The Landscape of lncRNAs in Multiple Myeloma: Implications in the “Hallmarks of Cancer”, Clinical Perspectives and Therapeutic Opportunities
Abstract
Simple Summary
Abstract
1. Introduction
2. LncRNAs Biogenesis and Functions
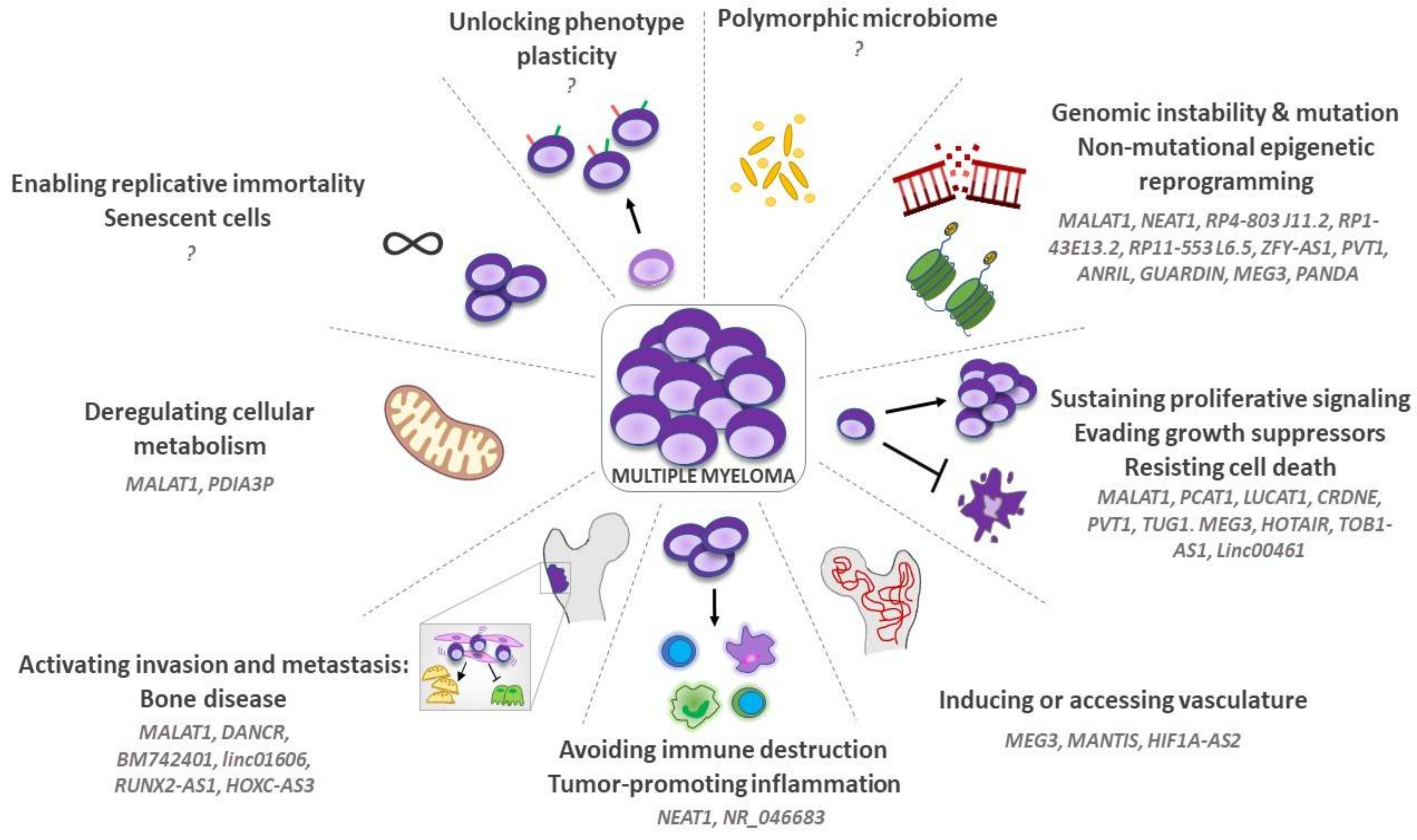
3. LncRNAs and “Hallmarks of Cancer” in MM
3.1. Genomic Instability, Mutation, and Non-Mutational Epigenetic Reprogramming
3.2. Sustaining Proliferative Signaling, Evading Growth Suppressors and Resisting Cell Death
3.3. Inducing or Accessing Vasculature
3.4. Tumor-Promoting Inflammation and Avoiding Immune Destruction
3.5. Activating Invasion and Metastasis: The Osteolytic Bone Disease (OBD)
3.6. Deregulating Cellular Metabolism
3.7. Enabling Replicative Immortality and Senescent Cells
3.8. Unlocking Phenotype Plasticity
3.9. Polymorphic Microbiomes
4. Clinical Perspectives and Therapeutic Opportunities of lncRNA
4.1. LncRNAs and Patient Stratification
4.2. Resistance to Therapy
4.3. LncRNAs as Therapeutic Targets: Exploring New Therapies
5. Limitations and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, L.; Desantis, V.; Solimando, A.G.; Ruggieri, S.; Annese, T.; Nico, B.; Fumarulo, R.; Vacca, A.; Frassanito, M.A. Microenvironment drug resistance in multiple myeloma: Emerging new players. Oncotarget 2016, 7, 60698–60711. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Anderson, K.C. Signaling Pathway Mediating Myeloma Cell Growth and Survival. Cancers 2021, 13, 216. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, N.; Colla, S.; Rizzoli, V. Angiogenic switch in multiple myeloma. Hematology 2004, 9, 377–381. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Neuse, C.J.; Lomas, O.C.; Schliemann, C.; Shen, Y.J.; Manier, S.; Bustoros, M.; Ghobrial, I.M. Genome instability in multiple myeloma. Leukemia 2020, 34, 2887–2897. [Google Scholar] [CrossRef]
- Taiana, E.; Gallo Cantafio, M.E.; Favasuli, V.K.; Bandini, C.; Viglietto, G.; Piva, R.; Neri, A.; Amodio, N. Genomic Instability in Multiple Myeloma: A "Non-Coding RNA" Perspective. Cancers 2021, 13, 2127. [Google Scholar] [CrossRef]
- Diamantopoulos, M.A.; Tsiakanikas, P.; Scorilas, A. Non-coding RNAs: The riddle of the transcriptome and their perspectives in cancer. Ann. Transl. Med. 2018, 6, 241. [Google Scholar] [CrossRef]
- Lu, M.; Hu, Y.; Wu, Y.; Zhou, H.; Jian, Y.; Tian, Y.; Chen, W. Genome-wide discovery and characterization of long noncoding RNAs in patients with multiple myeloma. BMC Med. Genomics 2019, 12, 135. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Carrasco-Leon, A.; Ezponda, T.; Meydan, C.; Valcarcel, L.V.; Ordonez, R.; Kulis, M.; Garate, L.; Miranda, E.; Segura, V.; Guruceaga, E.; et al. Characterization of complete lncRNAs transcriptome reveals the functional and clinical impact of lncRNAs in multiple myeloma. Leukemia 2021, 35, 1438–1450. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Lanzos, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; Drivers, P.; Functional Interpretation, G.; Johnson, R.; Consortium, P. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun. Biol. 2020, 3, 56. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Choi, S.W.; Kim, H.W.; Nam, J.W. The small peptide world in long noncoding RNAs. Brief Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: LncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 2018, 34, 142–157. [Google Scholar] [CrossRef]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647. [Google Scholar] [CrossRef]
- Lopez-Urrutia, E.; Bustamante Montes, L.P.; Ladron de Guevara Cervantes, D.; Perez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kumar, S.; Jin, D.Y.; Calin, G.A.; Chng, W.J.; Siu, K.L.; Poon, M.W.; Chim, C.S. Epigenetic silencing of long non-coding RNA BM742401 in multiple myeloma: Impact on prognosis and myeloma dissemination. Cancer Cell Int. 2020, 20, 403. [Google Scholar] [CrossRef]
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262. [Google Scholar] [CrossRef]
- Taiana, E.; Favasuli, V.; Ronchetti, D.; Todoerti, K.; Pelizzoni, F.; Manzoni, M.; Barbieri, M.; Fabris, S.; Silvestris, I.; Gallo Cantafio, M.E.; et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 2020, 34, 234–244. [Google Scholar] [CrossRef]
- Zhou, M.; Zhao, H.; Wang, Z.; Cheng, L.; Yang, L.; Shi, H.; Yang, H.; Sun, J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J. Exp. Clin. Cancer Res. 2015, 34, 102. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Hou, P.F.; Meng, S.; Chen, F.; Jiang, T.; Li, M.L.; Shi, M.L.; Liu, J.J.; Zheng, J.N.; Bai, J. Emerging Roles of p53 Related lncRNAs in Cancer Progression: A Systematic Review. Int. J. Biol. Sci. 2019, 15, 1287–1298. [Google Scholar] [CrossRef]
- Meng, H.; Han, L.; Hong, C.; Ding, J.; Huang, Q. Aberrant lncRNA Expression in Multiple Myeloma. Oncol. Res. 2018, 26, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Handa, H.; Kuroda, Y.; Kimura, K.; Masuda, Y.; Hattori, H.; Alkebsi, L.; Matsumoto, M.; Kasamatsu, T.; Kobayashi, N.; Tahara, K.I.; et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br. J. Haematol. 2017, 179, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, H.; Wu, B.; Yao, K.; Liao, A.; Miao, M.; Li, Y.; Yang, W. Down-regulation of long non-coding RNA MALAT1 by RNA interference inhibits proliferation and induces apoptosis in multiple myeloma. Clin. Exp. Pharmacol. Physiol. 2017, 44, 1032–1041. [Google Scholar] [CrossRef]
- Gao, D.; Lv, A.E.; Li, H.P.; Han, D.H.; Zhang, Y.P. LncRNA MALAT-1 Elevates HMGB1 to Promote Autophagy Resulting in Inhibition of Tumor Cell Apoptosis in Multiple Myeloma. J. Cell Biochem. 2017, 118, 3341–3348. [Google Scholar] [CrossRef]
- Di Lernia, G.; Leone, P.; Solimando, A.G.; Buonavoglia, A.; Saltarella, I.; Ria, R.; Ditonno, P.; Silvestris, N.; Crudele, L.; Vacca, A.; et al. Bortezomib Treatment Modulates Autophagy in Multiple Myeloma. J. Clin. Med. 2020, 9, 552. [Google Scholar] [CrossRef]
- Shen, X.; Shen, P.; Yang, Q.; Yin, Q.; Wang, F.; Cong, H.; Wang, X.; Ju, S. Knockdown of long non-coding RNA PCAT-1 inhibits myeloma cell growth and drug resistance via p38 and JNK MAPK pathways. J. Cancer 2019, 10, 6502–6510. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, H.; Peng, Q.; Yang, Y. Long Noncoding RNA LUCAT1 Promotes Multiple Myeloma Cell Growth by Regulating the TGF-beta Signaling Pathway. Technol. Cancer Res. Treat. 2020, 19, 1533033820945770. [Google Scholar] [CrossRef] [PubMed]
- David, A.; Zocchi, S.; Talbot, A.; Choisy, C.; Ohnona, A.; Lion, J.; Cuccuini, W.; Soulier, J.; Arnulf, B.; Bories, J.C.; et al. The long non-coding RNA CRNDE regulates growth of multiple myeloma cells via an effect on IL6 signalling. Leukemia 2021, 35, 1710–1721. [Google Scholar] [CrossRef]
- Harmer, D.; Falank, C.; Reagan, M.R. Interleukin-6 Interweaves the Bone Marrow Microenvironment, Bone Loss, and Multiple Myeloma. Front. Endocrinol. 2018, 9, 788. [Google Scholar] [CrossRef]
- Meng, Y.B.; He, X.; Huang, Y.F.; Wu, Q.N.; Zhou, Y.C.; Hao, D.J. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol. Res. 2017, 25, 1207–1214. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, L.; Wang, X.; Zhou, Y.; Wu, S. Down-regulation of miR-203a by lncRNA PVT1 in multiple myeloma promotes cell proliferation. Arch. Med. Sci. 2018, 14, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, D.; Li, S.; Xiao, M.; Zhou, H.; Yang, S.; Hao, Y.; Dong, S. Long non-coding RNA TUG1 knockdown hinders the tumorigenesis of multiple myeloma by regulating the microRNA-34a-5p/NOTCH1 signaling pathway. Open Life Sci. 2020, 15, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Bai, H.; Zhu, H.; Yan, Q.; Yang, Y.; Yu, W.; Shi, Q.; Wang, J.; Li, J.; Chen, L. Long Non-Coding RNA MEG3 Functions as a Competing Endogenous RNA to Regulate HOXA11 Expression by Sponging miR-181a in Multiple Myeloma. Cell Physiol. Biochem. 2018, 49, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Saltarella, I.; Lamanuzzi, A.; Desantis, V.; Di Marzo, L.; Melaccio, A.; Curci, P.; Annese, T.; Nico, B.; Solimando, A.G.; Bartoli, G.; et al. Myeloma cells regulate miRNA transfer from fibroblast-derived exosomes by expression of lncRNAs. J. Pathol. 2022, 256, 402–413. [Google Scholar] [CrossRef]
- Deng, M.; Yuan, H.; Liu, S.; Hu, Z.; Xiao, H. Exosome-transmitted LINC00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microRNA/BCL-2 expression. Cytotherapy 2019, 21, 96–106. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell Mol Life Sci 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Wang, S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics 2018, 8, 3654–3675. [Google Scholar] [CrossRef]
- Shi, C.; Yang, Q.; Pan, S.; Lin, X.; Xu, G.; Luo, Y.; Zheng, B.; Xie, X.; Yu, M. LncRNA OIP5-AS1 promotes cell proliferation and migration and induces angiogenesis via regulating miR-3163/VEGFA in hepatocellular carcinoma. Cancer Biol. Ther. 2020, 21, 604–614. [Google Scholar] [CrossRef]
- Zhao, J.; Du, P.; Cui, P.; Qin, Y.; Hu, C.; Wu, J.; Zhou, Z.; Zhang, W.; Qin, L.; Huang, G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018, 37, 4094–4109. [Google Scholar] [CrossRef]
- Li, W.; Fu, Q.; Man, W.; Guo, H.; Yang, P. LncRNA OR3A4 participates in the angiogenesis of hepatocellular carcinoma through modulating AGGF1/akt/mTOR pathway. Eur. J. Pharmacol. 2019, 849, 106–114. [Google Scholar] [CrossRef]
- Song, J.; Shu, H.; Zhang, L.; Xiong, J. Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wnt/beta-catenin signaling pathway. J. Cell Biochem. 2019, 120, 6937–6951. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Vacca, A.; De Falco, G.; Roccaro, A.; Roncali, L.; Dammacco, F. Angiogenesis, angiogenic factor expression and hematological malignancies. Anticancer Res. 2001, 21, 4333–4339. [Google Scholar]
- Allegra, A.; Mania, M.; D’Ascola, A.; Oteri, G.; Siniscalchi, E.N.; Avenoso, A.; Innao, V.; Scuruchi, M.; Allegra, A.G.; Musolino, C.; et al. Altered Long Noncoding RNA Expression Profile in Multiple Myeloma Patients with Bisphosphonate-Induced Osteonecrosis of the Jaw. Biomed. Res. Int. 2020, 2020, 9879876. [Google Scholar] [CrossRef]
- Ruan, W.; Zhao, F.; Zhao, S.; Zhang, L.; Shi, L.; Pang, T. Knockdown of long noncoding RNA MEG3 impairs VEGF-stimulated endothelial sprouting angiogenesis via modulating VEGFR2 expression in human umbilical vein endothelial cells. Gene 2018, 649, 32–39. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Fork, C.; Josipovic, I.; Richter, F.M.; Preussner, J.; Hu, J.; Miller, M.J.; Epah, J.; Hofmann, P.; Gunther, S.; et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation 2017, 136, 65–79. [Google Scholar] [CrossRef]
- Li, L.; Wang, M.; Mei, Z.; Cao, W.; Yang, Y.; Wang, Y.; Wen, A. lncRNAs HIF1A-AS2 facilitates the up-regulation of HIF-1alpha by sponging to miR-153-3p, whereby promoting angiogenesis in HUVECs in hypoxia. Biomed. Pharmacother 2017, 96, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Corrado, C.; Costa, V.; Giavaresi, G.; Calabrese, A.; Conigliaro, A.; Alessandro, R. Long Non Coding RNA H19: A New Player in Hypoxia-Induced Multiple Myeloma Cell Dissemination. Int. J. Mol. Sci. 2019, 20, 801. [Google Scholar] [CrossRef] [PubMed]
- Ria, R.; Vacca, A.; Russo, F.; Cirulli, T.; Massaia, M.; Tosi, P.; Cavo, M.; Guidolin, D.; Ribatti, D.; Dammacco, F. A VEGF-dependent autocrine loop mediates proliferation and capillarogenesis in bone marrow endothelial cells of patients with multiple myeloma. Thromb. Haemost. 2004, 92, 1438–1445. [Google Scholar] [CrossRef]
- Ria, R.; Catacchio, I.; Berardi, S.; De Luisi, A.; Caivano, A.; Piccoli, C.; Ruggieri, V.; Frassanito, M.A.; Ribatti, D.; Nico, B.; et al. HIF-1alpha of bone marrow endothelial cells implies relapse and drug resistance in patients with multiple myeloma and may act as a therapeutic target. Clin. Cancer Res. 2014, 20, 847–858. [Google Scholar] [CrossRef]
- Saltarella, I.; Frassanito, M.A.; Lamanuzzi, A.; Brevi, A.; Leone, P.; Desantis, V.; Di Marzo, L.; Bellone, M.; Derudas, D.; Ribatti, D.; et al. Homotypic and Heterotypic Activation of the Notch Pathway in Multiple Myeloma-Enhanced Angiogenesis: A Novel Therapeutic Target? Neoplasia 2019, 21, 93–105. [Google Scholar] [CrossRef]
- Palano, M.T.; Giannandrea, D.; Platonova, N.; Gaudenzi, G.; Falleni, M.; Tosi, D.; Lesma, E.; Citro, V.; Colombo, M.; Saltarella, I.; et al. Jagged Ligands Enhance the Pro-Angiogenic Activity of Multiple Myeloma Cells. Cancers 2020, 12, 2600. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Smyth, M.J.; Martinet, L. Cancer immunoediting and immune dysregulation in multiple myeloma. Blood 2020, 136, 2731–2740. [Google Scholar] [CrossRef]
- Shalapour, S.; Karin, M. Immunity, inflammation, and cancer: An eternal fight between good and evil. J. Clin. Invest. 2015, 125, 3347–3355. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, Y.; Zhang, J.; Liu, Y.; Qi, Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Lian, G.Y.; Song, Y.; Huang, Y.F.; Gong, Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019, 231, 116335. [Google Scholar] [CrossRef] [PubMed]
- Eptaminitaki, G.C.; Wolff, N.; Stellas, D.; Sifakis, K.; Baritaki, S. Long Non-Coding RNAs (lncRNAs) in Response and Resistance to Cancer Immunosurveillance and Immunotherapy. Cells 2021, 10, 3313. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, W.; Li, A.; Chen, Y.; Ou, Q.; He, Z.; Zhang, Y.; Liu, R.; Yao, H.; Song, E. Association of Long Noncoding RNA Biomarkers With Clinical Immune Subtype and Prediction of Immunotherapy Response in Patients With Cancer. JAMA Netw. Open 2020, 3, e202149. [Google Scholar] [CrossRef]
- Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef]
- Dong, H.; Jiang, S.; Fu, Y.; Luo, Y.; Gui, R.; Liu, J. Upregulation of lncRNA NR_046683 Serves as a Prognostic Biomarker and Potential Drug Target for Multiple Myeloma. Front. Pharmacol. 2019, 10, 45. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, P.; Li, W.J.; Zhang, J.; Wang, G.P.; Jiang, D.F.; Chen, F.P. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol. Immunol. 2020, 117, 20–28. [Google Scholar] [CrossRef]
- Miyamoto, T.; Murakami, R.; Hamanishi, J.; Tanigaki, K.; Hosoe, Y.; Mise, N.; Takamatsu, S.; Mise, Y.; Ukita, M.; Taki, M.; et al. B7-H3 Suppresses Antitumor Immunity via the CCL2-CCR2-M2 Macrophage Axis and Contributes to Ovarian Cancer Progression. Cancer Immunol. Res. 2022, 10, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Dimopoulos, M.A. Myeloma bone disease: From biology findings to treatment approaches. Blood 2019, 133, 1534–1539. [Google Scholar] [CrossRef]
- Bi, H.; Chen, X.; Gao, S.; Yu, X.; Xiao, J.; Zhang, B.; Liu, X.; Dai, M. Key Triggers of Osteoclast-Related Diseases and Available Strategies for Targeted Therapies: A Review. Front. Med. 2017, 4, 234. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Mitsiades, C.; Tonon, G.; Richardson, P.G.; Anderson, K.C. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat. Rev. Cancer 2007, 7, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Aurilia, C.; Donati, S.; Palmini, G.; Miglietta, F.; Iantomasi, T.; Brandi, M.L. The Involvement of Long Non-Coding RNAs in Bone. Int. J. Mol. Sci. 2021, 22, 3909. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Fan, X.; Zhang, B.; Wu, L.; Wu, X. Expression of LINC01606 in multiple myeloma and its effect on cell invasion and migration. Am. J. Transl. Res. 2021, 13, 8777–8786. [Google Scholar]
- Luo, B.; Yang, J.F.; Wang, Y.H.; Qu, G.B.; Hao, P.D.; Zeng, Z.J.; Yuan, J.; Yang, R.; Yuan, Y. MicroRNA-579-3p promotes the progression of osteoporosis by inhibiting osteogenic differentiation of mesenchymal stem cells through regulating Sirt1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6791–6799. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wong, K.Y.; Li, Z.H.; Chim, C.S. Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget 2016, 7, 82400–82410. [Google Scholar] [CrossRef]
- Chen, T.; Moscvin, M.; Bianchi, G. Exosomes in the Pathogenesis and Treatment of Multiple Myeloma in the Context of the Bone Marrow Microenvironment. Front. Oncol. 2020, 10, 608815. [Google Scholar] [CrossRef]
- Saltarella, I.; Lamanuzzi, A.; Apollonio, B.; Desantis, V.; Bartoli, G.; Vacca, A.; Frassanito, M.A. Role of Extracellular Vesicle-Based Cell-to-Cell Communication in Multiple Myeloma Progression. Cells 2021, 10, 3185. [Google Scholar] [CrossRef]
- Li, B.; Xu, H.; Han, H.; Song, S.; Zhang, X.; Ouyang, L.; Qian, C.; Hong, Y.; Qiu, Y.; Zhou, W.; et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene 2018, 37, 5508–5519. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Kidwai, F.K.; Kopher, R.A.; Motl, J.; Kellum, C.A.; Westendorf, J.J.; Kaufman, D.S. Use of RUNX2 expression to identify osteogenic progenitor cells derived from human embryonic stem cells. Stem Cell Reports 2015, 4, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Han, H.; Song, S.; Fan, G.; Xu, H.; Zhou, W.; Qiu, Y.; Qian, C.; Wang, Y.; Yuan, Z.; et al. HOXC10 Regulates Osteogenesis of Mesenchymal Stromal Cells Through Interaction with Its Natural Antisense Transcript lncHOXC-AS3. Stem Cells 2019, 37, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, P.; Qu, J.; Shi, L.; Zhuang, W.; Fu, J.; Li, J.; Zhang, X.; Sun, Y.; Zhuang, W. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J. Biol. Chem. 2014, 289, 29365–29375. [Google Scholar] [CrossRef] [PubMed]
- Robertson, I.B.; Horiguchi, M.; Zilberberg, L.; Dabovic, B.; Hadjiolova, K.; Rifkin, D.B. Latent TGF-beta-binding proteins. Matrix Biol. 2015, 47, 44–53. [Google Scholar] [CrossRef]
- Matsumoto, T.; Abe, M. TGF-beta-related mechanisms of bone destruction in multiple myeloma. Bone 2011, 48, 129–134. [Google Scholar] [CrossRef]
- Peng, F.; Yan, S.; Liu, H.; Liu, Z.; Jiang, F.; Cao, P.; Fu, R. Roles of LINC01473 and CD74 in osteoblasts in multiple myeloma bone disease. J. Investig Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Rizzieri, D.; Paul, B.; Kang, Y. Metabolic alterations and the potential for targeting metabolic pathways in the treatment of multiple myeloma. J. Cancer Metastasis Treat 2019, 5, 26. [Google Scholar] [CrossRef]
- Liu, N.; Feng, S.; Li, H.; Chen, X.; Bai, S.; Liu, Y. Long non-coding RNA MALAT1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13 axis. J. Cancer Res. Clin. Oncol. 2020, 146, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, H.; He, M.; Zhou, X.; Sun, N.; Guo, W.; Lin, X.; Huang, H.; Lin, Y.; Yao, R.; et al. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem. Biophys. Res. Commun. 2018, 498, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Gala, K.; Khattar, E. Long non-coding RNAs at work on telomeres: Functions and implications in cancer therapy. Cancer Lett 2021, 502, 120–132. [Google Scholar] [CrossRef]
- Yang, J.; Liu, M.; Hong, D.; Zeng, M.; Zhang, X. The Paradoxical Role of Cellular Senescence in Cancer. Front. Cell Dev. Biol. 2021, 9, 722205. [Google Scholar] [CrossRef]
- Liu, S.; Otsuyama, K.; Ma, Z.; Abroun, S.; Shamsasenjan, K.; Amin, J.; Asaoku, H.; Kawano, M.M. Induction of multilineage markers in human myeloma cells and their down-regulation by interleukin 6. Int. J. Hematol. 2007, 85, 49–58. [Google Scholar] [CrossRef]
- Spisek, R.; Kukreja, A.; Chen, L.C.; Matthews, P.; Mazumder, A.; Vesole, D.; Jagannath, S.; Zebroski, H.A.; Simpson, A.J.; Ritter, G.; et al. Frequent and specific immunity to the embryonal stem cell-associated antigen SOX2 in patients with monoclonal gammopathy. J. Exp. Med. 2007, 204, 831–840. [Google Scholar] [CrossRef]
- Bataille, R.; Pellat-Deceunynck, C.; Robillard, N.; Avet-Loiseau, H.; Harousseau, J.L.; Moreau, P. CD117 (c-kit) is aberrantly expressed in a subset of MGUS and multiple myeloma with unexpectedly good prognosis. Leuk Res. 2008, 32, 379–382. [Google Scholar] [CrossRef]
- Svachova, H.; Kryukov, F.; Kryukova, E.; Sevcikova, S.; Nemec, P.; Greslikova, H.; Rihova, L.; Kubiczkova, L.; Hajek, R. Nestin expression throughout multistep pathogenesis of multiple myeloma. Br. J. Haematol. 2014, 164, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhou, L.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Zhou, Y.; Du, Z. Downregulation of lncRNA-MALAT1 Affects Proliferation and the Expression of Stemness Markers in Glioma Stem Cell Line SHG139S. Cell Mol. Neurobiol. 2016, 36, 1097–1107. [Google Scholar] [CrossRef]
- Liang, L.; Ai, L.; Qian, J.; Fang, J.Y.; Xu, J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 2015, 5, 11763. [Google Scholar] [CrossRef]
- Dempsey, J.; Zhang, A.; Cui, J.Y. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genomics 2018, 19, 834. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhou, J.; Qi, H.; Wei, J.; Yang, Y.; Yue, J.; Liu, X.; Zhang, Y.; Yang, R. LncRNA lncLy6C induced by microbiota metabolite butyrate promotes differentiation of Ly6C(high) to Ly6C(int/neg) macrophages through lncLy6C/C/EBPbeta/Nr4A1 axis. Cell Discov. 2020, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Zhu, Y.; Ouyang, J.; Wang, Y.; Lei, Q.; Xia, J.; Guan, Y.; Zhang, J.; Guo, J.; He, Y.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Alkharabsheh, O.; Sidiqi, M.H.; Aljama, M.A.; Gertz, M.A.; Frankel, A.E. The Human Microbiota in Multiple Myeloma and Proteasome Inhibitors. Acta Haematol. 2020, 143, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, H. LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR-193a/MCL1 pathway. J. Biochem. Mol. Toxicol. 2018, 32, e22008. [Google Scholar] [CrossRef]
- Foltz, S.M.; Gao, Q.; Yoon, C.J.; Sun, H.; Yao, L.; Li, Y.; Jayasinghe, R.G.; Cao, S.; King, J.; Kohnen, D.R.; et al. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat. Commun. 2020, 11, 2666. [Google Scholar] [CrossRef]
- Sedlarikova, L.; Gromesova, B.; Kubaczkova, V.; Radova, L.; Filipova, J.; Jarkovsky, J.; Brozova, L.; Velichova, R.; Almasi, M.; Penka, M.; et al. Deregulated expression of long non-coding RNA UCA1 in multiple myeloma. Eur. J. Haematol. 2017, 99, 223–233. [Google Scholar] [CrossRef]
- Liu, H.; Shen, Y.; Xu, Y.; Wang, L.; Zhang, C.; Jiang, Y.; Hong, L.; Huang, H.; Liu, H. lncRNA transcription factor 7 is related to deteriorating clinical features and poor prognosis in multiple myeloma, and its knockdown suppresses disease progression by regulating the miR-203-mediated Jagged1-Notch1 signaling pathway. Oncol. Lett. 2021, 21, 412. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Ruan, J.; Zheng, W.; Yang, Z.; Pan, W. Long non-coding RNA OIP5-AS1 suppresses multiple myeloma progression by sponging miR-27a-3p to activate TSC1 expression. Cancer Cell Int. 2020, 20, 155. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Agnelli, L.; Pietrelli, A.; Todoerti, K.; Manzoni, M.; Taiana, E.; Neri, A. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci. Rep. 2018, 8, 107. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, J.; Zhou, Z.G. NEAT1 promotes cell proliferation in multiple myeloma by activating PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6403–6411. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Gao, L.; Ma, X.; Huang, J.J.; Chen, J.; Zeng, L.; Ashby, C.R., Jr.; Zou, C.; Chen, Z.S. Long non-coding RNAs regulate drug resistance in cancer. Mol. Cancer 2020, 19, 54. [Google Scholar] [CrossRef]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 2016, 7, 14814–14830. [Google Scholar] [CrossRef]
- Samur, M.K.; Minvielle, S.; Gulla, A.; Fulciniti, M.; Cleynen, A.; Aktas Samur, A.; Szalat, R.; Shammas, M.; Magrangeas, F.; Tai, Y.T.; et al. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia 2018, 32, 2626–2635. [Google Scholar] [CrossRef]
- Karreth, F.A.; Pandolfi, P.P. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013, 3, 1113–1121. [Google Scholar] [CrossRef]
- Ronchetti, D.; Manzoni, M.; Todoerti, K.; Neri, A.; Agnelli, L. In Silico Characterization of miRNA and Long Non-Coding RNA Interplay in Multiple Myeloma. Genes 2016, 7, 107. [Google Scholar] [CrossRef]
- Zhu, F.X.; Wang, X.T.; Ye, Z.Z.; Gan, Z.P.; Lai, Y.R. Construction of a prognosisassociated long noncoding RNAmRNA network for multiple myeloma based on microarray and bioinformatics analysis. Mol. Med. Rep. 2020, 21, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wu, Y.; Gao, W.; Tian, Y.; Wang, G.; Liu, A.; Chen, W. Novel Non-coding RNA Analysis in Multiple Myeloma Identified Through High-Throughput Sequencing. Front. Genet 2021, 12, 625019. [Google Scholar] [CrossRef]
- Kumar, S.K.; Lee, J.H.; Lahuerta, J.J.; Morgan, G.; Richardson, P.G.; Crowley, J.; Haessler, J.; Feather, J.; Hoering, A.; Moreau, P.; et al. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: A multicenter international myeloma working group study. Leukemia 2012, 26, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Pinto, V.; Bergantim, R.; Caires, H.R.; Seca, H.; Guimaraes, J.E.; Vasconcelos, M.H. Multiple Myeloma: Available Therapies and Causes of Drug Resistance. Cancers 2020, 12, 407. [Google Scholar] [CrossRef] [PubMed]
- Malek, E.; Kim, B.G.; Driscoll, J.J. Identification of Long Non-Coding RNAs Deregulated in Multiple Myeloma Cells Resistant to Proteasome Inhibitors. Genes 2016, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.H.; Du, P.; Liu, W.; An, L.K.; Li, J.; Zhu, W.Y.; Yuan, S.; Wang, L.; Zang, L. LncRNA ANRIL promotes multiple myeloma progression and bortezomib resistance by EZH2-mediated epigenetically silencing of PTEN. Neoplasma 2021, 68, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Wang, W.; Fu, B.; Pang, Y.; Lou, Y.; Li, H. Increased lncRNA HOTAIR expression promotes the chemoresistance of multiple myeloma to dexamethasone by regulating cell viability and apoptosis by mediating the JAK2/STAT3 signaling pathway. Mol. Med. Rep. 2019, 20, 3917–3923. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, Y.; Liu, W.; Huang, Y.; Shen, X.; Jing, R.; Pu, J.; Wang, X.; Ju, S.; Cong, H.; et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019, 10, 106. [Google Scholar] [CrossRef]
- Lu, D.; Yang, C.; Zhang, Z.; Cong, Y.; Xiao, M. Knockdown of Linc00515 Inhibits Multiple Myeloma Autophagy and Chemoresistance by Upregulating miR-140-5p and Downregulating ATG14. Cell Physiol. Biochem. 2018, 48, 2517–2527. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, G.; Liu, R.; Wang, Y.; Wang, Y.; Qiu, X.; Gao, X. RNase non-sensitive and endocytosis independent siRNA delivery system: Delivery of siRNA into tumor cells and high efficiency induction of apoptosis. Nanoscale 2013, 5, 7256–7264. [Google Scholar] [CrossRef]
- Shi, Y.; Parag, S.; Patel, R.; Lui, A.; Murr, M.; Cai, J.; Patel, N.A. Stabilization of lncRNA GAS5 by a Small Molecule and Its Implications in Diabetic Adipocytes. Cell Chem. Biol. 2019, 26, 319–330.e6. [Google Scholar] [CrossRef] [PubMed]
- Donlic, A.; Morgan, B.S.; Xu, J.L.; Liu, A.; Roble, C., Jr.; Hargrove, A.E. Discovery of Small Molecule Ligands for MALAT1 by Tuning an RNA-Binding Scaffold. Angew Chem. Int. Ed. Engl. 2018, 57, 13242–13247. [Google Scholar] [CrossRef] [PubMed]
- Abulwerdi, F.A.; Xu, W.; Ageeli, A.A.; Yonkunas, M.J.; Arun, G.; Nam, H.; Schneekloth, J.S., Jr.; Dayie, T.K.; Spector, D.; Baird, N.; et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. ACS Chem. Biol. 2019, 14, 223–235. [Google Scholar] [CrossRef] [PubMed]

| LncRNA | Expression | Clinical Correlation | Ref. |
|---|---|---|---|
| MALAT1 | Upregulated | PFS (shorter) | [32] |
| NEAT1 | Upregulated | OS (shorter) | [107] |
| CRNDE | Upregulated | OS (shorter) | [38] |
| PVT1 | Upregulated | PFS (shorter) | [108] |
| UCA1 | Upregulated | OS (shorter) | [109] |
| LINC00461 | Upregulated | OS (shorter) | [45] |
| TCF7 | Upregulated | OS (shorter) | [110] |
| MEG3 | Downregulated | OS (shorter) | [43] |
| OIP-AS1 | Downregulated | OS (shorter) | [111] |
| AC005307.4 AC005307.1 AC005616.1, RP11-161M6.2, RP11-23P13.6, AC005616.1, MIAT | Downregulated | t(11;14) | [112] |
| RP11-343J3.2, RP11-17M16.2, LINC01102, RP11-345J18.2, ST8SIA6-AS1 | Upregulated: RP11-343J3.2, RP11-17M16.2, LINC01102, RP11-345J18.2Downregulated: ST8SIA6-AS1 | t(4;14) | [112] |
| RP11-1085N6.5, RP5-887A10.1, RP11-212I21.4, MIR222HG, LINC00158 | Upregulated: RP11-1085N6.5, RP11-212I21.4, LINC00158 Downregulated: RP5-887A10.1, MIR222HG | MAF translocations | [112] |
| PDLIM1P4, ENSG00000249988, ENSG00000254343 | Upregulated | PFS (shorter) | [10] |
| PDLIM1P4, SMILO, ENSG00000249988 | Upregulated: PDLIM1P4 Downregulated: SMILO, ENSG00000249988 | OS (shorter) | [10] |
| RP4-803J11.2, RP1-43E13.2, ZFY-AS1, RP11-553 L6.5 | Upregulated in high-risk patients: RP4-803 J11.2, RP1-43E13.2 Upregulated in low-risk patients: ZFY-AS1, RP11-553 L6.5 | Survival (shorter in high-risk, longer in low-risk) | [29] |
| LncRNA Target | Method | Platform | Effect | Ref. |
|---|---|---|---|---|
| HOXC-AS3 | siRNA | In vitro | MM-MSCs osteogenic differentiation | [84] |
| HOXC-AS3 | In vivo grade siRNA | In vivo | Decreased bone loss | [84] |
| MALAT1 | ASO | In vitro | Reduced tumor cell viability and motility | [128] |
| MALAT1 | ASO | In vivo | Reduced tumor growth | [128] |
| MALAT1 | ASO | In vitro | Reduced tumor cell viability, increased sensitivity to Bortezomib, Melphalan, Doxorubicin | [27] |
| MALAT1 | SWCNT-ASO | In vivo | Reduced tumor growth, increased survival | [27] |
| NEAT1 | ASO | In vitro | Reduced tumor cell viability, increased sensitivity to Bortezomib, Melphalan, Carfilzomib | [28] |
| NEAT1 | ASO | In vivo | Reduced tumor growth | [28] |
| CRNDE | CRISPR-Cas9 | In vitro | Reduced IL6R expression, Reduced tumor cell proliferation, increased sensitivity to Bortezomib, Dexamethasone | [38] |
| CRNDE | CRISPR-Cas9 | In vivo | Reduced tumor growth | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saltarella, I.; Apollonio, B.; Lamanuzzi, A.; Desantis, V.; Mariggiò, M.A.; Desaphy, J.-F.; Vacca, A.; Frassanito, M.A. The Landscape of lncRNAs in Multiple Myeloma: Implications in the “Hallmarks of Cancer”, Clinical Perspectives and Therapeutic Opportunities. Cancers 2022, 14, 1963. https://doi.org/10.3390/cancers14081963
Saltarella I, Apollonio B, Lamanuzzi A, Desantis V, Mariggiò MA, Desaphy J-F, Vacca A, Frassanito MA. The Landscape of lncRNAs in Multiple Myeloma: Implications in the “Hallmarks of Cancer”, Clinical Perspectives and Therapeutic Opportunities. Cancers. 2022; 14(8):1963. https://doi.org/10.3390/cancers14081963
Chicago/Turabian StyleSaltarella, Ilaria, Benedetta Apollonio, Aurelia Lamanuzzi, Vanessa Desantis, Maria Addolorata Mariggiò, Jean-François Desaphy, Angelo Vacca, and Maria Antonia Frassanito. 2022. "The Landscape of lncRNAs in Multiple Myeloma: Implications in the “Hallmarks of Cancer”, Clinical Perspectives and Therapeutic Opportunities" Cancers 14, no. 8: 1963. https://doi.org/10.3390/cancers14081963
APA StyleSaltarella, I., Apollonio, B., Lamanuzzi, A., Desantis, V., Mariggiò, M. A., Desaphy, J.-F., Vacca, A., & Frassanito, M. A. (2022). The Landscape of lncRNAs in Multiple Myeloma: Implications in the “Hallmarks of Cancer”, Clinical Perspectives and Therapeutic Opportunities. Cancers, 14(8), 1963. https://doi.org/10.3390/cancers14081963








