Ontologies and Knowledge Graphs in Oncology Research
Simple Summary
Abstract
1. Introduction
2. Background
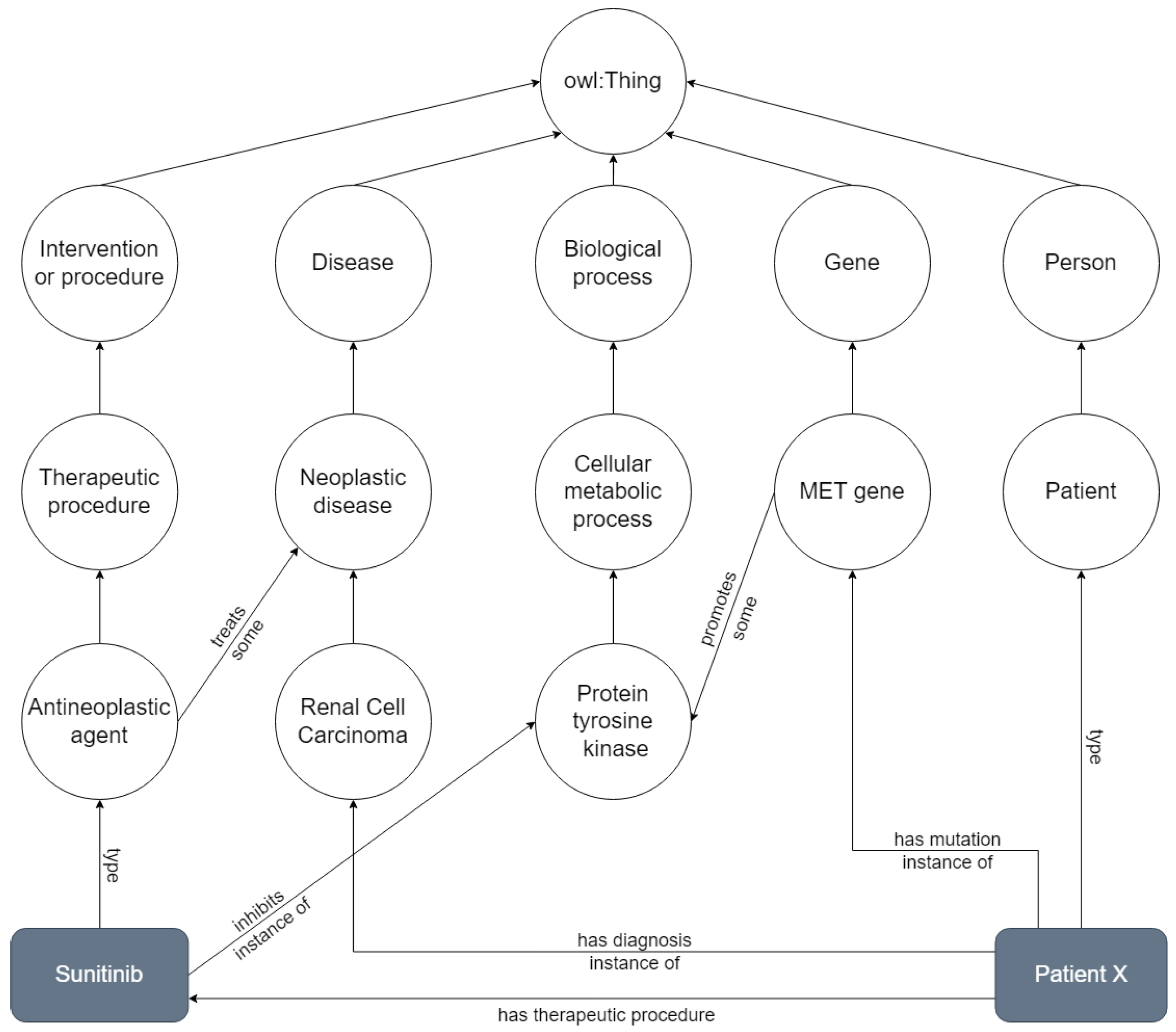
2.1. Ontologies
2.2. Ontologies in Cancer Research
3. Materials and Methods
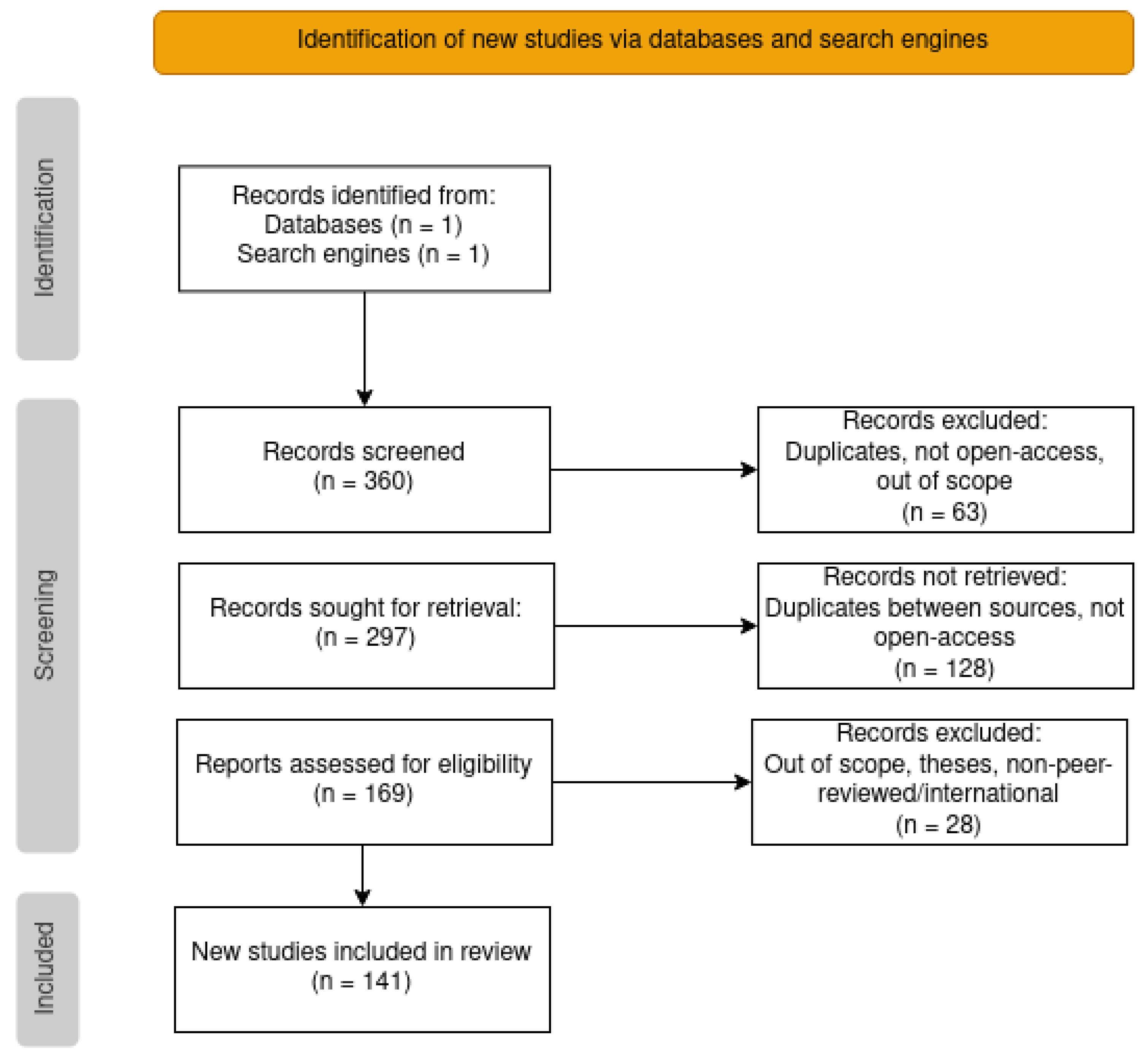
3.1. Initial Search and Screening
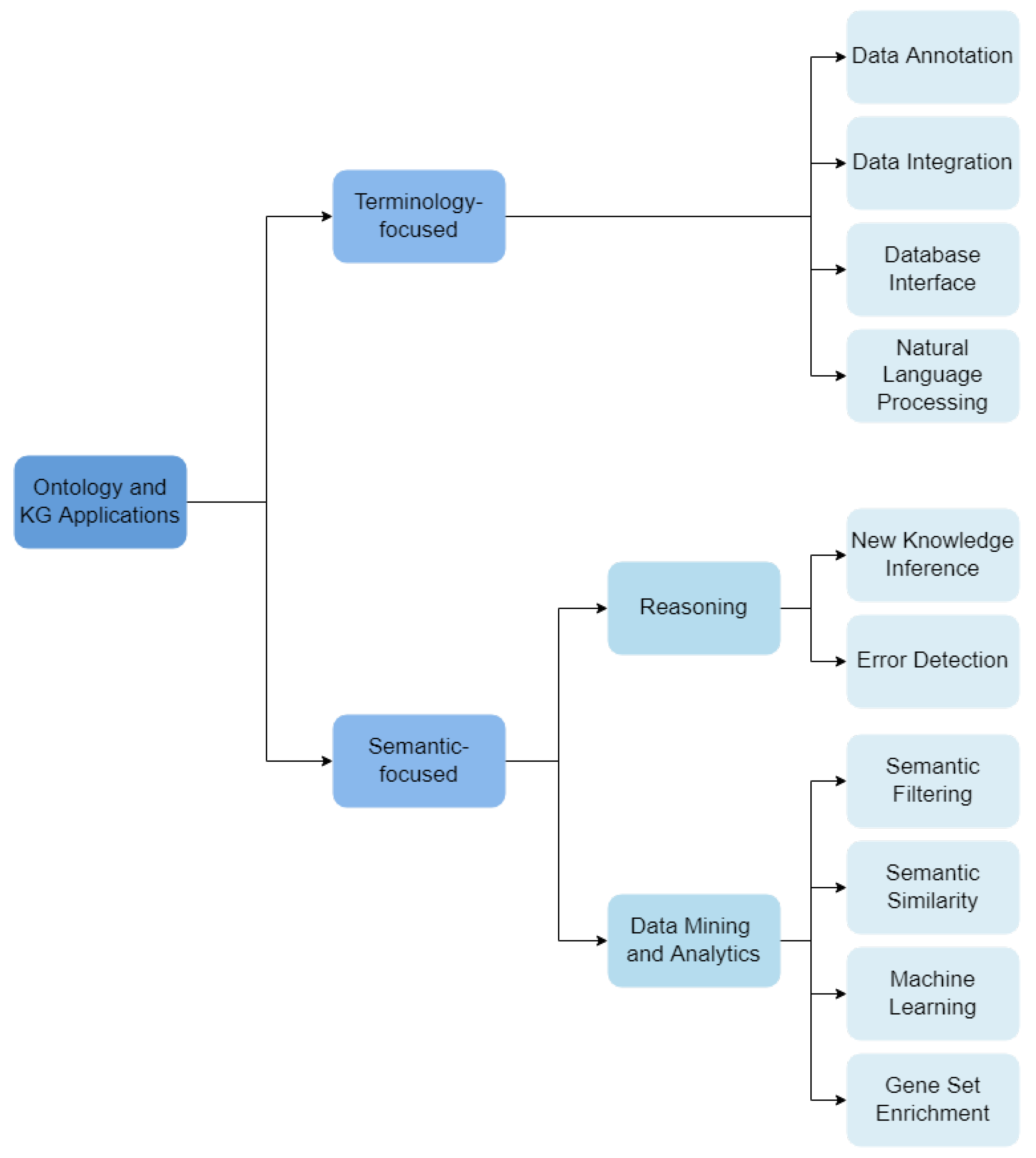
3.2. Categorization
- Data Annotation: ontologies are used to describe data under a common schema, linking data objects to ontology classes that describe them.
- Data Integration: ontologies support the integration of different data sets or databases.
- Database Interface: ontologies are used to support user interfaces for databases, where labels of ontology classes and relations allow text annotation. These interfaces are notably useful in dealing with medical data, for integration and querying of different knowledge resources.
- NLP: ontologies are used as the vocabulary source for Natural Language Processing (NLP) methods, where entities, events or relations in a text are identified through the corresponding ontology labels.
- Reasoning: Automatic reasoners process ontologies’ axioms and their formal definitions.
- –
- Inference of New Knowledge: complex reasoning-based queries can reveal novel biological knowledge based on the already defined axioms.
- –
- Error Detection: reasoning applied to check for consistency (or contradictions) in the ontology.
- Data Mining and Analytics: ontologies are used to support data mining and analytics tasks.
- –
- Semantic Filtering: ontology-based annotations are used to filter and process data.
- –
- Semantic Similarity: ontology-based annotations are used to compare data entities.
- –
- Machine Learning: ontologies and KGs are explored by machine learning algorithms.
- –
- Gene Set Enrichment: statistical analysis of gene set ontology-annotations.
4. Ontologies in Oncology
4.1. Ontologies Used in the Reviewed Applications
4.2. Ontologies Created for the Reviewed Applications
| Ref | Objective | Ontology Name | Domain | Reused Ontologies | Language |
|---|---|---|---|---|---|
| [51] | Model lung cancer for the clinical decision support application Lung Cancer Assistant | LUCADA ontology | Clinical | SNOMED-CT | OWL |
| [33] | Use a hybrid approach to build a breast cancer ontology | N/A | Breast Cancer | N/A | OWL |
| [32] | Describe cancer cells and capture the properties of tumorigenesis | OncoCL | Cell Lines | CL, UBERON, BTO, Pathway Ontology, PATO, CPO, SO | OWL |
| [11] | Represent the project domain and link the NeoMark data to other domains | NeoMark ontology | Clinical | BFO, RO | OWL |
| [50] | Cancer reclassification and drug inference | N/A | Farmacology | N/A | N/A |
| [54] | Drug target prediction | CRC ontology | Colorectal Cancer | PharmGKB | OWL |
| [63] | Assist medical students and professionals in the breast cancer domain | OntoMama | Clinical | N/A | N/A |
| [34] | Development of an ontology-driven survivor engagement framework for mobile apps | POCS | Social | FOAF | OWL |
| [46] | Creation of TNM-O | TNM-O | Anatomical | FMA, BioTopLite 2 | OWL |
| [41] | Represent obesity-related cancer (ORC) ontology to organize information and allow data querying | FOORC | Obesity Related Cancer | DOID | OWL |
| [49] | Extraction of association rules from large datasets on gastric cancer patients | Gastric cancer ontology | Clinical | N/A | N/A |
| [55] | Aid data integration; enable association between SE variables and health outcomes | OCRSEV | Social-Ecological Factors | BFO | OWL |
| [45] | Interoperability across quantitative histopathological imaging data sets | QHIO | Imaging | OBI | OWL |
| [39] | Design of a semantic model for local cancer registries | N/A | Epidemiology | SIO, OBI | OWL |
| [40] | Development of ontologies for the public health domain | N/A | Public Health | N/A | OWL |
| [61] | Understand cellular responses to different perturbations | LINCS-CLOview | Cell Lines | CLO | OWL |
| [53] | Integrate heterogeneous datasets | OCRV | Cancer Outcomes | BFO, NCIt, TEO | OWL |
| [47] | Define a specific terminological system to standardized data collection for head and neck cancer patients | ENT COBRA ontology | Clinical | N/A | N/A |
| [10] | Use structured knowledge representation with concepts of treatment end points | CCTOO | Clinical | NCIt, CTCAE | OBO |
| [62] | Represent the data elements identified by the synoptic worksheets of College of American Pathologists | SNOMED CT observable ontology | Clinical | SNOMED CT, LOINC | N/A |
| [35] | Create a standardized hierarchic ontology of cancer treatments, mapped to standard nomenclatures | N/A | Cancer Treatments | HemOnc | OWL |
| [56] | Increase interoperability between data sources to allow the creation of Big Data studies involving several treatment centers | ROS | Radiation Oncology | FMA | OWL |
| [36] | Create temporal ontology of survival outcome measures of clinical trials in oncology | TOCSOC | Clinical | EFO, CCTOO, IOBC, NCIT | OWL |
| [60] | Provide an ontological representation of immunophenotyping cell types found in hematologic malignancies | CCL | Hematologic Malignancies | CL | OWL |
| [42] | Semi-automatic development of CHV for breast cancer | MuEVo | Clinical | MeSH, MedDRA, SNOMEDint | SKOS |
| [44] | Offer ontology-based approach modeling HCC tumors | OntHCC | Liver Cancer | N/A | OWL |
| [57] | Support integrative data analysis in cancer outcomes research | ODVDS | Risk Factors | BFO | OWL |
| [58] | Cytological tissue image analysis of cervical cancer | CCOWL | Cervical Cancer | N/A | OWL |
| [31] | Standardize the terminology used in the selection and integration steps of RF variables and data sources | OD-ATTEST | Risk Factors | BFO, others in NCBO (not specified) | OWL |
| [48] | Standardize data collection for non-melanoma skin cancer patients treated with brachytherapy | SKIN-COBRA ontology | Clinical | N/A | N/A |
| [43] | Analyze social media data to identify information needs and emotions related to cancer | N/A | Social | LCO, BCO, GCO, SOSW | N/A |
| [37] | Solve the heterogeneity and diversity of different data types related to prostate cancer by establishing a standardized lifestyle ontology | PCLiON | Risk Factors | NCIT, WordNet, SNOMED CT, The Cochrane Library, FooDB, CheBI | OWL |
| [59] | Build a knowledge graph that represents causal associations between incidence of breast cancer and risk factors | RiskExplorer | Clinical | UMLS | N/A |
| [30] | Facilitate the integrity and maintenance of ENCR core data set. | ENCR core-data | Epidemiology | N/A | OWL |
| [14] | Minimizing vagueness in the formalization of medical knowledge | BCFO | Clinical | DO | OWL |
| [52] | Predict side effects of bladder cancer treatments | N/A | Bladder Cancer | N/A | OWL |
| [38] | Provide a generalizing pattern of more concise definitions to correctly classify all tumor configurations | N/A | Gastrointestinal Tumors | BioTopLite2 | N/A |
5. Ontologies and Knowledge Graph Applications in Cancer Research
5.1. Terminology-Focused Applications
5.1.1. Data Annotation
5.1.2. Data Integration
5.1.3. Database Interfaces
5.1.4. Natural Language Processing
| Ref | Summary | Ontologies | Data | Tag | Cancer Type |
|---|---|---|---|---|---|
| [51] | Ontology for a clinical decision support system to produce treatment recommendations | SNOMED-CT, New ontology | N/A | Database Interface | Lung |
| [74] | Ontology-based querying for cancer research data | NCIt | N/A | Database Interface | Various |
| [77] | Mining of genetic marker data in a journal | SNOMED-CT, HUGO | NEJM | NLP | Various |
| [11] | Automatic translation of NeoMark relational database | BFO, RO, OBI, OGMS, HDO | NeoMark database | Data Integration | OSCC |
| [15] | Manual identification and inference of associations between breast cancer drugs | New ontology | PharmGKB, NCI | Data Annotation | Breast |
| [65] | Genome-wide functional predictions of lncRNAs | GO | Gencode, Ensembl, ENCODE project LncRNA Ontology | Data Integration | Various |
| [66] | Extraction of semantic entities in eligibility criteria and annotation | UMLS | CTG | Data Integration, Database Interface, NLP | Breast |
| [34] | Development of an ontology-driven survivor engagement framework for mobile apps | FOAF | N/A | Database Interface, Data Annotation | POCS |
| [67] | Prediction of clinical outcomes from a graph-based approach with multi-omics and genetic data | GO | TCGA | Data Integration | Ovarian |
| [68] | Development of a focused view within the DO from cancer datasets | DO | COSMIC, TCGA, ICGC, TARGET, IO, EDRN | Data Integration | Various |
| [39] | Development of a platform for analysis and visualization of data | ICD10, ICD-O-3, TNM staging, SIO, OBI, OQuaRE | NCRI | Data Annotation, Database Interface | Various |
| [13] | Automatic annotation of cancer hallmarks on biomedical literature | MeSH | N/A | Data Annotation, NLP | Various |
| [70] | Connection of predictors with cancer survival with a use-case ontology | OCRV | FCDS 2000 U.S. census, BRFSS | Data Integration | Various |
| [69] | Data integration of several databases with ontologies to enable querying of patient data | DO, UBERON | TCIA, TCGA, LIDC-IDRI, Head-Neck-PET-CT | Data Integration | Various |
| [78] | Construcion of OCRV based on data analysis needs | NCIt, TEO, ICD-O-3, ICD-9-CM | UF Health CCCA, FCDS, ATSDR, USCB, BRFSS, County Health Ranking & Roadmaps | Data Integration | Various |
| [64] | Manual representation of semantic temporal components of CDEs | TEO | NCI, caDSR | Data Annotation | Various |
| [44] | Ontology built following the MethOntology methodology [79] | DICOM | University Hospital of Clermont-Ferrand | Data Annotation | HCC |
| [42] | Semi-automatic development of CHV for breast cancer | INDC dictionary | N/A | NLP | Various |
| [71] | KG of cancer registry data, with data analysis and visualization | New ontology | LTR | Data Integration, Database Interface | Various |
| [43] | Development of an ontology to understand information needs and emotions | LCO, BCO, GCO, SOSW | N/A | NLP | Various |
| [72] | KGHC is a KG constructed from clinical data available publicly | UMLS | PubMed, UpToDate, CTG, SemMedDB | Data Integration | HCC |
| [76] | Functional annotation of circRNAs obtained from sequencing lung cell lines | GO | Lung cell lines sequencing data | Database Interface | Lung |
| [12] | IMI is a web-based system that creates mappings from the NAACCR data dictionary to NCIt | NAACCR data dictionary, NCIt | KCR | Data Integration, Database Interface | Various |
| [73] | Comparative analysis of cancer hallmark mapping strategies | GO | MSigDB, KEGG, cancer hallmark mapping schemes, TCGA | Data Integration | Various |
5.2. Semantic-Focused Applications
5.2.1. Formalized Definitions and Axioms: Reasoning with Ontologies
| Ref | Objective | Input Ontologies | Reasoner | Tag | Cancer Type |
|---|---|---|---|---|---|
| [81] | Determine cancer type and stage of the patient to recommend treatments | LuCO, BCO, LCO | FaCT++ | New Knowledge Inference | Various |
| [15] | Identification of new indications for existing drugs | New ontology | Automated semantic inference (Protégé) | New knowledge Inference | Breast |
| [82] | Prediction of new drug targets | New ontology | Pellet (Protégé) | New knowledge Inference | Colorectal |
| [49] | Extraction of association rules from large datasets on gastric cancer patients | GCO | Apriori algorithm | New Knowledge Inference | Gastric |
| [38] | Provide a generalizing pattern of more concise definitions to correctly classify all tumor configurations | New ontology | HermiT DL (Protégé) | Error Detection | Various |
| [46] | Creation of TNM-O | FMA, BioTopLite 2 | HermIT DL | Error Detection | Various |
| [52] | Predict side effects of bladder cancer treatments | New ontology | Pellet (Protégé) | New knowledge Inference + Error Detection | Bladder |
| [83] | Signal rule violations in a validation process of multiple primary tumors international rules | ICD-O-3 | FaCT++, HermiT | New knowledge Inference + Error Detection | Multiple primary tumors |
| [30] | Facilitate the integrity and maintenance of ENCR core data set | New ontology | FaCT++ (Protégé) | Error Detection | Various |
| [14] | Minimizing vagueness in the formalization of medical knowledge | DO | Fuzzy DL, HermiT/Pellet (Protégé) | Error Detection | Breast |
5.2.2. Mining and Analyzing Multimodal Data with Ontologies
| Ref | Objective | Method | Input Ontologies | Input Data | Tag | Cancer Type |
|---|---|---|---|---|---|---|
| [77] | Mining of genetic marker data in a journal | MCVS NLP engine | SNOMED CT, HUGO | NEJM | ML | Various |
| [74] | Ontology-based querying for cancer research data | Construction of a OWL Generation facility | NCIt | caGrid | ML | Various |
| [11] | Represent the project domain and link the NeoMark data to other domains | Bayesian Networks, ANN, SVMs, Decision Trees, Random Forests | BFO, RO, OBI, OGMS, HDO | N/A | ML | OSCC |
| [50] | Cancer reclassification and drug inference | Vazquez Bayesian clustering algorithm | N/A | HemOnc.org | ML | Various |
| [19] | Ontological application in Clinical Decision Support | CBR and MAS | UML | Patient Health Records | ML | Gastric |
| [82] | Prediction of new drug targets | KEGG functional PharmGKB drug annotation. Network neighborhood modeling ranking | New ontology, ATC | PharmGKB, GAD, CGC, OMIM, NCI, DrugBank, TTD | ML | Colorectal |
| [39] | Design of a semantic model for local cancer registries | Ontology-driven search filters and aggregates properties of interest | ICD10, ICD-O-3, TNM staging, SIO, OBI, OQuaRE | NCRI | Filtering | Various |
| [90] | Discover patterns related to the patients’ ability to perform daily living activities | AQ21—multi-task ML and data mining system | UMLS | Surveillance, Epidemiology, and End Results—Medicare HOS | ML | Various |
| [13] | Automatic annotation of cancer hallmarks on biomedical literature | United Decision Tree and Random Forest | MeSH | Pubmed abstracts | ML | Various |
| [85] | Prediction of microRNA related to glucocorticoid resistance | Manual background literature search. Semantic searches in resulting subset | OMIT, NCRO, MeSH | PubMed | Filtering | Pediatric ALL |
| [17] | Cancer-related gene prioritization | Fuzzy similarity | GO | GSEA website, TCGA, SNP4Disease | Similarity | PAC, Breast |
| [161] | Predict drug synergy in cancer treatment | Stacked Restricted Boltzmann machine | GO, Ontology Fingerprints | AstraZeneca-Sanger Drug Combination Prediction Challenge, GDSC, KEGG | ML | Various |
| [18] | Identification of cancer driver genes with role distinction | Neuro-symbolic deep learning on semantic knowledge representation on genetic information | CMPO, GO, MP | Uniprot, MGI database, Mutational Cancer Drivers Database, CPD | ML | Naso-pharyngeal, Colorectal |
| [87] | Identification of relevant, expression data non-redundant cancer gene markers | Unsupervised Multi-View Multi-Objective clustering | GO | Gene expression datasets from own lab | ML | Prostate, DLBCL, FL |
| [58] | Predict cervical cancer cells from cytological tissue images | DNN | New ontology | hospital cervical cancer data, kaggle data repository | ML | Cervical |
| [88] | Complement system role inference from immunofunctionome analysis | SVMs | GO | GEO database | ML | OCCC |
| [89] | Cancer detection based on gene expression data | Multilayer Perceptrons | GO | Affymetrix HG-U133Plus2 chip arrays, TCGA | ML | Various |
| [91] | Tolerating data missing in breast cancer diagnosis from clinical ultrasound reports | KG embeddings | BI-RADS | Ultrasound reports | ML | Breast |
| [92] | Real-time inference on a lung KG | GAT | New ontology | KEGG, Uniprot, DrugBank, TCGA | ML | Lung |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ATC | Anatomical Therapeutic Chemical |
| ATSDR | Agency for Toxic Substances and Disease Registry |
| ALL | Acute Lymphoblastic Leukemia |
| ANN | Artificial Neural Network |
| BCFO | Breast Cancer Fuzzy Ontology |
| BCO | Breast Cancer Ontology |
| BFO | Basic Formal Ontology |
| BRFSS | Behavioral Risk Factor Surveillance System |
| BTL2 | BioTopLite 2 |
| caDSR | Cancer Data Standards Repository |
| CBR | Case-Based Reasoning |
| CCL | Cancer Cell Ontology |
| CCTOO | Cancer Care Treatment Outcome Ontology |
| CDEs | Common Data Elements |
| CGC | Cancer Gene Census |
| CHV | Consumer Health Vocabulary |
| CL | Cell Ontology |
| CLO | Cell Line Ontology |
| CMPO | Cellular Microscopy Phenotype Ontology |
| COBRA | COnsortium for BRachytherapy data Analysis |
| COnQueSt | Cancer Ontology Querying System |
| CPD | Cellular Phenotype Database |
| CTCAE | Common Terminology Criteria for Adverse Events |
| CTG | ClinicalTrials.gov |
| DICOM | Digital Imaging and Communications in Medicine |
| DL | Description Logic |
| DLBCL | Diffuse Large B Cell Lymphoma |
| DO | Disease Ontology |
| EFO | Experimental Factor Ontology |
| ENCR | European Network of Cancer Registries |
| ENCR core-data | European Cancer-Registry core-data ontology |
| FCDS | Florida Cancer Data System |
| FL | Follicular Lymphoma |
| FMA | Foundational Model of Anatomy |
| FOAF | Friend of a Friend ontology |
| FOORC | Fuzzy Ontology for Obesity-Related Cancer |
| GAD | Genetic Association Database |
| GCO | Gastric Cancer Ontology |
| GDSC | Genomics of Drug Sensitivity in Cancer |
| GO | Gene Ontology |
| HDO | Human Disease Ontology |
| HCC | Hepatocellular Carcinoma |
| HOS | Health Outcomes Survey |
| HUGO Gene Nomenclature | Human Genome Organization Gene Nomenclature |
| ICD-9-CM | International Classification of Diseases Ninth Revision Clinical |
| Modification | |
| ICD-O-3 | International Classification of Disease for Oncology 3rd edition |
| IMI | Interactive Mapping Interface |
| IOBC | Interlinking Ontology for Biological Concepts |
| KCR | Kentucky Cancer Registry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KG | Knowledge Graph |
| LCO | Liver Cancer Ontology |
| LCKGO | Lung Cancer Knowledge Graph Ontology |
| LINCS | Library of Integrated Network-based Cellular Signatures |
| lncRNAs | long non-coding RNAs |
| LOINC | Logical Observation Identifier Names and Codes |
| LTR | Louisiana Tumor Registry |
| LuCO | Lung Cancer Ontology |
| MAS | Multi-Agent System |
| MCVS | Multi-threaded Clinical Vocabulary Server |
| MedDRA | Medical Dictionary for Regulatory Activities |
| MeSH | Medical Subject Headings |
| MGI | Mouse Genome Informatics |
| ML | Machine Learning |
| MP | Mammalian Phenotype ontology |
| MuEVo | Multi-Expertise Vocabulary |
| NAACCR | North American Association of Central Cancer Registries |
| NCI | National Cancer Institute |
| NCIt | National Cancer Institute Thesaurus |
| NCRI | National Cancer Registry Ireland |
| NCRO | Non-Coding RNA Ontology |
| NEJM | New England Journal of Medicine |
| NLP | Natural Language Processing |
| OBDA | Ontology-Based Data Access |
| OBI | Ontology for Biomedical Investigators |
| OCCC | Ovarian clear cell carcinoma |
| OCRV | Ontology for Cancer Research Variables |
| OCRSEV | Ontology of Cancer Related Social-Ecological Variables |
| OD-ATTEST | Ontology for the Documentation of vAriable selecTion and daTa |
| sourcE Selection and inTegration | |
| ODVDS | Ontology for Documentation of Variable and Data Source |
| OGMS | Ontology of General Medical Science |
| OIE | Open Information Extraction |
| OMIM | Online Mendelian Inheritance in Man |
| OMIT | Ontology for MicroRNA Target |
| OntHCC | Ontology of Hepatocellular Carcinoma |
| OQuaRE | Ontology Quality Evaluation Framework |
| OSCC | Oral Squamous Cell Carcinoma |
| OWL | Web Ontology Language |
| PAC | Prostatic Adenocarcinoma |
| POCS | Profile Ontology for Cancer Survivors |
| QHIO | Quantitative Histopathological Imaging Ontology |
| RO | Relation Ontology |
| ROS | Radiation Oncology Structures |
| SCRS | Semantic Cancer Registry System |
| SEER-MHOS | Surveillance, Epidemiology, and End Results—Medicare Health |
| Outcomes Survey | |
| SIO | Semanticscience Integrated Ontology |
| SKOS | Simple Knowledge Organization System |
| SNOMED CT | Systematized Nomenclature of Medicine Clinical Terms |
| SNOMEDint | SNOMED International |
| SOSW | Sentiment Ontology for Social Web |
| SVMs | Support Vector Machines |
| SWIT | Semantic Web Integration Tool |
| TCGA | The Cancer Genome Atlas |
| TEO | Time Event Ontology |
| TNM | Tumor–Node–Metastasis |
| TNM-O | Tumor–Node–Metastasis Ontology |
| TOCSOC | Temporal Ontology for Comparing the Survival Outcomes |
| TTD | Therapeutic Target Database |
| UMLS | Unified Medical Language System |
| USCB | United States Census Bureau |
References
- SNOMED International. Available online: https://www.snomed.org/ (accessed on 25 March 2022).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Whetzel, P.L.; Noy, N.F.; Shah, N.H.; Alexander, P.R.; Nyulas, C.; Tudorache, T.; Musen, M.A. BioPortal: Enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011, 39, W541–W545. [Google Scholar] [CrossRef]
- Golbeck, J.; Fragoso, G.; Hartel, F.; Hendler, J.; Oberthaler, J.; Parsia, B. The National Cancer Institute’s thesaurus and ontology. J. Web Semant. First Look 2003, 1, 4. [Google Scholar]
- Chin, L.; Andersen, J.N.; Futreal, P.A. Cancer genomics: From discovery science to personalized medicine. Nat. Med. 2011, 17, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Gruber, T.R. Toward principles for the design of ontologies used for knowledge sharing? Int. J. Hum.-Comput. Stud. 1995, 43, 907–928. [Google Scholar] [CrossRef]
- McGuinness, D.L. Ontologies come of age. In Spinning the Semantic Web: Bringing the World Wide Web to Its Full Potential; MIT Press: Cambridge, MA, USA, 2002; pp. 171–194. [Google Scholar]
- OWL 2 Web Ontology Language Document Overview (Second Edition). Available online: https://www.w3.org/TR/owl2-overview/ (accessed on 25 March 2022).
- Gutiérrez, C.; Sequeda, J.F. Knowledge graphs. Commun. ACM 2021, 64, 96–104. [Google Scholar] [CrossRef]
- Lin, F.P.; Groza, T.; Kocbek, S.; Antezana, E.; Epstein, R.J. Cancer Care Treatment Outcome Ontology: A novel computable ontology for profiling treatment outcomes in patients with solid tumors. JCO Clin. Cancer Inform. 2018, 2, 1–14. [Google Scholar] [CrossRef]
- Salvi, D.; Picone, M.; Arredondo, M.T.; Cabrera-Umpierrez, M.F.; Esteban, Á.; Steger, S.; Poli, T. Merging person-specific bio-markers for predicting oral cancer recurrence through an ontology. IEEE Trans. Biomed. Eng. 2013, 60, 216–220. [Google Scholar] [CrossRef]
- Tao, S.; Zeng, N.; Hands, I.; Hurt-Mueller, J.; Durbin, E.B.; Cui, L.; Zhang, G.Q. Web-based interactive mapping from data dictionaries to ontologies, with an application to cancer registry. BMC Med. Inform. Decis. Mak. 2020, 20, 271. [Google Scholar] [CrossRef]
- Yan, S.; Wong, K. Elucidating high-dimensional cancer hallmark annotation via enriched ontology. J. Biomed. Inform. 2017, 73, 84–94. [Google Scholar] [CrossRef]
- Oyelade, O.N.; Ezugwu, A.E.; Adewuyi, S.A. Enhancing reasoning through reduction of vagueness using fuzzy OWL-2 for representation of breast cancer ontologies. Neural Comput. Appl. 2021, 34, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Tao, C.; Shen, F.; Chute, C.G. Exploring the pharmacogenomics knowledge base (PharmGKB) for repositioning breast cancer drugs by leveraging Web ontology language (OWL) and cheminformatics approaches. Pac. Symp. Biocomput. 2014, 2014, 172–182. [Google Scholar]
- Agioutantis, P.C.; Loutrari, H.; Kolisis, F.N. Computational analysis of transcriptomic and proteomic data for deciphering molecular heterogeneity and drug responsiveness in model human hepatocellular carcinoma cell lines. Genes 2020, 11, 623. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Liu, G.; Bai, T.; Meng, X.; Ma, Q. MGOGP: A gene module-based heuristic algorithm for cancer-related gene prioritization. BMC Bioinform. 2018, 19, 215. [Google Scholar] [CrossRef]
- Althubaiti, S.; Karwath, A.; Dallol, A.; Noor, A.; Alkhayyat, S.S.; Alwassia, R.; Mineta, K.; Gojobori, T.; Beggs, A.D.; Schofield, P.N.; et al. Ontology-based prediction of cancer driver genes. Sci. Rep. 2019, 9, 17405. [Google Scholar] [CrossRef]
- Shen, Y.; Colloc, J.; Jacquet-Andrieu, A.; Lei, K. Emerging medical informatics with case-based reasoning for aiding clinical decision in multi-agent system. J. Biomed. Inform. 2015, 56, 307–317. [Google Scholar] [CrossRef]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 10 January 2022).
- Google Scholar. Available online: https://ncit.nci.nih.gov/ncitbrowser/ (accessed on 21 March 2022).
- NCI Thesaurus. Available online: https://scholar.google.com/ (accessed on 25 March 2022).
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef]
- Medical Subject Headings. Available online: https://www.nlm.nih.gov/mesh/meshhome.html (accessed on 25 March 2022).
- Schriml, L.M.; Mitraka, E.; Munro, J.; Tauber, B.; Schor, M.; Nickle, L.; Felix, V.; Jeng, L.; Bearer, C.; Lichenstein, R.; et al. Human Disease Ontology 2018 update: Classification, content and workflow expansion. Nucleic Acids Res. 2019, 47, D955–D962. [Google Scholar] [CrossRef]
- World Health Organization (WHO). International Classification of Diseases for Oncology (ICD-O), 3rd ed.; 1st Revision; World Health Organization (WHO): Geneva, Switzerland, 2013.
- Bandrowski, A.; Brinkman, R.; Brochhausen, M.; Brush, M.H.; Bug, B.; Chibucos, M.C.; Clancy, K.; Courtot, M.; Derom, D.; Dumontier, M.; et al. The ontology for biomedical investigations. PLoS ONE 2016, 11, e0154556. [Google Scholar] [CrossRef]
- Sarntivijai, S.; Lin, Y.; Xiang, Z.; Meehan, T.F.; Diehl, A.D.; Vempati, U.D.; Schürer, S.C.; Pang, C.; Malone, J.; Parkinson, H.; et al. CLO: The cell line ontology. J. Biomed. Semant. 2014, 5, 1–10. [Google Scholar] [CrossRef]
- Li, F.; Du, J.; He, Y.; Song, H.Y.; Madkour, M.; Rao, G.; Xiang, Y.; Luo, Y.; Chen, H.W.; Liu, S.; et al. Time event ontology (TEO): To support semantic representation and reasoning of complex temporal relations of clinical events. J. Am. Med. Inform. Assoc. 2020, 27, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, N.C.; Giusti, F.; Bettio, M.; Negrao Carvalho, R.; Dimitrova, N.; Dyba, T.; Flego, M.; Neamtiu, L.; Randi, G.; Martos, C. An ontology-based approach for developing a harmonised data-validation tool for European cancer registration. J. Biomed. Semant. 2021, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guo, Y.; Prosperi, M.; Bian, J. An ontology-based documentation of data discovery and integration process in cancer outcomes research. BMC Med. Inform. Decis. Mak. 2020, 20, 292. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.E.; Dolan, M.E. OncoCL: A Cancer Cell Ontology; ICBO: Lansing, MI, USA, 2013; p. 126. [Google Scholar]
- Jusoh, F.; Ibrahim, R.; Othman, M.S.; Omar, N. Development of breast cancer ontology based on hybrid approach. Int. J. Innov. Comput. 2013, 3, 1. [Google Scholar]
- Myneni, S.; Amith, M.; Geng, Y.; Tao, C. Towards an ontology-driven framework to enable development of personalized mHealth solutions for cancer survivors’ engagement in healthy living. Stud. Health Technol. Inform. 2015, 216, 113–117. [Google Scholar]
- Malty, A.M.; Jain, S.K.; Yang, P.C.; Harvey, K.; Warner, J.L. Computerized approach to creating a systematic ontology of hematology/oncology regimens. JCO Clin. Cancer Inform. 2018, 2, 1–11. [Google Scholar] [CrossRef]
- Dinakarpandian, D.; Liedtke, M.; Musen, M.A.; Dinakar, B. TOCSOC: A Temporal Ontology for Comparing the Survival Outcomes of Clinical Trials in Oncology; ICBO: Lansing, MI, USA, 2018. [Google Scholar]
- Chen, Y.; Yu, C.; Liu, X.; Xi, T.; Xu, G.; Sun, Y.; Zhu, F.; Shen, B. PCLiON: An ontology for data standardization and sharing of prostate cancer associated lifestyles. Int. J. Med Inform. 2021, 145, 104332. [Google Scholar] [CrossRef]
- Herrmann, J.; Zabka, S.; Boeker, M.; Schulz, S. Ontology Patterns for Tubular or Spherical Layered Structures. A Case Study from Oncology. In Proceedings of the Joint Ontology Workshop, Graz, Austria, 23–25 September 2019; Volume 2518. [Google Scholar]
- Esteban-Gil, A.; Fernández-Breis, J.T.; Boeker, M. Analysis and visualization of disease courses in a semantically-enabled cancer registry. J. Biomed. Semant. 2017, 8, 46. [Google Scholar] [CrossRef]
- Amith, M.; Song, H.Y.; Zhang, Y.; Xu, H.; Tao, C. Lightweight predicate extraction for patient-level cancer information and ontology development. BMC Med. Inform. Decis. Mak. 2017, 17, 73. [Google Scholar] [CrossRef]
- Elhefny, M.; Elmogy, M.; Elfetouh, A.; Badria, F. FOORC: A Fuzzy Ontology-Based Representation for Obesity Related Cancer Knowledge. Int. J. Intell. Comput. Inf. Sci. 2016, 16, 15–36. [Google Scholar] [CrossRef]
- Tapi Nzali, M.D.; Aze, J.; Bringay, S.; Lavergne, C.; Mollevi, C.; Optiz, T. Reconciliation of patient/doctor vocabulary in a structured resource. Health Inform. J. 2019, 25, 1219–1231. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, H.A.; Park, S.K.; Song, T.M. Using social media data to understand consumers’ information needs and emotions regarding cancer: Ontology-based data analysis study. J. Med. Internet Res. 2020, 22, e18767. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, R.; Jaziri, F.; Mtibaa, A.; Grand-Brochier, M.; Ali, H.M.; Amouri, A.; Fourati, H.; Chabrot, P.; Gargouri, F.; Vacavant, A. Ontology-based approach for liver cancer diagnosis and treatment. J. Digit. Imaging 2019, 32, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Gurcan, M.N.; Tomaszewski, J.; Overton, J.A.; Doyle, S.; Ruttenberg, A.; Smith, B. Developing the Quantitative Histopathology Image Ontology (QHIO): A case study using the hot spot detection problem. J. Biomed. Inform. 2017, 66, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Boeker, M.; França, F.; Bronsert, P.; Schulz, S. TNM-O: Ontology support for staging of malignant tumors. J. Biomed. Semant. 2016, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, L.; Budrukkar, A.; Lenkowicz, J.; Cambeiro, M.; Bussu, F.; Guinot, J.L.; Hildebrandt, G.; Johansson, B.; Meyer, J.E.; Niehoff, P.; et al. ENT COBRA ONTOLOGY: The covariates classification system proposed by the Head & Neck and Skin GEC-ESTRO Working Group for interdisciplinary standardized data collection in head and neck patient cohorts treated with interventional radiotherapy (brachytherapy). J. Contemp. Brachyther. 2018, 10, 260–266. [Google Scholar]
- Lancellotta, V.; Guinot, J.L.; Fionda, B.; Rembielak, A.; Di Stefani, A.; Gentileschi, S.; Federico, F.; Rossi, E.; Guix, B.; Chyrek, A.J.; et al. SKIN-COBRA (Consortium for Brachytherapy data Analysis) ontology: The first step towards interdisciplinary standardized data collection for personalized oncology in skin cancer. J. Contemp. Brachyther. 2020, 12, 105–110. [Google Scholar] [CrossRef]
- Mahmoodi, S.A.; Mirzaie, K.; Mahmoudi, S.M. A new algorithm to extract hidden rules of gastric cancer data based on ontology. Springerplus 2016, 5, 312. [Google Scholar] [CrossRef][Green Version]
- Gao, M.; Warner, J.; Yang, P.; Alterovitz, G. On the Bayesian derivation of a treatment-based cancer ontology. AMIA Summits Transl. Sci. Proc. 2014, 2014, 209–217. [Google Scholar]
- Sesen, M.B.; Banares-Alcántara, R.; Fox, J.; Kadir, T.; Brady, J.M. Lung Cancer Assistant: An ontology-driven, online decision support prototype for lung cancer treatment selection. In Proceedings of the OWL: Experiences and Directions Workshop (OWLED), Heraklion, Greece, 27–28 May 2012. [Google Scholar]
- Barki, C.; Rahmouni, H.B.; Labidi, S. Prediction of Bladder Cancer Treatment Side Effects Using an Ontology-Based Reasoning for Enhanced Patient Health Safety. Informatics 2021, 8, 55. [Google Scholar] [CrossRef]
- Zhang, L.; Geng, Z.; Meng, X.; Meng, F.; Wang, L. Screening for key lncRNAs in the progression of gallbladder cancer using bioinformatics analyses. Mol. Med. Rep. 2018, 17, 6449–6455. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Sun, J.; Zheng, W.J.; Chen, J.; Xu, H. Drug Target Prediction for Colorectal Cancer by Combining Ontology and Network Approaches; ICBO: Lansing, MI, USA, 2014; p. 67. [Google Scholar]
- Balasubramanian, D.K.; Khan, J.Z.; Bian, J.; Guo, Y.; Hogan, W.R.; Hicks, A. Ontology of Cancer Related Social-Ecological Variables; ICBO: Lansing, MI, USA, 2017. [Google Scholar]
- Bibault, J.E.; Zapletal, E.; Rance, B.; Giraud, P.; Burgun, A. Labeling for Big Data in radiation oncology: The Radiation Oncology Structures ontology. PLoS ONE 2018, 13, e0191263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guo, Y.; Bian, J. Ontology for Documentation of Variable and Data Source Selection Process to Support Integrative Data Analysis in Cancer Outcomes Research. In Proceedings of the SEPDA@ ISWC, Aukland, New Zealand, 27 October 2019; pp. 63–67. [Google Scholar]
- Divakar, H.; Ramesh, D.; Prakash, B.; Tumkur, M.T. Prediction of Cervical Cancer with Ontology Based Deep Learning Approach. Int. J. Comput. Sci. Commun. 2020, 60–66. [Google Scholar]
- Daowd, A.; Barrett, M.; Abidi, S.; Abidi, S.S.R. Building a Knowledge Graph Representing Causal Associations Between Risk Factors and Incidence of Breast Cancer. In Public Health and Informatics; IOS Press: Amsterdam, The Netherlands, 2021; pp. 724–728. [Google Scholar]
- Serra, L.M.; Duncan, W.D.; Diehl, A.D. An ontology for representing hematologic malignancies: The cancer cell ontology. BMC Bioinform. 2019, 20, 181. [Google Scholar] [CrossRef]
- Ong, E.; Xie, J.; Ni, Z.; Liu, Q.; Sarntivijai, S.; Lin, Y.; Cooper, D.; Terryn, R.; Stathias, V.; Chung, C.; et al. Ontological representation, integration, and analysis of LINCS cell line cells and their cellular responses. BMC Bioinform. 2017, 18, 556. [Google Scholar] [CrossRef]
- Campbell, W.S.; Karlsson, D.; Vreeman, D.J.; Lazenby, A.J.; Talmon, G.A.; Campbell, J.R. A computable pathology report for precision medicine: Extending an observables ontology unifying SNOMED CT and LOINC. J. Am. Med. Inform. Assoc. 2018, 25, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.T.D.; Gonçalves, V.; Costa, H.; Braga, D.; Gomide, L.; Alves, C.; Brasil, L.M. OntoMama: An Ontology Applied to Breast Cancer. In MEDINFO 2015: eHealth-Enabled Health; IOS Press: Amsterdam, The Netherlands, 2015; p. 1104. [Google Scholar]
- Chen, H.W.; Du, J.; Song, H.Y.; Liu, X.; Jiang, G.; Tao, C. Representation of time-relevant common data elements in the Cancer Data Standards Repository: Statistical evaluation of an ontological approach. JMIR Med. Inform. 2018, 6, e7. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; Pan, T.; Jiang, C.; Zhao, Z.; Wang, Z.; Zhang, J.; Xu, J.; Li, X. LncRNA ontology: Inferring lncRNA functions based on chromatin states and expression patterns. Oncotarget 2015, 6, 39793–39805. [Google Scholar] [CrossRef]
- Milian, K.; Hoekstra, R.; Bucur, A.; Ten Teije, A.; van Harmelen, F.; Paulissen, J. Enhancing reuse of structured eligibility criteria and supporting their relaxation. J. Biomed. Inform. 2015, 56, 205–219. [Google Scholar] [CrossRef]
- Kim, D.; Joung, J.G.; Sohn, K.A.; Shin, H.; Park, Y.R.; Ritchie, M.D.; Kim, J.H. Knowledge boosting: A graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J. Am. Med. Inform. Assoc. 2015, 22, 109–120. [Google Scholar] [CrossRef]
- Wu, T.J.; Schriml, L.M.; Chen, Q.R.; Colbert, M.; Crichton, D.J.; Finney, R.; Hu, Y.; Kibbe, W.A.; Kincaid, H.; Meerzaman, D.; et al. Generating a focused view of disease ontology cancer terms for pan-cancer data integration and analysis. Database 2015, 2015, bav032. [Google Scholar] [CrossRef] [PubMed]
- Bona, J.P.; Nolan, T.S.; Brochhausen, M. Ontology-enhanced representations of non-image data in The Cancer Imaging Archive. In Proceedings of the 9th International Conference on Biological Ontology (ICBO 2018), Corvallis, OR, USA, 7–10 August 2018. [Google Scholar]
- Zhang, L.; Hao, C.; Li, J.; Qu, Y.; Bao, L.; Li, Y.; Yue, Z.; Zhang, M.; Yu, X.; Chen, H.; et al. Bioinformatics methods for identifying differentially expressed genes and signaling pathways in nano-silica stimulated macrophages. Tumour Biol. 2017, 39, 1010428317709284. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.M.S.; Rivera, D.; Wu, X.C.; Durbin, E.B.; Christian, J.B.; Tourassi, G. Knowledge graph-enabled cancer data analytics. IEEE J. Biomed. Health Inform. 2020, 24, 1952–1967. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, Z.; Luo, L.; Wang, L.; Zhang, Y.; Lin, H.; Wang, J. KGHC: A knowledge graph for hepatocellular carcinoma. BMC Med. Inform. Decis. Mak. 2020, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Verbeek, F.J.; Wolstencroft, K. Establishing a consensus for the hallmarks of cancer based on gene ontology and pathway annotations. BMC Bioinform. 2021, 22, 178. [Google Scholar] [CrossRef] [PubMed]
- González-Beltrán, A.; Tagger, B.; Finkelstein, A. Federated ontology-based queries over cancer data. BMC Bioinform. 2012, 13 (Suppl. 1), S9. [Google Scholar] [CrossRef]
- Oster, S.; Langella, S.; Hastings, S.; Ervin, D.; Madduri, R.; Phillips, J.; Kurc, T.; Siebenlist, F.; Covitz, P.; Shanbhag, K.; et al. caGrid 1.0: An enterprise Grid infrastructure for biomedical research. J. Am. Med Inform. Assoc. 2008, 15, 138–149. [Google Scholar] [CrossRef]
- Lyu, Y.; Caudron-Herger, M.; Diederichs, S. Circ2GO: A database linking circular RNAs to gene function. Cancers 2020, 12, 2975. [Google Scholar] [CrossRef]
- Elkin, P.L.; Frankel, A.; Liebow-Liebling, E.H.; Elkin, J.R.; Tuttle, M.S.; Brown, S.H. Bioprospecting the bibleome: Adding evidence to support the inflammatory basis of cancer. Metabolomics 2012, 2, 6451. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, Y.; Li, Q.; George, T.J.; Shenkman, E.; Modave, F.; Bian, J. An ontology-guided semantic data integration framework to support integrative data analysis of cancer survival. BMC Med. Inform. Decis. Mak. 2018, 18, 41. [Google Scholar] [CrossRef]
- Fernández-López, M.; Gómez-Pérez, A.; Juristo, N. Methontology: From ontological art towards ontological engineering. In Proceedings of the Ontological Engineering AAAI-97 Spring Symposium, Stanford, CA, USA, 24–26 March 1997. [Google Scholar]
- Musen, M.; Protégé Team. The protégé project: A look back and a look forward. AI Matters 2015, 1, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Alfonse, M.; Aref, M.M.; Salem, A.B.M. An ontology-based system for cancer diseases knowledge management. Int. J. Inf. Eng. Electron. Bus. 2014, 6, 55–63. [Google Scholar] [CrossRef]
- Tao, C.; Sun, J.; Zheng, W.J.; Chen, J.; Xu, H. Colorectal cancer drug target prediction using ontology-based inference and network analysis. Database 2015, 2015, bav015. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, N.C.; Giusti, F.; Bettio, M.; Negrao Carvalho, R.; Dimitrova, N.; Dyba, T.; Flego, M.; Neamtiu, L.; Randi, G.; Martos, C. An ontology to model the international rules for multiple primary malignant tumours in cancer registration. Appl. Sci. 2021, 11, 7233. [Google Scholar] [CrossRef]
- Rebholz-Schuhmann, D.; Oellrich, A.; Hoehndorf, R. Text-mining solutions for biomedical research: Enabling integrative biology. Nat. Rev. Genet. 2012, 13, 829–839. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, D.; Zhang, G.; Li, X.; Liang, Y.; Kasukurthi, M.V.; Li, S.; Borchert, G.M.; Huang, J. A semantics-oriented computational approach to investigate microRNA regulation on glucocorticoid resistance in pediatric acute lymphoblastic leukemia. BMC Med. Inform. Decis. Mak. 2018, 18, 57. [Google Scholar] [CrossRef]
- Pesquita, C.; Faria, D.; Falcao, A.O.; Lord, P.; Couto, F.M. Semantic similarity in biomedical ontologies. PLoS Comput. Biol. 2009, 5, e1000443. [Google Scholar] [CrossRef]
- Acharya, S.; Cui, L.; Pan, Y. Multi-view feature selection for identifying gene markers: A diversified biological data driven approach. BMC Bioinform. 2020, 21, 483. [Google Scholar] [CrossRef]
- Su, K.M.; Lin, T.W.; Liu, L.C.; Yang, Y.P.; Wang, M.L.; Tsai, P.H.; Wang, P.H.; Yu, M.H.; Chang, C.M.; Chang, C.C. The potential role of complement system in the progression of ovarian clear cell carcinoma inferred from the Gene Ontology-based immunofunctionome analysis. Int. J. Mol. Sci. 2020, 21, 2824. [Google Scholar] [CrossRef]
- Bourgeais, V.; Zehraoui, F.; Ben Hamdoune, M.; Hanczar, B. Deep GONet: Self-explainable deep neural network based on Gene Ontology for phenotype prediction from gene expression data. BMC Bioinform. 2021, 22, 455. [Google Scholar] [CrossRef]
- Min, H.; Mobahi, H.; Irvin, K.; Avramovic, S.; Wojtusiak, J. Predicting activities of daily living for cancer patients using an ontology-guided machine learning methodology. J. Biomed. Semant. 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Ye, L.; Huang, Q.; Li, X. Tolerating data missing in breast cancer diagnosis from clinical ultrasound reports via knowledge graph inference. In Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, Singapore, 14–18 August 2021; pp. 3756–3764. [Google Scholar]
- Zhang, M.Y.; Du, R.Z. A Real-time Inference Method of Graph Attention Network Based on Knowledge Graph for Lung Cancer. In Proceedings of the 5th International Conference on Digital Signal Processing, Chengdu, China, 26–28 February 2021; pp. 326–331. [Google Scholar]
- Kim, J. In silico analysis of differentially expressed genesets in metastatic breast cancer identifies potential prognostic biomarkers. World J. Surg. Oncol. 2021, 19, 188. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Y.; Gao, X.; Du, M.; Gao, M.; Zhong, X.; Wei, X. Analysis of LncRNA-mRNA co-expression profiles in patients with polycystic ovary syndrome: A pilot study. Front. Immunol. 2021, 12, 669819. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, J.; Ma, J.; Zang, L.; Dong, F.; Sun, J.; Zheng, M. Identification of key genes and signaling pathways associated with the progression of gastric cancer. Pathol. Oncol. Res. 2020, 26, 1903–1919. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, B.; Quan, Z. Identification of aberrantly methylated-differentially expressed genes and gene ontology in prostate cancer. Mol. Med. Rep. 2020, 21, 744–758. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, S.; Ma, K. Bioinformatics analysis of different candidate genes involved in hepatocellular carcinoma induced by HepG2 cells or tumor cells of patients. J. Int. Med. Res. 2020, 48, 300060520932112. [Google Scholar] [CrossRef]
- Tang, M.; Dai, W.; Wu, H.; Xu, X.; Jiang, B.; Wei, Y.; Qian, H.; Han, L. Transcriptome analysis of tongue cancer based on high-throughput sequencing. Oncol. Rep. 2020, 43, 2004–2016. [Google Scholar] [CrossRef]
- Wei, S.; Chen, J.; Huang, Y.; Sun, Q.; Wang, H.; Liang, X.; Hu, Z.; Li, X. Identification of hub genes and construction of transcriptional regulatory network for the progression of colon adenocarcinoma hub genes and TF regulatory network of colon adenocarcinoma. J. Cell. Physiol. 2020, 235, 2037–2048. [Google Scholar] [CrossRef]
- Anukriti; Dhasmana, A.; Uniyal, S.; Somvanshi, P.; Bhardwaj, U.; Gupta, M.; Haque, S.; Lohani, M.; Kumar, D.; Ruokolainen, J.; et al. Investigation of precise molecular mechanistic action of tobacco-associated carcinogen ‘NNK´ induced carcinogenesis: A system biology approach. Genes 2019, 10, 564. [Google Scholar] [CrossRef]
- Rendleman, M.C.; Buatti, J.M.; Braun, T.A.; Smith, B.J.; Nwakama, C.; Beichel, R.R.; Brown, B.; Casavant, T.L. Machine learning with the TCGA-HNSC dataset: Improving usability by addressing inconsistency, sparsity, and high-dimensionality. BMC Bioinform. 2019, 20, 339. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, L.; Chen, J.; Su, J.; Shen, W.; Liu, B.; Zhou, J.; Yu, S.; Qian, J. A four-gene signature for prognosis in breast cancer patients with hypermethylated IL15RA. Oncol. Lett. 2019, 17, 4245–4254. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Wang, C.Y.; Wang, S.; Zhang, J.; Yan, Y.J.; Guan, Z.Y.; Meng, F.J. Alteration in gene expression profile of thymomas with or without myasthenia gravis linked with the nuclear factor-kappaB/autoimmune regulator pathway to myasthenia gravis pathogenesis. Thorac. Cancer 2019, 10, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.H.; Yang, H.; He, R.Q.; Lu, J.N.; Lin, X.G.; Liang, H.W.; Dang, Y.W.; Feng, Z.B.; Chen, G.; Luo, D.Z. Analysis of microarrays of miR-34a and its identification of prospective target gene signature in hepatocellular carcinoma. BMC Cancer 2018, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Bi, M.; Li, S.; Wang, Q.; Teng, D. Determination of core pathways for oral squamous cell carcinoma via the method of attract. J. Cancer Res. Ther. 2018, 14, S1029–S1034. [Google Scholar]
- Xu, X.; Li, M.; Hu, J.; Chen, Z.; Yu, J.; Dong, Y.; Sun, C.; Han, J. Expression profile analysis identifies a two-gene signature for prediction of head and neck squamous cell carcinoma patient survival. J. Cancer Res. Ther. 2018, 14, 1525–1534. [Google Scholar]
- Shen, Y.; Feng, Y.; Chen, H.; Huang, L.; Wang, F.; Bai, J.; Yang, Y.; Wang, J.; Zhao, W.; Jia, Y.; et al. Focusing on long non-coding RNA dysregulation in newly diagnosed multiple myeloma. Life Sci. 2018, 196, 133–142. [Google Scholar] [CrossRef]
- Yang, M.; Li, H.; Li, Y.; Ruan, Y.; Quan, C. Identification of genes and pathways associated with MDR in MCF-7/MDR breast cancer cells by RNA-seq analysis. Mol. Med. Rep. 2018, 17, 6211–6226. [Google Scholar] [CrossRef]
- She, S.; Jiang, L.; Zhang, Z.; Yang, M.; Hu, H.; Hu, P.; Liao, Y.; Yang, Y.; Ren, H. Identification of the C-reactive protein interaction network using a bioinformatics approach provides insights into the molecular pathogenesis of hepatocellular carcinoma. Cell. Physiol. Biochem. 2018, 48, 741–752. [Google Scholar] [CrossRef]
- Wang, S.; Cai, Y. Identification of the functional alteration signatures across different cancer types with support vector machine and feature analysis. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2218–2227. [Google Scholar] [CrossRef]
- Yang, Z.; Li, H.; Wang, Z.; Yang, Y.; Niu, J.; Liu, Y.; Sun, Z.; Yin, C. Microarray expression profile of long non-coding RNAs in human lung adenocarcinoma. Thorac. Cancer 2018, 9, 1312–1322. [Google Scholar] [CrossRef]
- Yu, C.; Xue, P.; Zhang, L.; Pan, R.; Cai, Z.; He, Z.; Sun, J.; Zheng, M. Prediction of key genes and pathways involved in trastuzumab-resistant gastric cancer. World J. Surg. Oncol. 2018, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.M.; Yang, Y.P.; Chuang, J.H.; Chuang, C.M.; Lin, T.W.; Wang, P.H.; Yu, M.H.; Chang, C.C. Discovering the deregulated molecular functions involved in malignant transformation of endometriosis to endometriosis-associated ovarian carcinoma using a data-driven, function-based analysis. Int. J. Mol. Sci. 2017, 18, 2345. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhang, Y.Y.; Han, B.; Bai, Y.; Xiong, Y.; Song, Y.; Zhou, L.M. Suppression subtractive hybridization identified differentially expressed genes in colorectal cancer: microRNA-451a as a novel colorectal cancer-related gene. Tumour Biol. 2017, 39, 1010428317705504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Fan, X.; Song, L.; Ren, L.; Ma, E.; Zhang, S.; Ren, L.; Zheng, Y.; Zhang, J. c-Fos is involved in inhibition of human bladder carcinoma T24 cells by brazilin. IUBMB Life 2015, 67, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, S.; Bagler, G. An approach for the identification of targets specific to bone metastasis using cancer genes interactome and gene ontology analysis. PLoS ONE 2012, 7, e49401. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Lee, S.Y.; Kang, H.S.; Sung, J.S.; Cho, C.K.; Yoo, H.S.; Shin, S.; Choi, J.S.; Lee, Y.W.; Jang, I.S. Differential expression of gene profiles in MRGX-treated lung cancer. J. Pharmacopunct. 2013, 16, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, J.; Jiang, A.; Liu, Q.; Li, C.; Yang, C.; Xiu, J. Expression profile of long non-coding RNAs is altered in endometrial cancer. Int. J. Clin. Exp. Med. 2015, 8, 5010–5021. [Google Scholar]
- Valavanis, I.; Pilalis, E.; Georgiadis, P.; Kyrtopoulos, S.; Chatziioannou, A. Cancer biomarkers from genome-scale DNA methylation: Comparison of evolutionary and semantic analysis methods. Microarrays 2015, 4, 647–670. [Google Scholar] [CrossRef]
- Lo, Y.H.; Chung, E.; Li, Z.; Wan, Y.W.; Mahe, M.M.; Chen, M.S.; Noah, T.K.; Bell, K.N.; Yalamanchili, H.K.; Klisch, T.J.; et al. Transcriptional regulation by ATOH1 and its target SPDEF in the intestine. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 51–71. [Google Scholar] [CrossRef]
- Liu, M.Y.; Zhang, H.; Hu, Y.J.; Chen, Y.W.; Zhao, X.N. Identification of key genes associated with cervical cancer by comprehensive analysis of transcriptome microarray and methylation microarray. Oncol. Lett. 2016, 12, 473–478. [Google Scholar] [CrossRef]
- Yang, F.; Lyu, S.; Dong, S.; Liu, Y.; Zhang, X.; Wang, O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco. Targets Ther. 2016, 9, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wang, S.; Zhang, Y.H.; Cai, Y.D.; Liu, H. Analysis of important gene ontology terms and biological pathways related to pancreatic cancer. Biomed Res. Int. 2016, 2016, 7861274. [Google Scholar] [CrossRef] [PubMed]
- Shangkuan, W.C.; Lin, H.C.; Chang, Y.T.; Jian, C.E.; Fan, H.C.; Chen, K.H.; Liu, Y.F.; Hsu, H.M.; Chou, H.L.; Yao, C.T.; et al. Risk analysis of colorectal cancer incidence by gene expression analysis. PeerJ 2017, 5, e3003. [Google Scholar] [CrossRef] [PubMed]
- Khayer, N.; Zamanian-Azodi, M.; Mansouri, V.; Ghassemi-Broumand, M.; Rezaei-Tavirani, M.; Heidari, M.H.; Rezaei Tavirani, M. Oral squamous cell cancer protein-protein interaction network interpretation in comparison to esophageal adenocarcinoma. Gastroenterol. Hepatol. Bed Bench 2017, 10, 118–124. [Google Scholar] [PubMed]
- Vaseghi Maghvan, P.; Rezaei-Tavirani, M.; Zali, H.; Nikzamir, A.; Abdi, S.; Khodadoostan, M.; Asadzadeh-Aghdaei, H. Network analysis of common genes related to esophageal, gastric, and colon cancers. Gastroenterol. Hepatol. Bed Bench 2017, 10, 295–302. [Google Scholar]
- Ding, Y.; Yang, D.Z.; Zhai, Y.N.; Xue, K.; Xu, F.; Gu, X.Y.; Wang, S.M. Microarray expression profiling of long non-coding RNAs in epithelial ovarian cancer. Oncol. Lett. 2017, 14, 2523–2530. [Google Scholar] [CrossRef]
- Kumar, R.; Samal, S.K.; Routray, S.; Dash, R.; Dixit, A. Identification of oral cancer related candidate genes by integrating protein-protein interactions, gene ontology, pathway analysis and immunohistochemistry. Sci. Rep. 2017, 7, 2472. [Google Scholar] [CrossRef]
- Valizadeh, R.; Bahadorimonfared, A.; Rezaei-Tavirani, M.; Norouzinia, M.; Ehsani Ardakani, M.I. Evaluation of involved proteins in colon adenocarcinoma: An interactome analysis. Gastroenterol. Hepatol. Bed Bench 2017, 10, S129–S138. [Google Scholar]
- Attar, R.; Cincin, Z.B.; Bireller, E.S.; Cakmakoglu, B. Apoptotic and genomic effects of corilagin on SKOV3 ovarian cancer cell line. Onco. Targets Ther. 2017, 10, 1941–1946. [Google Scholar] [CrossRef]
- Tian, P.; Liang, C. Transcriptome profiling of cancer tissues in Chinese patients with gastric cancer by high-throughput sequencing. Oncol. Lett. 2018, 15, 2057–2064. [Google Scholar] [CrossRef]
- Deng, Y.; He, R.; Zhang, R.; Gan, B.; Zhang, Y.; Chen, G.; Hu, X. The expression of HOXA13 in lung adenocarcinoma and its clinical significance: A study based on The Cancer Genome Atlas, Oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol. Lett. 2018, 15, 8556–8572. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gong, M.; Zhao, M.; Wang, X.; Cheng, W.; Xia, Y. LncRNAs KB-1836B5, LINC00566 and FAM27L are associated with the survival time of patients with ovarian cancer. Oncol. Lett. 2018, 16, 3735–3745. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhao, Y.; Liu, Y.; Yang, X.; Yan, M.; Min, Y.; Pan, Z.; Qiu, S.; Xia, S.; Yu, J.; et al. Identifying miRNA-mRNA regulation network of major depressive disorder in ovarian cancer patients. Oncol. Lett. 2018, 16, 5375–5382. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Wang, X.; Wang, H.L.; Zhang, X.L.; Gan, T.Q.; Chen, G.; Luo, D.Z. A comprehensive analysis of the predicted targets of miR-642b-3p associated with the long non-coding RNA HOXA11-AS in NSCLC cells. Oncol. Lett. 2018, 15, 6147–6160. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hua, T.; Chi, S.; Wang, H. Identification of key pathways and genes in endometrial cancer using bioinformatics analyses. Oncol. Lett. 2019, 17, 897–906. [Google Scholar] [CrossRef]
- Qi, F.; Qin, W.X.; Zang, Y.S. Molecular mechanism of triple-negative breast cancer-associated BRCA1 and the identification of signaling pathways. Oncol. Lett. 2019, 17, 2905–2914. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Tan, X.; Mao, X.; Wei, D.; Yao, Y.; Jiang, P.; Mo, D.; Wang, T.; Yan, F. Identification of tRNA-derived fragments expression profile in breast cancer tissues. Curr. Genom. 2019, 20, 199–213. [Google Scholar] [CrossRef]
- Jin, L.; Zhu, C.; Qin, X. Expression profile of tRNA-derived fragments in pancreatic cancer. Oncol. Lett. 2019, 18, 3104–3114. [Google Scholar] [CrossRef]
- Guo, W.; Yu, H.; Zhang, L.; Chen, X.; Liu, Y.; Wang, Y.; Zhang, Y. Effect of hyperoside on cervical cancer cells and transcriptome analysis of differentially expressed genes. Cancer Cell Int. 2019, 19, 235. [Google Scholar] [CrossRef]
- Asadzadeh-Aghdaei, H.; Okhovatian, F.; Razzaghi, Z.; Heidari, M.; Vafaee, R.; Nikzamir, A. Radiation therapy in patients with brain cancer: Post-proteomics interpretation. J. Lasers Med. Sci. 2019, 10, S59–S63. [Google Scholar] [CrossRef]
- Han, B.; Wang, H.; Zhang, J.; Tian, J. FNDC3B is associated with ER stress and poor prognosis in cervical cancer. Oncol. Lett. 2020, 19, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Vallino, L.; Ferraresi, A.; Vidoni, C.; Secomandi, E.; Esposito, A.; Dhanasekaran, D.N.; Isidoro, C. Modulation of non-coding RNAs by resveratrol in ovarian cancer cells: In silico analysis and literature review of the anti-cancer pathways involved. J. Tradit. Complement. Med. 2020, 10, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.P.; Saha, I.; Lancucki, A.; Ghosh, N.; Wlasnowolski, M.; Bokota, G.; Dey, A.; Lipinski, P.; Plewczynski, D. Identification of miRNA biomarkers for diverse cancer types using statistical learning methods at the whole-genome scale. Front. Genet. 2020, 11, 982. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yang, X.; Zhu, R.; Yu, L. Identifying discriminative biological function features and rules for cancer-related long non-coding RNAs. Front. Genet. 2020, 11, 598773. [Google Scholar] [CrossRef] [PubMed]
- Hermawan, A.; Ikawati, M.; Jenie, R.I.; Khumaira, A.; Putri, H.; Nurhayati, I.P.; Angraini, S.M.; Muflikhasari, H.A. Identification of potential therapeutic target of naringenin in breast cancer stem cells inhibition by bioinformatics and in vitro studies. Saudi Pharm. J. 2021, 29, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Q.; Li, W.L.; Ma, S.M.; Liang, L.; Kou, Z.Y.; Yang, J. Discovery of core gene families associated with liver metastasis in colorectal cancer and regulatory roles in tumor cell immune infiltration. Transl. Oncol. 2021, 14, 101011. [Google Scholar] [CrossRef]
- Abeni, E.; Grossi, I.; Marchina, E.; Coniglio, A.; Incardona, P.; Cavalli, P.; Zorzi, F.; Chiodera, P.L.; Paties, C.T.; Crosatti, M.; et al. DNA methylation variations in familial female and male breast cancer. Oncol. Lett. 2021, 21, 468. [Google Scholar] [CrossRef]
- Pedroza, D.A.; Ramirez, M.; Rajamanickam, V.; Subramani, R.; Margolis, V.; Gurbuz, T.; Estrada, A.; Lakshmanaswamy, R. MiRNome and functional network analysis of PGRMC1 regulated miRNA target genes identify pathways and biological functions associated with triple negative breast cancer. Front. Oncol. 2021, 11, 710337. [Google Scholar] [CrossRef]
- Wu, S.; Lv, X.; Zhang, Y.; Xu, X.; Zhao, F.; Zhang, Y.; Chen, L.; Ou-Yang, H.; Ti, X. Microarray analysis of genes with differential expression of m6A methylation in lung cancer. Biosci. Rep. 2021, 41, BSR20210523. [Google Scholar] [CrossRef]
- Siavoshi, A.; Taghizadeh, M.; Dookhe, E.; Piran, M. Gene expression profiles and pathway enrichment analysis to identification of differentially expressed gene and signaling pathways in epithelial ovarian cancer based on high-throughput RNA-seq data. Genomics 2021, 114, 161–170. [Google Scholar] [CrossRef]
- Ai, X.; Jia, Z.M.; Wang, J.; Di, G.P.; Zhang, X.U.; Sun, F.; Zang, T.; Liao, X. Bioinformatics analysis of the target gene of fibroblast growth factor receptor 3 in bladder cancer and associated molecular mechanisms. Oncol. Lett. 2015, 10, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Ung, T.H.; Madsen, H.J.; Hellwinkel, J.E.; Lencioni, A.M.; Graner, M.W. Exosome proteomics reveals transcriptional regulator proteins with potential to mediate downstream pathways. Cancer Sci. 2014, 105, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.G.; Koh, Y.; Kim, J.K.; Jung, J.; Kim, H.L.; Yoon, S.S.; Park, J.W. Identification of somatic mutations using whole-exome sequencing in Korean patients with acute myeloid leukemia. BMC Med. Genet. 2017, 18, 23. [Google Scholar] [CrossRef]
- Makler, A.; Narayanan, R. Mining exosomal genes for pancreatic cancer targets. Cancer Genom. Proteom. 2017, 14, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Wu, C.; Chen, Y.; Guo, L.; Chen, W.; Pan, Y.; Fu, X.; Wang, G.; Ding, Y. Spectrum of gene mutations identified by targeted next-generation sequencing in Chinese leukemia patients. Mol. Genet. Genom. Med. 2020, 8, e1369. [Google Scholar] [CrossRef]
- Hindumathi, V.; Kranthi, T.; Rao, S.; Manimaran, P. The prediction of candidate genes for cervix related cancer through gene ontology and graph theoretical approach. Mol. BioSyst. 2014, 10, 1450–1460. [Google Scholar] [CrossRef]
- Simjanoska, M.; Madevska Bogdanova, A.; Panov, S. Gene ontology analysis of colorectal cancer biomarkers probed with affymetrix and illumina microarrays. In Proceedings of the 5th International Joint Conference on Computational Intelligence, Algarve, Portugal, 25–27 October 2013. [Google Scholar]
- Chen, L.; Zhang, Y.H.; Lu, G.; Huang, T.; Cai, Y.D. Analysis of cancer-related lncRNAs using gene ontology and KEGG pathways. Artif. Intell. Med. 2017, 76, 27–36. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chen, G.; Tsoi, A.; Xu, H.; Zheng, W.J. Predict effective drug combination by deep belief network and ontology fingerprints. J. Biomed. Inform. 2018, 85, 149–154. [Google Scholar] [CrossRef]
- Vesteghem, C.; Brøndum, R.F.; Sønderkær, M.; Sommer, M.; Schmitz, A.; Bødker, J.S.; Dybkær, K.; El-Galaly, T.C.; Bøgsted, M. Implementing the FAIR Data Principles in precision oncology: Review of supporting initiatives. Brief. Bioinform. 2020, 21, 936–945. [Google Scholar] [CrossRef]
- Seneviratne, O.; Rashid, S.M.; Chari, S.; McCusker, J.P.; Bennett, K.P.; Hendler, J.A.; McGuinness, D.L. Knowledge integration for disease characterization: A breast cancer example. In Proceedings of the International Semantic Web Conference, Monterey, CA, USA, 8–12 October 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 223–238. [Google Scholar]
- Pesquita, C.; Faria, D.; Santos, E.; Couto, F.M. To repair or not to repair: Reconciling correctness and coherence in ontology reference alignments. In Proceedings of the 8th ISWC Ontology Matching Workshop (OM), Sydney, Australia, 25 October 2013; Volume 3. [Google Scholar]
- Lecue, F. On the role of knowledge graphs in explainable AI. Semant. Web 2020, 11, 41–51. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, M.C.; Eugénio, P.; Faria, D.; Pesquita, C. Ontologies and Knowledge Graphs in Oncology Research. Cancers 2022, 14, 1906. https://doi.org/10.3390/cancers14081906
Silva MC, Eugénio P, Faria D, Pesquita C. Ontologies and Knowledge Graphs in Oncology Research. Cancers. 2022; 14(8):1906. https://doi.org/10.3390/cancers14081906
Chicago/Turabian StyleSilva, Marta Contreiras, Patrícia Eugénio, Daniel Faria, and Catia Pesquita. 2022. "Ontologies and Knowledge Graphs in Oncology Research" Cancers 14, no. 8: 1906. https://doi.org/10.3390/cancers14081906
APA StyleSilva, M. C., Eugénio, P., Faria, D., & Pesquita, C. (2022). Ontologies and Knowledge Graphs in Oncology Research. Cancers, 14(8), 1906. https://doi.org/10.3390/cancers14081906






