 57.87 ± 8.30 | 0.0087 |
| Gender (F/M) | 116/111 | 93/80 | 23/31 | 0.1515 |
| BMI | 25.33 ± 3.60 | 25.40 ± 3 | 25.30 ± 3.05 | 0.9484 |
| Total Viral loads log10 (IU/mL) # | 5.62 ± 0.85 | 5.68 ± 0.85 | 5.42 ± 0.81 | 0.0494 |
| HCV genotype (type1/non-type1/unclassified) | (123/99/5) | (95/76/2) | (28/23/3) | 0.2510 |
| FIB-4 > 3.25 (yes/no) | (114/122) | (67/106) | (37/16) | <0.0001 |
| SVR (Yes/No) | (130/97) | (102/71) | (28/26) | 0.4924 |
| AFP (ng/mL) | 22.88 ± 55.43 | 16.91 ± 37.02 | 41.62 ± 89.95 | 0.0063 |
| Cr | 0.83 ± 0.25 | 0.81 ± 0.23 | 0.88 ± 0.31 | 0.0609 |
| AST ± SD (IU/L) | 90.36 ± 53.72 | 81.37 ± 43.58 | 119.72 ± 71.12 | <0.0001 |
| ALT ± SD (IU/L) | 123.04 ± 69.95 | 114.41 ± 59.71 | 150.70 ± 0.94 | 0.0008 |
| GGT (IU/L) | 68.16 ± 66.36 | 58.82 ± 57.99 | 96.17 ± 81.12 | 0.0003 |
| WBC | 5437.45 ± 1639.53 | 5503.24 ± 1722.15 | 5226.67 ± 1333.40 | 0.2946 |
| Platelet count (×103/µL, mean ± SD) | 153.20 ± 61.61 | 162.88 ± 63.07 | 122.20 ± 44.68 | <0.0001 |
| Hemoglobin (g/dL) | 14.03 ± 1.47 | 14.09 ± 1.50 | 13.85 ± 1.36 | 0.3457 |
| Follow-up period (year; mean ± SD) | 9.30 ± 4.18 | 10.98 ± 3.06 | 4.14 ± 2.63 | <0.0001 |
| Ct valube_pre_Cel-39 | 27.16 ± 1.03 | 27.12 ± 1.09 | 27.30 ± 0.80 | 0.3629 |
| Ct valube_post_Cel-39 | 27.25 ± 1.13 | 27.20 ± 1.11 | 27.39 ± 1.20 | 0.2533 |
| Log 102^−Delta Ct | Procedure | Without HCC Development (n = 173) | De Novo HCC (n = 54) | p |
|---|---|---|---|---|
| let7a | Pre | −1.89 ± 0.59 | −1.98 ± 0.50 | 0.2951 |
| Post | −1.65 ± 0.67 | −1.97 ± 0.45 | 0.0011 * | |
| Δ post−pre | 0.24 ± 0.73 | 0.01 ± 0.59 | 0.0405 | |
| let7b | Pre | −1.55 ± 0.59 | −1.55 ± 0.61 | 0.9684 |
| Post | −1.31 ± 0.73 | −1.66 ± 0.54 | 0.0015 * | |
| Δ post−pre | 0.24 ± 0.82 | −0.11 ± 0.77 | 0.0062 | |
| let7c | Pre | −2.17 ± 0.44 | −2.20 ± 0.38 | 0.6392 |
| Post | −1.94 ± 0.48 | −1.77 ± 0.95 | 0.0787 | |
| Δ post−pre | 0.23 ± 0.61 | 0.43 ± 1.04 | 0.0726 | |
| let7d | Pre | −1.90 ± 0.52 | −1.99 ± 0.44 | 0.2448 |
| Post | −1.66 ± 0.66 | −2.04 ± 0.39 | 0.0078 | |
| Δ post−pre | 0.23 ± 0.72 | −0.05 ± 0.48 | 0.0092 | |
| let7e | Pre | −1.68 ± 0.64 | −1.68 ± 0.56 | 0.9623 |
| Post | −1.48 ± 0.71 | −1.72 ± 0.85 | 0.0557 | |
| Δ post−pre | 0.18 ± 0.80 | −0.04 ± 1.00 | 0.0979 | |
| let7f | Pre | −2.17 ± 0.42 | −2.25 ± 0.30 | 0.2065 |
| Post | −2.04 ± 0.53 | −2.20 ± 0.36 | 0.0353 | |
| Δ post−pre | 0.13 ± 0.57 | 0.04 ± 0.47 | 0.3212 | |
| let7g | Pre | −1.90 ± 0.57 | −1.83 ± 0.55 | 0.4272 |
| Post | −1.68 ± 0.67 | −1.95 ± 0.46 | 0.0053 * | |
| Δ post−pre | 0.22 ± 0.73 | −0.12 ± 0.69 | 0.0025 * | |
| let7i | Pre | −1.80 ± 0.57 | −1.83 ± 0.59 | 0.7959 |
| Post | −1.53 ± 0.68 | −1.96 ± 0.52 | <0.0001 * | |
| Δ post−pre | 0.28 ± 0.80 | −0.13 ± 0.81 | 0.0013 * | |
| miR−98 | Pre | −2.29 ± 0.34 | −2.29 ± 0.26 | 0.8889 |
| Post | −2.17 ± 0.44 | −2.29 ± 0.41 | 0.0803 | |
| Δ post−pre | 0.12 ± 0.52 | −0.003 ± 0.48 | 0.1180 |
| Log 102^−Delta Ct | AST | p | ALT | p | GGT | p | PLT | p | FIB−4 Score | p |
|---|---|---|---|---|---|---|---|---|---|---|
| let7a | −0.22 * | 0.0027 | −0.1476* | 0.0317 | −0.1587 | 0.0946 | 0.317 * | <0.0001 | −0.3088 * | <0.0001 |
| let7b | −0.116 | 0.1176 | −0.0334 | 0.6287 | −0.2202 * | 0.0197 | 0.3357 * | <0.0001 | −0.3108 * | <0.0001 |
| let7c | −0.123 | 0.0975 | −0.1062 | 0.1233 | 0.0623 | 0.5143 | 0.0602 | 0.382 | −0.0414 | 0.5797 |
| let7d | −0.182 * | 0.0134 | −0.1123 | 0.1031 | −0.1846 | 0.0513 | 0.4361 * | <0.0001 | −0.3644 * | <0.0001 |
| let7e | −0.206 * | 0.005 | −0.1126 | 0.1019 | −0.0648 | 0.4974 | 0.3336 * | <0.0001 | −0.3391 * | <0.0001 |
| let7f | −0.183 * | 0.0129 | −0.1017 | 0.1399 | −0.1486 | 0.1178 | 0.3247 * | <0.0001 | −0.3144 * | <0.0001 |
| let7g | −0.156 * | 0.0348 | −0.067 | 0.3313 | −0.1136 | 0.2332 | 0.2838 * | <0.0001 | −0.3035 * | <0.0001 |
| let7i | −0.162 * | 0.028 | −0.1127 | 0.1017 | −0.2294 * | 0.015 | 0.3915 * | <0.0001 | −0.3714 * | <0.0001 |
| miR98 | −0.209 * | 0.0044 | −0.1553* | 0.0237 | −0.1972 * | 0.0372 | 0.3238 * | <0.0001 | −0.2736 * | 0.0002 |
| Log 102^−Delta Ct | Procedure | HR(95%CI) | p | Adjusted HR (95%CI) | p |
|---|---|---|---|---|---|
| let7a | M6 median: ≥−1.88 vs. <−1.88 | 0.47 (0.26–0.81) | 0.0067 | 0.68 (0.25–1.80) | 0.4490 |
| Δ post(M6)-pre(baseline) median: ≥0.18 vs. <0.18 | 0.50 (0.28–0.86) | 0.0128 | 0.42 (0.16–1.06) | 0.0669 | |
| let7b | M6 median: ≥−1.44 vs. <−1.44 | 0.48 (0.21–0.67) | 0.0007 * | 0.30 (0.08–0.89) | 0.0296 * |
| Δ post(M6)-pre(baseline) Median: ≥0.14 vs. <0.14 | 0.70 (0.40–1.20) | 0.1959 | 0.93 (0.37–2.31) | 0.8752 | |
| let7c | M6 median: ≥−2.02 vs. <−2.02 | 1.02 (0.59–1.74) | 0.9521 | 1.62 (0.57–5.32) | 0.3763 |
| Δ post(M6)-pre(baseline) median: ≥0.198 vs. <0.198 | 1.16 (0.68–2.00) | 0.5809 | 1.72 (0.64–5.44) | 0.2878 | |
| let7d | M6 median: ≥−1.90 vs. <−1.90 | 0.35 (0.18–0.61) | 0.0002 * | 0.90 (0.29–2.78) | 0.8591 |
| Δ post(M6)-pre(baseline) median: ≥0.170 vs. <0.170 | 0.42 (0.23–0.73) | 0.0022 | 0.62 (0.22–1.66) | 0.3405 | |
| let7e | M6 median: ≥−1.67 vs. <−1.67 | 0.47 (0.26–0.82) | 0.0072 | 1.25 (0.43–3.57) | 0.6721 |
| Δ post(M6)-pre(baseline) median: ≥0.150 vs. <0.150 | 0.37 (0.20–0.65) | 0.0005 * | 0.55 (0.20–1.43) | 0.2259 | |
| let7f | M6 median: ≥−2.16 vs. <−2.16 | 0.69 (0.40–1.18) | 0.1778 | 1.84 (0.67–5.35) | 0.2395 |
| Δ post(M6)-pre(baseline) median: ≥0.0918 vs. <0.0918 | 0.83 (0.48–1.41) | 0.4954 | 1.42 (0.55–3.85) | 0.4658 | |
| let7g | M6 median: ≥−1.91 vs. <−1.91 | 0.58 (0.33–1.00) | 0.0537 | 0.95 (0.35–2.49) | 0.9208 |
| Δ post(M6)-pre(baseline) Median: ≥0.12 vs. <0.12 | 0.60 (0.34–1.04) | 0.0690 | 1.34 (0.52–3.57) | 0.5473 | |
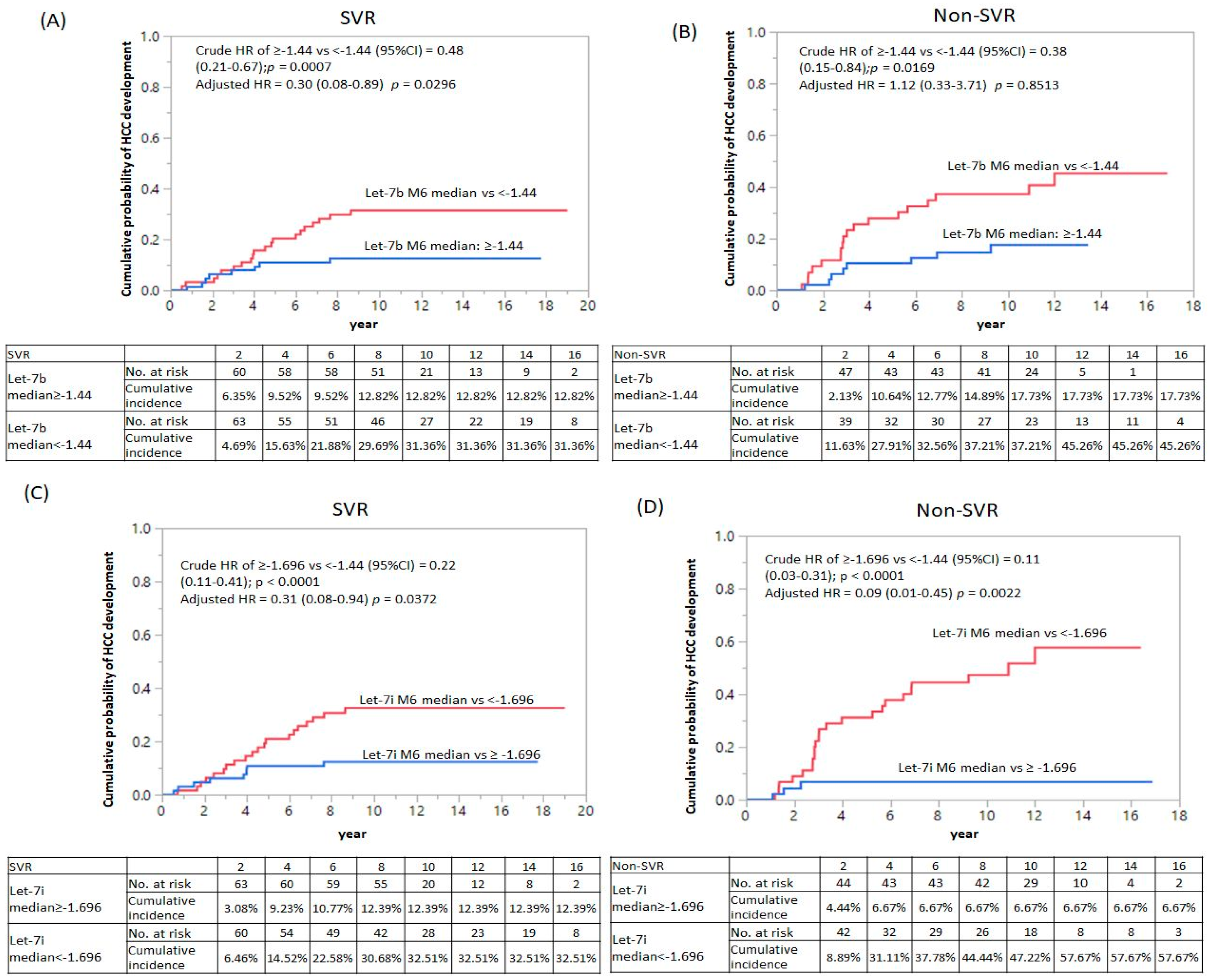
| let7i | M6 median: ≥−1.696 vs. <−1.696 | 0.22 (0.11–0.41) | <0.0001 * | 0.31(0.08–0.94) | 0.0372 * |
| Δ post(M6)-pre(baseline) median: ≥0.177 vs. <0.177 | 0.53 (0.30–0.92) | 0.0237 | 1.08 (0.41–2.83) | 0.8793 | |
| miR-98 | M6 median: ≥−2.235 vs. <−2.235 | 0.85(0.49–1.45) | 0.5538 | 1.81 (0.69–5.08) | 0.2293 |
| Δ post(M6)-pre(baseline) median: ≥0.069 vs. <0.069 | 0.76(0.44–1.30) | 0.3191 | 1.49 (0.57–4.01) | 0.4121 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, Y.-S.; Huang, C.-I.; Tsai, P.-C.; Yeh, M.-L.; Huang, C.-F.; Hsieh, M.-H.; Liu, T.-W.; Lin, Y.-H.; Liang, P.-C.; Lin, Z.-Y.; et al. Circulating Let-7 Family Members as Non-Invasive Biomarkers for Predicting Hepatocellular Carcinoma Risk after Antiviral Treatment among Chronic Hepatitis C Patients. Cancers 2022, 14, 2023. https://doi.org/10.3390/cancers14082023
Tsai Y-S, Huang C-I, Tsai P-C, Yeh M-L, Huang C-F, Hsieh M-H, Liu T-W, Lin Y-H, Liang P-C, Lin Z-Y, et al. Circulating Let-7 Family Members as Non-Invasive Biomarkers for Predicting Hepatocellular Carcinoma Risk after Antiviral Treatment among Chronic Hepatitis C Patients. Cancers. 2022; 14(8):2023. https://doi.org/10.3390/cancers14082023
Chicago/Turabian StyleTsai, Yi-Shan, Ching-I Huang, Pei-Chien Tsai, Ming-Lun Yeh, Chung-Feng Huang, Meng-Hsuan Hsieh, Ta-Wei Liu, Yi-Hung Lin, Po-Cheng Liang, Zu-Yau Lin, and et al. 2022. "Circulating Let-7 Family Members as Non-Invasive Biomarkers for Predicting Hepatocellular Carcinoma Risk after Antiviral Treatment among Chronic Hepatitis C Patients" Cancers 14, no. 8: 2023. https://doi.org/10.3390/cancers14082023
APA StyleTsai, Y.-S., Huang, C.-I., Tsai, P.-C., Yeh, M.-L., Huang, C.-F., Hsieh, M.-H., Liu, T.-W., Lin, Y.-H., Liang, P.-C., Lin, Z.-Y., Chen, S.-C., Huang, J.-F., Chuang, W.-L., Dai, C.-Y., & Yu, M.-L. (2022). Circulating Let-7 Family Members as Non-Invasive Biomarkers for Predicting Hepatocellular Carcinoma Risk after Antiviral Treatment among Chronic Hepatitis C Patients. Cancers, 14(8), 2023. https://doi.org/10.3390/cancers14082023









